Abstract
Copper proteins play key roles in biological processes such as electron transfer and dioxygen activation; the active site of each of these proteins is classified as either type 1, 2, or 3, depending on its optical and electron paramagnetic resonance properties. We have built a new type of site that we call “type zero copper” by incorporating leucine, isoleucine, or phenylalanine in place of methionine at position 121 in C112D Pseudomonas aeruginosa azurin. X-ray crystallographic analysis shows that these sites adopt distorted tetrahedral geometries, with an unusually short Cu-O(G45 carbonyl) bond (2.35–2.55 Å). Relatively weak absorption near 800 nm and narrow parallel hyperfine splittings in EPR spectra are the spectroscopic signatures of type zero copper. Copper K-edge x-ray absorption spectra suggest elevated Cu(II) 4p character in the d-electron ground state. Cyclic voltammetric experiments demonstrate that the electron transfer reactivities of type zero azurins are enhanced relative to that of the corresponding type 2 (C112D) protein.
Introduction
One of the hallmarks of bioinorganic chemistry is the adaptation by proteins of relatively few transition metals to a host of functions spanning the range from structural to reactive1. Copper proves especially adept at meeting these biochemical demands and typically requires only ligands provided by the host protein. In a wide range of coordination environments, copper acts as an electron transfer agent, activates dioxygen, and scavenges reactive oxygen species to cite only a few of its biological roles2–4.
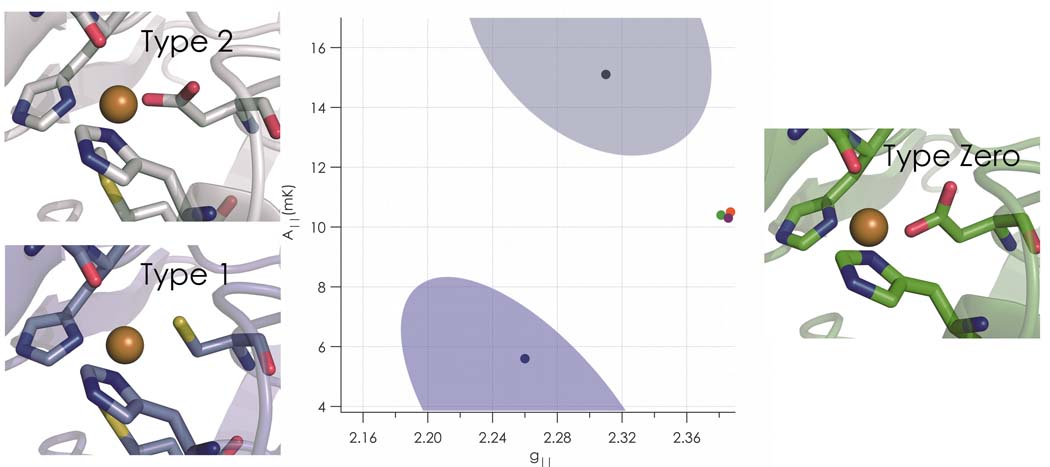
Copper sites in proteins have been classified according to their spectroscopic and functional properties5. A type 1 or blue copper site is so-named for an intense absorption band near 600 nm (ε ~ 5000 M−1cm−1 ) attributable to cysteine S π to Cu(II) dx2−y2 charge transfer. The highly covalent interaction between this thiolate ligand and Cu(II) also gives rise to drastically reduced parallel hyperfine splitting (A‖) in the EPR spectrum of a type 1 protein6. Blue copper sites behave as highly efficient electron transfer agents, and their reduction potentials span a wide range (200 to 1000 mV vs NHE). By contrast, type 2 sites do not exhibit intense bands in their visible absorption spectra, display A‖ values resembling aqueous Cu(II) ions, and often have low reduction potentials. They do, however, combine with antiferromagnetically coupled (type 3) dicopper sites to form catalytically active trinuclear clusters capable of dioxygen activation4,7. CuA represents a case of delocalized binuclear copper related to type 1 but involving two cysteine ligands; it functions as another copper-based electron transfer site in several proteins8.
We have been testing the proposition that sulfur ligation is essential for the widespread electron transfer functions of type 1 proteins, as successful replacement of soft sulfur donors with hard ligands could greatly enhance protein lifetimes in catalytic processes involving dioxygen or other powerful oxidants. In preliminary work on the Mizoguchi Pseudomonas aeruginosa C112D azurin scaffold9,10, we found that the additional mutation M121L generates a protein with a high Cu(II/I) reduction potential as well as type 1 EPR parameters11. We also constructed C112D/M121F and C112D/M121I mutants that exhibit similar EPR behavior, though as in C112D/M121L, the intense charge transfer absorption is absent, owing to coordination that heretofore has not appeared in the catalog of copper sites. We call these unique constructs “type zero” copper proteins.
Results
Structures
Crystals of C112D/M121X (X=L,F,I) azurins diffracted to high resolution (reflections visible to 1.6 Å) despite the limitations of laboratory X-ray sources. We took advantage of this unexpected windfall by collecting higher-resolution data for C112D to facilitate more meaningful comparisons among the series of mutants. In all cases, only one crystal proved necessary to collect > 95% complete datasets to high resolution. Structures were solved readily by molecular replacement, and coordinates were refined with the resolution limit set by evaluation of Wilson plots and scaling/merging statistics (Table S1). Coordinate error may be obtained from the estimated standard uncertainty (e.s.u.) in atomic positions as calculated by REFMAC5 from Rwork and Rfree 12. As the e.s.u.’s represent an average uncertainty for each structure, Cu(II) to ligand bond distances are expected to exhibit increased precision owing to lower thermal disorder (B-factors) relative to the entire atomic ensemble13. Coordinates and structure factors have been deposited in the PDB. Cu oxidation state was verified by comparing inner-sphere bond distances from Cu K-edge EXAFS to the crystallographic coordinates (Table S2, Fig. S1).
