Abstract

We report the molecular design and synthesis of EG00229, 2, the first small molecule ligand for the VEGF-A receptor neuropilin 1 (NRP1) and the structural characterization of NRP1−ligand complexes by NMR spectroscopy and X-ray crystallography. Mutagenesis studies localized VEGF-A binding in the NRP1 b1 domain and a peptide fragment of VEGF-A was shown to bind at the same site by NMR, providing the basis for small molecule design. Compound 2 demonstrated inhibition of VEGF-A binding to NRP1 and attenuated VEGFR2 phosphorylation in endothelial cells. Inhibition of migration of endothelial cells was also observed. The viability of A549 lung carcinoma cells was reduced by 2, and it increased the potency of the cytotoxic agents paclitaxel and 5-fluorouracil when given in combination. These studies provide the basis for design of specific small molecule inhibitors of ligand binding to NRP1.
Introduction
Neuropilin 1 (NRP1a)(1) is a receptor for vascular endothelial growth factor A165 (VEGF-A165) and the neuronal guidance molecule semaphorin 3A (SEMA3A)(2) with key roles in vascular and neuronal development (Figure 1). In endothelial cells, NRP1 enhances the biological signals of VEGF-A mediated by binding to its receptor vascular endothelial growth factor 2 (VEGFR2). NRP1 has also been implicated in tumor growth and angiogenesis; inhibition by a blocking antibody that prevents VEGF-A binding to NRP1 enhanced the antitumor effects of the inhibitory anti-VEGF-A antibody, bevacizumab, in mouse xenograft models.(3) As an alternative to biological therapeutics, small molecule inhibitors of NRP1 function would be desirable, but development of protein−protein interaction inhibitors is not a trivial task.4,5 We utilized the bicyclic peptide 1, corresponding to the C-terminal 28 amino acids of VEGF-A165 (Figure 2) as a starting point for small molecule design. From this peptide we developed EG00229, 2 (Figure 2), a small molecule designed to interact with the VEGF-A165 binding pocket of NRP1. Mutational analysis, NMR, and X-ray crystallography establish that the interaction with NRP1 of peptide ligands (and by inference VEGF-A) and the new small molecules described herein is with the same binding site formed by the loops at the end of the b1 domain.(6) These molecules act as inhibitors of VEGF-A function, reducing VEGF-A receptor phosphorylation and endothelial cell migration. The in vitro cytotoxic effect of paclitaxel and 5-fluorouracil was enhanced in the presence of 2. Small molecule inhibitors of NRP1 have considerable potential as novel anticancer therapeutics.
Figure 1.

Model for binding of VEGF-A165 to NRP1. NRP1 has a large extracellular (Ex) domain comprising tandem a1/a2, b1/2, and a c domain, a single membrane-spanning domain, and a small cytosolic domain (Cyt). The VEGF-A165 C-terminal domain encoded by exons 7 and 8 (yellow and blue oblongs, respectively) binds to the extracellular NRP1 b1 domain. Concomitant binding of the VEGF homology domain of VEGF-A165 (solid red ovals) to VEGFR2 results in formation of a receptor complex of NRP1 with VEGF-A165 and VEGFR2 and enhanced intracellular signaling, essential for optimal migration and angiogenesis in development and in tumors.
Figure 2.