In all cases the proteins packed as monomers around a copper coordination complex consisting of the N-terminal alanine of one azurin molecule and H83 of a second, with the remaining coordination sites occupied by one molecule of Tris buffer and solvent. The protein tertiary structure remains unchanged among the four proteins, displaying the characteristic β-sandwich motif exhibited by members of the cupredoxin family14.
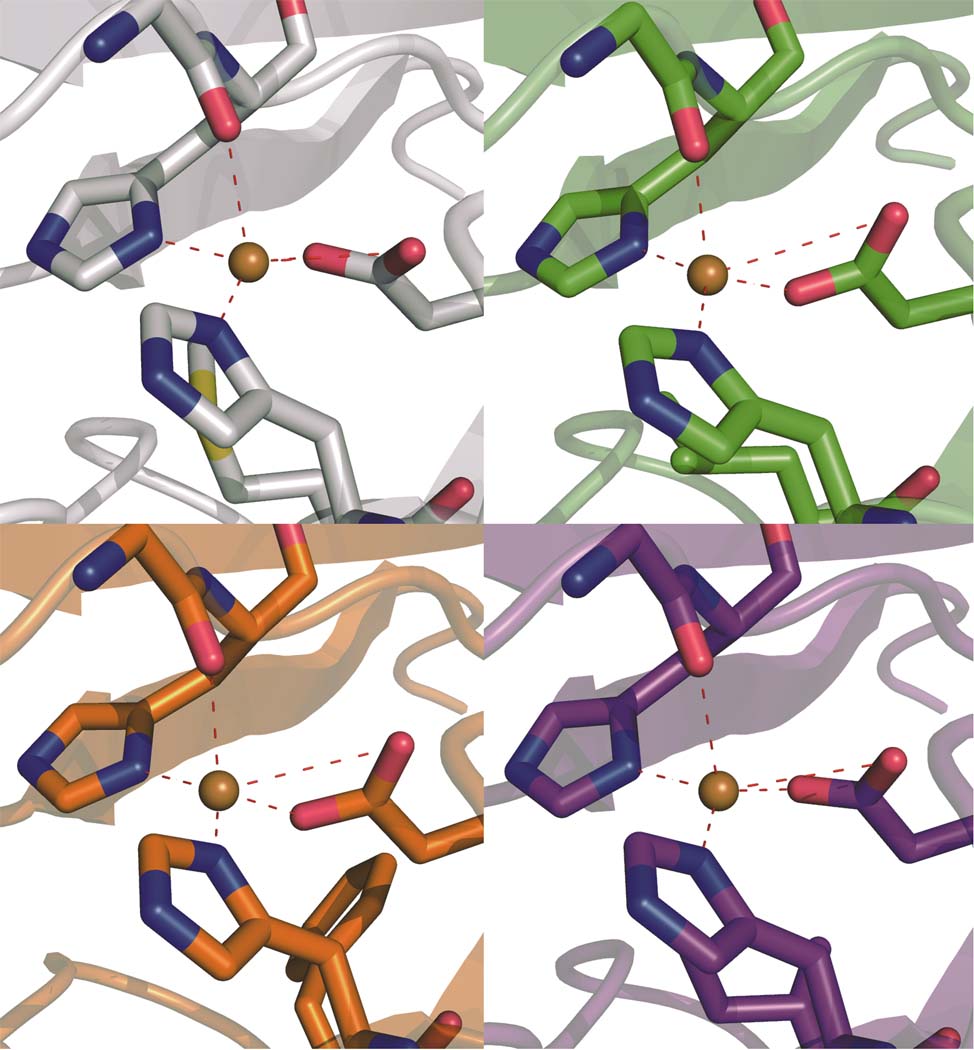
The double mutants display striking differences within both their inner and outer copper coordination spheres as compared to the C112D protein. In the inner-sphere case, the remarkable perturbations manifest themselves in the orientation and position of the oxygen-donor ligands. G45, heretofore regarded as weakly interacting due to its remoteness to Cu(II) (2.6 to 3.1 Å), now appears to function as a bona fide ligand to the metal, with distances ranging from 2.55 Å in C112D/M121I to a record 2.35 Å in C112D/M121L (Fig. 1). In this extreme case, the Cu(II) is distorted significantly out of the NNO (H46-H117-D112) trigonal plane. The absence of M121 thioether ligation likely promotes metal ion bonding to the G45 oxygen lone pair.
Figure 1. The distorted tetrahedral coordination sphere of C112D/M121X (X=L,F,I) azurins features a relatively short Cu-O(G45 carbonyl) bond.
The Cu(II) binding sites of C112D (a, 1.9 Å, PDBID: 3FQY), C112D/M121L (b, 2.1 Å, PDBID: 3FPY), C112D/M121F (c, 1.9 Å, PDBID: 3FQ2), and C112D/M121I (d, 1.9 Å, PDBID: 3FQ1) azurins are displayed with Cu-heteroatom bond distances indicated in Å. Oxygen atoms are red; nitrogen atoms are blue
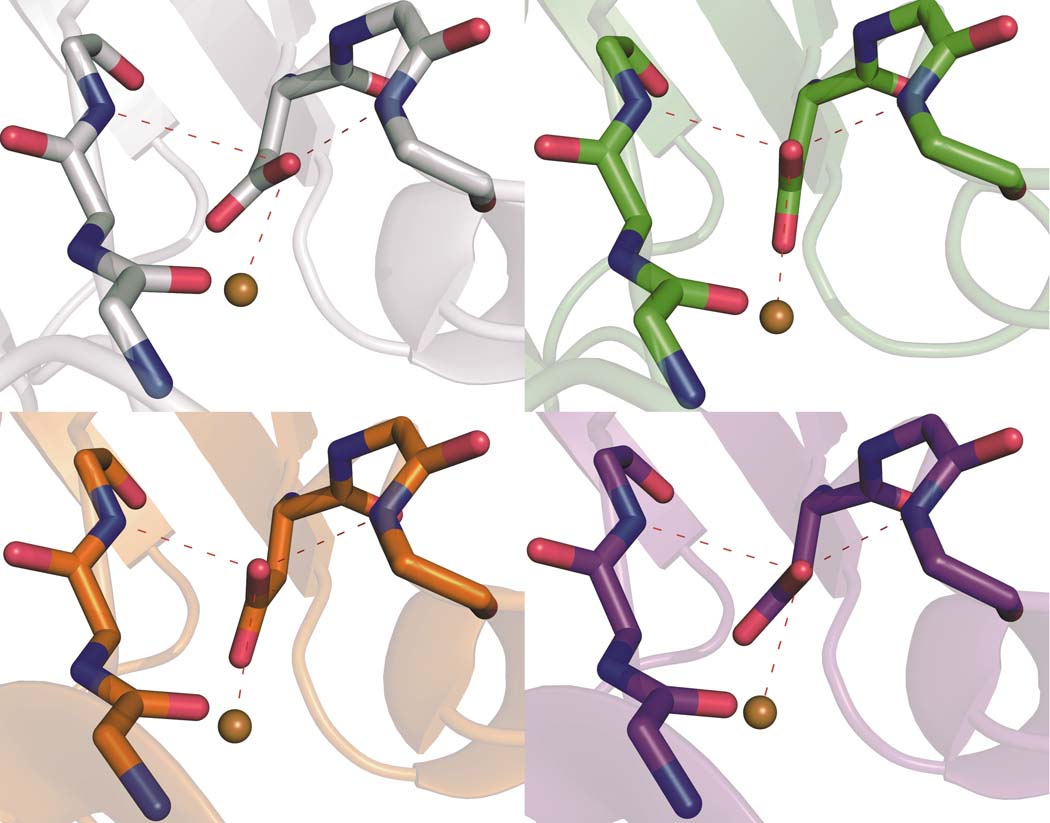
The orientation of the D112 carboxylate and by extension the distance of the second oxygen from copper precludes assignment of this residue as a true bidentate ligand in any of the four proteins (Fig. S3). We attribute this long-standing error to ambiguity stemming from the modest (2.4 Å) resolution of the earlier C112D structure15. Interestingly, major reorganization of the outer-sphere coordination structure accompanies positioning of the monodentate carboxylate (Fig. 2). In C112D/M121L, the nonligating carboxyl oxygen forms hydrogen bonds with the backbone amides of N47 and F114 in a near-perfect 120° arrangement. In contrast, this oxygen in C112D, C112D/M121I, and C112D/M121F makes a hydrogen bond only with F114. The two hydrogen bonds to D112 in C112D/M121L provide further impetus for Cu(II) to seek stabilization by bonding to the G45 carbonyl. This release of Cu(II) from interaction with position 121 and movement toward 45 allows D112 the freedom to rearrange and reconstitute the sterically contrained (“rack” or “entatic”) state responsible in part for the spectroscopic and reactivity properties of naturally occurring type 1 proteins16. Furthermore, it appears that this rearrangement of hydrogen bonds is modulated by the hydrophobic packing of residue 121 (Fig. S2). Specifically, I121 and F121 sterically clash with F15, causing a reorientation of this residue. In the extreme case of C112D/M121F, this clash forces F15 to adopt multiple conformations. By contrast, F15 remains unperturbed by L121. This interaction likely causes the structural rearrangement leading to reorientation of the N47-D112-F114 hydrogen bonding network.
Figure 2. The position of D112 shifts among the proteins, leading to variations in hydrogen bonding to the carboxylate.
Secondary coordination spheres of Cu(II) in C112D (a, 1.9 Å, PDBID: 3FQY), C112D/M121L (b, 2.1 Å, PDBID: 3FPY), C112D/M121F (c, 1.9 Å, PDBID: 3FQ2), and C112D/M121I (d, 1.9 Å, PDBID: 3FQ1) azurins are highlighted with bond distances shown in Å for heteroatoms involved in the hydrogen bonding “rack” network of the wild-type protein. Oxygen atoms are red; nitrogen atoms are blue.
Electronic Absorption Spectra
Each of the C112D and C112D/M121X (X= L,F,I) azurins displays a ligand-field (LF) absorption band near 800 nm, as well as a shoulder at ~310 nm corresponding to imidazole to Cu(II) charge transfer (LMCT) (Fig. S4, Table 1). The intensities of the LF features are very similar in the four proteins, though bandshapes and positions differ somewhat. Comparison of the hydrogen bond network between D112 and the backbone amide protons of N47 and F114 suggests that the withdrawal of electron density from the D112 carboxylate by these hydrogen bonds accounts for the lower LF splitting in the C112D/M121L protein. LMCT intensties vary across the series, suggestive of differential Cu-N orbital overlap owing to varying degrees of tetrahedral distortion among the proteins.
Table 1.
Electronic absorption data.
| LF λmax | LF εmax | LMCT ε310 | |
|---|---|---|---|
| C112D10 | 754 nm | 97 M−1cm−1 | 1950 M−1cm−1 |
| C112D/M121L | 798(2) | 96(1) | 1650(20) |
| C112D/M121I | 789(2) | 99(5) | 2000(30) |
| C112D/M121F | 789(2) | 90(10) | 1450(20) |
X-ray Absorption Spectra
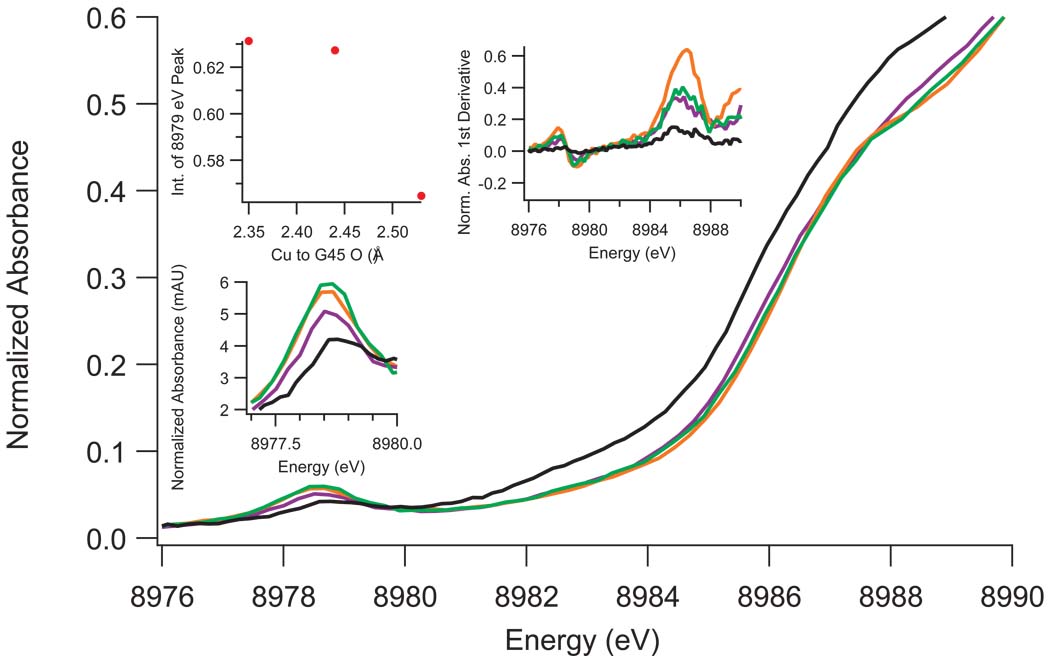
Two features are present in the Cu(II) XANES of C112D/M121X (X=L,F,I) azurins (Figure 3, S5). The first is a weak absorption at ~8979 eV that is assigned as a 1s to 3d transition. Formally forbidden in octahedral geometry, this feature, which gains intensity upon geometric distortion, has been used as a metric of 4p mixing into 3d ground states6. The intensity of this transition increases on going from C112D/M121I to C112D/M121F to C112D/M121L, which appears to correlate with the Cu(II) to G45 carbonyl oxygen bond distance. The data suggest that Cu(II)-O(G45) bonding promotes Cu(II) 4p mixing into the 3d ground state.
Figure 3. XANES of C112D (black) and C112D/M121X (X= L, green; F, orange; I, purple) azurins with focus on the 8979 eV and 8987 eV pre-edge features.
Intensity of the 1s to 3d 8979 eV absorption (highlighted by the bottom-left inset) appears to correlate with the Cu(II) to G45-O bond distance as shown in the upper left-hand inset. The first derivative of the absorption spectrum (upper right inset) indicates that the ~8987 eV “shakedown” energy does not vary within the resolution limits of the instrumentation.
The second feature is a shoulder near 8987 eV, which has been previously assigned as a “shakedown” transition due to an orbital contraction upon promotion of core electrons to valence shells17. The energy and intensity of this transition have been used as metrics of site covalency. Although the poorly resolved nature of this feature precludes quantitative analysis of any differences in band positions, the decreased intensities could indicate that there is a slight increase in site covalency for the C112D/M121X (X=L,F,I) proteins.
EPR Spectra
EPR spectra for C112D and C112D/M121X (X=L,F,I) azurins were recorded at X-band (9.5 GHz) (Table 2, Fig. S6). The C112D protein displays spin parameters characteristic of type 2 copper5. Previously reported superhyperfine coupling to 14N is visible as unresolved broadening in the perpendicular region of the spectrum. The C112D/M121X (X=L,F,I) proteins differ markedly from C112D, with elevated gz and decreased A‖ values. Such spin parameters are typical for tetrahedral Cu(II) complexes5. Despite narrow linewidths, C112D/M121X (X=L,F,I) derivatives appear not to exhibit superhyperfine structure in their EPR spectra, suggesting diminished coupling of the unpaired electron to the ligating imidazole nitrogens in each case.
Table 2.
EPR parameters from SpinCount simulations.
| gx | gy | gz | σgx | σgy | σgz | Ax,y (mK) |
Az (mK) |
σAx,y (mK) |
σAz (mK) |
Rg | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C112D | 2.063 | 2.063 | 2.311 | 0.008 | 0.008 | 0.002 | 0.07 | 14.89 | 0.06 | 0.17 | 0 |
| C112D/M121L | 2.047 | 2.116 | 2.381 | 0.015 | 0.008 | 0.007 | 1.65 | 10.32 | 0.001 | 0.10 | 0.87 |
| C112D/M121I | 2.043 | 2.101 | 2.386 | 0.013 | 0.021 | 0.005 | 0.25 | 10.18 | 0.003 | 0.10 | 0.83 |
| C112D/M121F | 2.054 | 2.100 | 2.386 | 0.018 | 0.011 | 0.006 | 0.18 | 10.36 | 0.009 | 0.09 | 0.62 |
| WT39 | 2.059 | 2.059 | 2.259 | 5.0 |
EPR spectra were simulated with g- and A-strain (σg, σA) included as fitting parameters to account for conformational heterogeneity in the glassed samples. Anisotropy in Ax,y was not modeled due to the resolution limits of operating at X-band. Select parameters for wild-type azurin are included as a type 1 reference. Units are dimensionless unless otherwise indicated.
The g⊥ anisotropy is given by18:
| (1) |
Factors affecting Rg include splitting between dxz and dyz, , differences in the orbital reduction factors, and mixing of dz2 into the Cu(II) ground state19. C112D/M121L displays the largest g⊥ anisotropy, followed by C112D/M121I and C112D/M121F.
The following expressions for g values apply to Cu(II) in a distorted tetrahedral ligand field20:
| (2a) |
| (2b) |
| (2c) |
where Δgi is the difference between observed g values and the free electron ge = 2.0023, Δdi is the energy difference between ground and excited states formed by transitions to orbital 3di, λ is the Cu(II) spin-orbit coupling constant (taken to be −830 cm−1), and κi represents the orbital reduction factor along axis i. This latter parameter provides a metric of Cu(II) character in the ground state. These expressions also hold for distorted Oh, 3dx2−y2 ground state systems, although Δdi terms are exchanged for Δgx and Δgy.
Assuming a perturbed D4h geometry for the C112D protein, a κ of ~0.78 is in agreement with the band at ~750 nm in the absorption spectrum (Fig. S7). Calculated LF splittings based on assignment of the corresponding bands in the spectra of the C112D/M121X (X=L,F,I) proteins as dx2−y2 to dxy transitions are in accord with experiment only if κ is near 0.85, which is consistent with the disappearance of superhyperfine broadening on going from the C112D protein to the double mutants. The LF transitions in C112D/M121F and C112D/M121I azurins should be lower in energy than in C112D/M121L, contrary to observation. However, as shown in the case of IrCl6 2–21,22, mixing of ligand orbitals into the metal ground state leads to angular momentum quenching and in turn a decrease in the orbital reduction factor. The shortening of the Cu(II)-O(G45) bond increases oxygen 2p mixing into the ground state, which would lead to a smaller κ in C112D/M121L azurin relative to the C112D/M121F and C112D/M121I proteins. Differences in covalency predicted by the Δκ of ~0.05 going from C112D to the double mutants and Δκ ~0.01 between C112D/M121L and C112D/M121F,I are likely to be small, in accord with the lack of substantial change in the ~8987 eV XANES shakedown transition.
The axial hyperfine splitting arises from Fermi contact, spin dipolar, and orbital dipolar terms. Mixing of 4pz into the electronic ground state of tetrahedral Cu(II) sites decreases the spin dipolar term, leading to smaller axial hyperfine splittings. In the absence of polarized single crystal XAS we cannot definitively assign the ~8979 eV feature to a transition to a specific 4p orbital. The lack of correlation between ~8979 eV intensity and the A‖ values suggests that other mechanisms account for the diversity among the azurins. Differing g⊥ anisotropies indicate varying degrees of dz2 mixing, possibly attributable to a low-lying LF excited state in each of the proteins.
Electron Transfer
We measured the Cu(II) reduction potentials of C112D/M121I and C112D/M121F azurins at pH 7.0 by direct electrochemistry on self-assembled monolayer (SAM) modified monocrystalline gold bead electrodes (Figure S8). From both cyclic voltammetry (CV) and square wave voltammetry (SWV), the potential of C112D/M121I is 310 mV vs NHE (Figure S9). For C112D/M121F, which couples poorly to the electrode, the SWV Cu(II) reduction potential also is 310 mV.
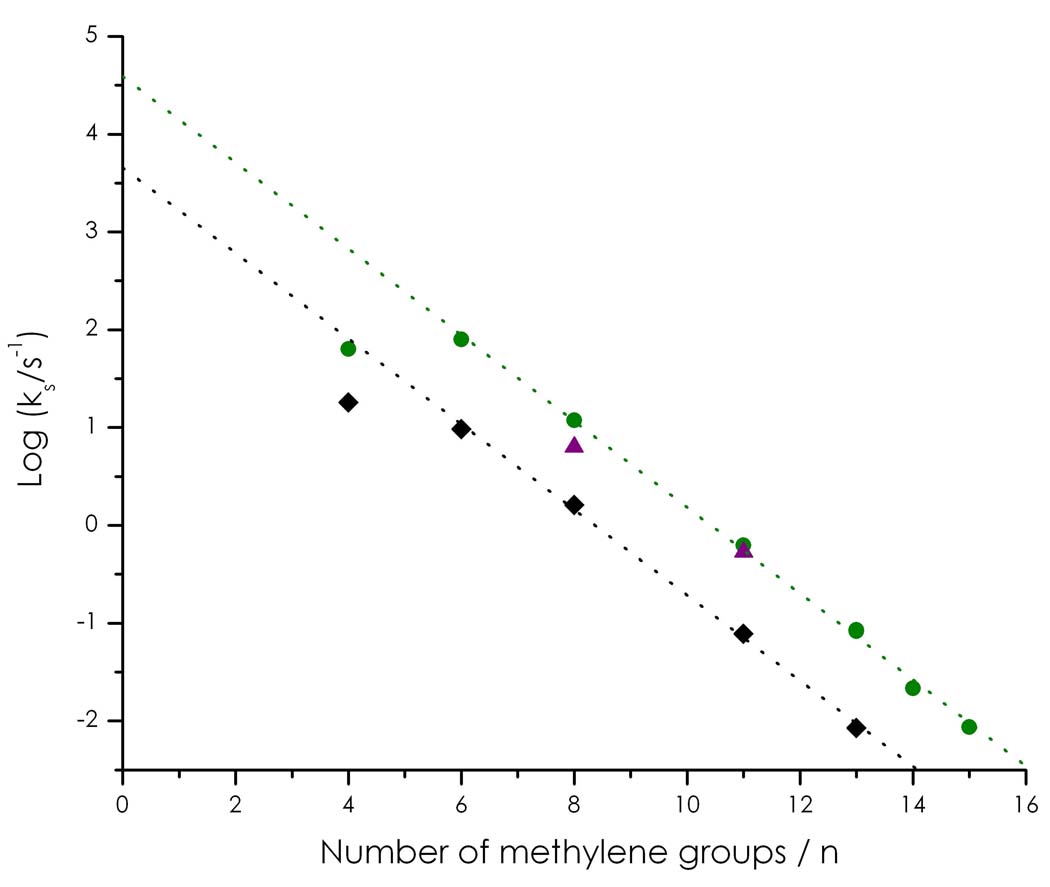
CV experiments using SAM-modified gold bead electrodes afford not only reduction potentials but also electron transfer (ET) rates23. Cu(II/I)-electrode ET rates (ΔG° = 0) were calculated for C112D, C112D/M121L, and C112D/M121I for several SAM chain lengths (Figure 4). We were not able to determine ET rates for the C112D/M121F protein (weak CV signals). Importantly, the copper centers of C112D/M121L and C112D/M121I exhibit enhanced ET reactivities relative to that of C112D, consistent with the lower Cu(II/I) reorganization associated with a sterically constrained active site16.
Figure 4. Cu(II/I)-electrode ET rates obtained for C112D (black), C112D/M121L (green), and C112D/M121I (purple) azurins from analysis of CV data (Au-SAM electrochemistry).
Type zero azurins demonstrate significant rate enhancement relative to the type 2 C112D protein.
Discussion
Type zero copper proteins display marked similarities to their type 1 counterparts, most especially in the striking resemblance of the F114-D112-N47 hydrogen bond network of C112D/M121L to that found in the sterically constrained native state16,24. The intensity of the 1s to 3d XAS transition and the narrow axial hyperfine splitting in EPR spectra are type 1 properties, while the absence of intense visible LMCT is a hallmark of type 2 proteins. Type zero copper features a relatively short Cu(II)-O(G45) bond as part of near tetrahedral coordination with accompanying narrow axial hyperfine splittings and relatively high reduction potentials. We suggest that 4p mixing accounts for much of the decreased axial hyperfine splitting relative to type 2 copper; and we conclude that the electronic ground state of type zero copper is Cu 3dxy coupled to a low-lying, dz2-based excited state.
Copper proteins have received much attention recently as components of fuel cells based on biological materials25–28. Many of these studies have evaluated the efficacy of dioxygen reduction by multicopper oxidases as cathodes in such devices. Microscopic reversibility suggests that tuning reduction potentials in engineered multicopper oxidases could facilitate the oxidation of water to dioxygen. Such systems would be susceptible to irreversible deactivation of catalytic activity by oxidation of cysteine in their type 1 copper centers. Type zero copper, with enhanced ET reactivity relative to type 2 copper, offers a very promising alternative if reduction potentials in the 1 V range can be achieved.
Materials and Methods
Crystallography
Crystal growth procedures are available as supplementary information. All X-ray diffraction experiments were carried out at the Caltech Molecular Observatory. Crystals were removed from wells and incubated in a cryoprotectant solution consisting of 30% ethylene glycol, 25% PEG 4000, 100 mM lithium nitrate, 10 mM copper sulfate, and 100 mM tris pH 7.0 for several minutes. During this time the large crystals under a 100 K nitrogen cryostream. 1.54 Å X-rays were generated using a Rigaku rotating anode source. Oscillation images were recorded following determination of crystal parameters in a manner to maximize completeness of the data sets. Images were recorded on a Rigaku Raxis IV++ detector operated with CRYSTALCLEAR.
Reflections were integrated and processed using iMOSFLM29. Data were scaled and merged using SCALA30. Structures were solved by the method of molecular replacement as implemented in program MOLREP12 using the 2.4 Å structure of azurin C112D (PDBID: 1AG0) as a search model. The MOLREP output was then used for model building with ARP/wARP31, with a single coordinate randomization included to alleviate any possible model bias. Maximum likelihood restrained refinement was carried out using program REFMAC512.
Electronic Absorption Spectroscopy
Samples for UV/VIS spectra were concentrated to ~2 mM and exchanged into 100 mM sodium phosphate pH 7.0, with spectra recorded on a HP 8453 diode array spectrophotometer. Extinction coefficients were determined by titration of apoprotein with analytically prepared copper(II) sulfate. For NIR measurements, proteins were exchanged into 100 mM sodium phosphate pH 7.0 (uncorrected) in D2O. NIR spectra were recorded on a Thermo Nicolet 6700 FTIR.
EPR Spectroscopy
Samples for X-band EPR experiments were prepared by exchanging concentrated (~2 mM) protein into 100 mM sodium phosphate pH 7.0 containing 38% glycerol. Samples were glassed by rapid freezing in liquid nitrogen. X-band data were collected at 77 K on a Bruker EMX Biospin fitted with a cold finger. EPR spin parameters were simulated using the SPINCOUNT32 package.
X-ray Spectroscopy
Cu K-edge XAS, including EXAFS data, were collected at the Stanford Synchrotron Radiation Lightsource at beam line 7-3 under ring condition of 3 GeV and 60–100 mA. A Si(220) double-crystal monochromator was used for energy selection and a Rh-coated mirror (set to an energy cutoff of 13 keV) was used for harmonic rejection. Internal energy calibration was performed by assigning the first inflection point of a Cu foil spectrum to 8980.3 eV. Samples were exchanged into 100 mM sodium phosphate pH 7.0 with 38% glycerol and concentrated to ~3 mM. Proteins were loaded into 2 mm Lucite XAS cells with 38 micron Kapton windows and glassed by rapid immersion in liquid nitrogen. Data were collected in fluorescence mode (using a Canberra Ge 30-element array detector) with the sample maintained at 10 K in an Oxford liquid helium flow cryostat. To minimize photoreduction of Cu(II), the incident beam intensity was attenuated by a factor of ~3 with a 4-layer aluminum Reynolds filter. Data were collected from 8900 to 9694 eV (k = 13.5 Å−1) to reduce collection time and thus sample photoreduction. Photoreduction was monitored by following the growth of the 8984 eV preedge feature indicative of Cu(I) formation. Four scans per spot were deemed to yield acceptably oxidized sample (~20% Cu(I)), with a total of 16 scans being performed per protein. Scans were averaged and processed using the MAVE and PROCESS modules of the EXAFSPAK software package33. Background subtractions were achieved using PYSPLINE34. For XANES analysis, only the first scans per spot were averaged. XRD coordinates for the azurins were used to generate models for path calculation by FEFF735,36. Relevant paths were then optimized by least-squares fitting in the OPT package of EXAFSPAK.
Electrochemistry
Detailed procedures for SAM-modified monocrystalline gold bead electrode preparation, azurin immobilization, and electrochemical measurements have been reported previously22,37. 20 mM sodium phosphate pH 7.0 served as the electrolyte for electrochemical measurements. ET rate constants were obtained by analysis of CV peak separations38.
Supplementary Material
Figure 5.
Figure 6.
Figure 7.
Figure 8.
Acknowledgements
We thank Bruce Brunschwig for assistance with FTIR spectroscopy, Zachary Gates and Leonard Thomas for assistance with X-ray diffraction data collection, and Mike Day and Jens Kaiser for discussions of crystal structural analyses. We thank Ed Solomon for helpful comments on electronic structural formulations, and Yuling Sheng for assistance with protein expression and purification. SSRL operations are funded by DOE(BES). The Structural Molecular Biology program is supported by NIH (NCRR BMTP) (Grant Number 5 P41 RR001209)N and DOE(BER). This work was supported by NIH DK019038(HBG), Stanford GCEP, and NSF CHE-0802907. The Caltech Molecular Observatory is supported by the Gordon and Betty Moore Foundation.
Footnotes
Author contributions:
K.M.L. and H.B.G. conceived and designed the experiments; K.M.L., S.D.G., and K.Y. performed the experiments; K.M.L., S.D.G., K.Y., and H.B.G. analyzed the data, K.M.L., S.D.G., J.H.R., and H.B.G. co-wrote the paper.
REFERENCES
- 1.Gray HB. Biological inorganic chemistry at the beginning of the 21st century. Proc. Natl. Acad. Sci. USA. 2003;100:3563–3568. doi: 10.1073/pnas.0730378100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gradinaru C, Crane BR, Abrahamsson ML, Gray HB. Electron transfer in metalloproteins (blue copper azurin) Biophys. J. 2004;86:473A–473A. [Google Scholar]
- 3.Roberts SA, et al. Crystal structure and electron transfer kinetics of CueO, a multicopper oxidase required for copper homeostasis in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:2766–2771. doi: 10.1073/pnas.052710499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer AE, et al. Spectroscopic characterization and O-2 reactivity of the trinuclear Cu cluster of mutants of the multicopper oxidase Fet3p. Biochemistry. 2002;41:6438–6448. doi: 10.1021/bi011979j. [DOI] [PubMed] [Google Scholar]
- 5.Solomon EI, Szilagyi RK, DeBeer George S, Basumallick L. Electronic structures of metal sites in proteins and models: Contributions to function in blue copper proteins. Chem. Rev. 2004;104:419–458. doi: 10.1021/cr0206317. [DOI] [PubMed] [Google Scholar]
- 6.Solomon EI. Spectroscopic methods in bioinorganic chemistry: Blue to green to red copper sites. Inorg. Chem. 2006;45:8012–8025. doi: 10.1021/ic060450d. [DOI] [PubMed] [Google Scholar]
- 7.Lee SK, et al. Nature of the intermediate formed in the reduction of O-2 to H2O at the trinuclear copper cluster active site in native laccase. J. Am. Chem. Soc. 2002;124:6180–6193. doi: 10.1021/ja0114052. [DOI] [PubMed] [Google Scholar]
- 8.Hay M, Richards JH, Lu Y. Construction and characterization of an azurin analog for the purple copper site in cytochrome c oxidase. Proc. Natl. Acad. Sci. U. S. A. 1996;93:461–464. doi: 10.1073/pnas.93.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mizoguchi TJ, Di Bilio AJ, Gray HB, Richards JH. Blue to type 2 binding. Copper(II) and cobalt (II) derivatives of a Cys112Asp mutant of Pseudomonas aeruginosa azurin. J. Am. Chem. Soc. 1992;114:10076–10078. [Google Scholar]
- 10.Mizoguchi TJ. Ph.D. Thesis. California Institute of Technology; 1996. [Google Scholar]
- 11.Lancaster KM, Yokoyama K, Richards JH, Winkler JR, Gray HB. High-potential C112D/M121X (X = M, E, H, L) Pseudomonas aeruginosa azurins. Inorg. Chem. 2009;48:1278–1280. doi: 10.1021/ic802322e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect D: Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 13.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr., Sect D: Biol. Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 14.Engman KC, Sandberg A, Leckner J, Karlsson BG. Probing the influence on folding behavior of structurally conserved core residues in P-aeruginosa apo-azurin. Protein Sci. 2004;13:2706–2715. doi: 10.1110/ps.04849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faham S, et al. Role of the active-site cysteine of Pseudomonas aeruginosa azurin. Crystal structure analysis of the CuII (Cys112Asp) protein. J. Biol. Inorg. Chem. 1997;2:464–469. [Google Scholar]
- 16.Gray HB, Malmstrom BG, Williams RJP. Copper coordination in blue proteins. J. Biol. Inorg. Chem. 2000;5:551–559. doi: 10.1007/s007750000146. [DOI] [PubMed] [Google Scholar]
- 17.DeBeer S, et al. X-ray absorption spectra of the oxidized and reduced forms of C112D azurin from Pseudomonas aeruginosa. Inorg. Chem. 1999;38:433–438. doi: 10.1021/ic9804622. [DOI] [PubMed] [Google Scholar]
- 18.Hitchman MA, Olson CD, Belford RL. Behavior of in-plane g-tensor in low-symmetry d1 and d9 systems with application to copper and vanadyl chelates. J. Chem. Phys. 1969;50 1195-&. [Google Scholar]
- 19.Gewirth AA, Cohen SL, Schugar HJ, Solomon EI. Spectroscopic and theoretical studies of the unusual electron-paramagnetic-resonance parameters of distorted tetrahedral cupric sites - correlations to X-ray spectral features of core levels. Inorg. Chem. 1987;26:1133–1146. [Google Scholar]
- 20.Mabbs FE, Machin DJ. Magnetism and transition metal complexes. xxii. Mineola, N.Y.: Dover Publications; 2008. p. 206. [Google Scholar]
- 21.Stevens KWH. On The Magnetic Properties of Covalent XY6 Complexes. Proc. R. Soc. London, Ser. A. 1953;219:542–555. [Google Scholar]
- 22.Griffiths JHE, Owen J. Complex Hyperfine Structures in Microwave Spectra of Covalent Iridium Compounds. Proc. R. Soc. London, Ser. A. 1954;226:96–111. [Google Scholar]
- 23.Fujita K, et al. Mimicking protein-protein electron transfer: Voltammetry of Pseudomonas aeruginosa azurin and the Thermus thermophilus Cu-A domain at omega-derivatized self-assembled-monolayer gold electrodes. J. Am. Chem. Soc. 2004;126:13954–13961. doi: 10.1021/ja047875o. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S, Xie X, Dey A, Sun Y, Scholes CP, Solomon EI. Thermodynamic equilibrium between blue and green copper sites and the role of the protein in controlling function. Proc. Natl. Acad. Sci. USA. 2009;106:4969–4974. doi: 10.1073/pnas.0900995106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miura Y, et al. Direct Electrochemistry of CueO and Its Mutants at Residues to and Near Type I Cu for Oxygen-Reducing Biocathode. Fuel Cells. 2009;9:70–78. [Google Scholar]
- 26.Tsujimura S, Miura Y, Kano K. CueO-immobilized porous carbon electrode exhibiting improved performance of electrochemical reduction of dioxygen to water. Electrochimica Acta. 2008;53:5716–5720. [Google Scholar]
- 27.Miura Y, et al. Bioelectrocatalytic reduction of O-2 catalyzed by CueO from Escherichia coli adsorbed on a highly oriented pyrolytic graphite electrode. Chem. Lett. 2007;36:132–133. [Google Scholar]
- 28.Blanford CF, Foster CE, Heath RS, Armstrong FA. Efficient electrocatalytic oxygen reduction by the 'blue' copper oxidase, laccase, directly attached to chemically modified carbons. Faraday Discussions. 2008;140:319–335. doi: 10.1039/b808939f. [DOI] [PubMed] [Google Scholar]
- 29.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 30.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr., Sect D: Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 31.Cohen SX, et al. ARP/wARP and molecular replacement: the next generation. Acta Crystallogr., Sect D: Biol. Crystallogr. 2008;64:49–60. doi: 10.1107/S0907444907047580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golombek AP, Hendrich MP. Quantitative analysis of dinuclear manganese(II) EPR spectra. J. Magn. Reson. 2003;165:33–48. doi: 10.1016/j.jmr.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 33.George GN. EXAFSPAK. Stanford, CA: Stanford Synchrotron Radiation Laboratory, Stanford Linear Accelerator Center, Stanford University; [Google Scholar]
- 34.Tenderhold A. PySpline. Stanford, CA: Stanford Synchrotron Radiation Laboratory, Stanford Linear Accelerator Center, Stanford University; [Google Scholar]
- 35.DeLeon JM, Rehr JJ, Zabinsky SI, Albers RC. Abinitio Curved-Wave X-Ray-Absorption Fine-Structure. Phys. Rev. B. 1991;44:4146–4156. doi: 10.1103/physrevb.44.4146. [DOI] [PubMed] [Google Scholar]
- 36.Rehr JJ, DeLeon JM, Zabinsky SI, Albers RC. Theoretical X-Ray Absorption Fine-Structure Standards. J. Am. Chem. Soc. 1991;113:5135–5140. [Google Scholar]
- 37.Yokoyama K, et al. Electron tunneling through Pseudomonas aeruginosa azurins on SAM gold electrodes. Inorg. Chim. Acta. 2008;361:1095–1099. doi: 10.1016/j.ica.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laviron E. General expression of the linear potential sweep voltammogram in the case of diffusionless electrochemical systems. J. of Electroanal. Chem. 1979;101:19–28. [Google Scholar]
- 39.Pascher T, Karlsson BG, Nordling M, Malmstrom BG, Vanngard T. Reduction potentials and their pH dependence in site-directed-mutant forms of azurin from Pseudomonas aeruginosa. Eur. J. Biochem. 1993;212:289–296. doi: 10.1111/j.1432-1033.1993.tb17661.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.