Bicyclic peptide 1 (C-terminus of VEGF) and small molecule neuropilin inhibitor 2.
Results and Discussion
Computational Prediction of the Binding Pocket on NRP1 and Mutational Analysis of VEGF-A Binding
The reported crystal structures6,7 and our own computational analysis of the NRP b1 domain using SYBYL SITEID identified the cleft formed by the loops at one end of the β-barrel as a potential binding site (Figure 3a). Residues clustered in this region(8) were conserved in mammalian NRP1 species and in human NRP2, a closely related receptor for VEGF-A1 (Figure 3b), implying an important functional role in VEGF-A binding. Mutational analysis of VEGF-A binding to NRP1 was therefore performed to confirm the identity of the binding pocket. Alanine substitution of amino acid Y297, W301, T316, D320, S346, T349, Y353, or W411 resulted in complete loss of high affinity biotinylated-VEGF-A binding to COS-7 cells transfected with mutant NRP1 cDNA constructs (Figure 4a). In addition, alanine substitution of K351 resulted in partial loss of VEGF-A binding, while mutation of T337, P398, and S416 caused modest decreases in binding and mutation of E319 had no effect (Figure 4a and Supporting Information Figure S1a). Loss of binding was not due to impaired expression of NRP1 mutants, as Western blot analysis of transfected COS-7 cells indicated similar levels of protein expression of all constructs (Supporting Information Figure S1b). A triple mutant b1 protein (S346A, E348A, T349A) was previously shown to prevent VEGF-A binding to rat NRP1.(7)
Figure 3.
(a) VEGF/tuftsin binding site of NRP1 b1 domain (black arrow), with the protein surface and the loops L1 (green), L2 (yellow), L3 (cyan), L4 (pink), L5 (red), and L6 (black) shown. Model constructed from PDB code 2ORZ. (b) Protein sequence alignment of human, mouse, and rat NRP1 (hNRP1, mNRP1, rNRP1) with human NRP2 (hNRP2). Highlighted residues were predicted to be in close contact with bound ligand from the model in panel a.
Figure 4.
(a) Mutational analysis of the NRP1 pocket. COS-7 cells were transfected with expression plasmids for wild-type (WT) or mutant NRP1 as indicated. Binding assays using bt-VEGF-A165 were performed 48 h after transfection. Values presented are the means ± SD obtained from three to six independent experiments each performed in duplicate. (b) An excerpted region of the 2D 15N,1H HSQC NMR spectrum of human NRP1 b1 in the free state (black contours) and with saturating concentrations of tuftsin (blue) and 1 (green). The cross-peak assignments for the free-state NRP1 b1 are indicated in bold. Arrows depict significant ligand-dependent chemical shift changes. (c) Solvent accessible surface representations of the structure NRP1 b1 with color highlighting showing those residues whose backbone amide NH cross-peak exhibits a significant chemical shift perturbation (compound Δδ > 0.15; see Experimental Section) upon addition of a saturating concentration of tuftsin. (d) As for (c) but with 1 as the ligand. The intensity of the color is scaled to the residue-by-residue value of the compound Δδ. (e) Pymol diagram showing positions of mutated residues on the NRP1 b1 protein surface. Mutations giving 100% inhibition of binding are shown as red, and moderate inhibition is shown as salmon (K351).
NMR Studies with the Peptide. 1, Tuftsin, and Ac-DKPRR-OH Provide Additional Evidence To Identify the NRP1 Binding Pocket for the C-terminus of VEGF-A
The binding of ligands was assessed by chemical shift perturbation mapping with 2D 15N,1H NMR of 15N-labeled NRP1 b1 (Figure 4b). In each case the residues with significant perturbations of the backbone NH cross-peak chemical shifts were identified (compound Δδ > 0.15; see Experimental Section). We employed tuftsin as a control ligand for these studies because its binding site has been established by X-ray crystallography.(7) Titration with 1, tuftsin (H-TKPR-OH), and the linear peptide Ac-DKPRR-OH (C-terminal five residues of VEGF-A) yielded highly similar patterns of chemical shift changes that mapped the VEGF-A interaction site (Figure 4c,d and Supporting Information Figure S2). The largest chemical shift changes were observed for the backbone NH groups of W301 (adjacent to C-terminal end of loop 1); T316, G318, Y322 (loop 5); A344, I345, K347, T349, K351 (loop 3); and G414, M417 (loop 4) (Supporting Information Figure S3). The binding site revealed by NMR was identical to that predicted(6) and previously shown for tuftsin(7) and very similar to a map of the key residues essential for binding (Figure 4e). These data clearly indicate that 1 and the peptide Ac-DKPRR-OH bind in a common mode to the same site as tuftsin and VEGF-A, providing a strong rationale for modeling the interaction of small molecule inhibitors with this binding pocket. These data imply that small molecules blocking this site could interfere with the binding (and function) of VEGF, and other molecules that bind this site.
Synthetic Chemistry
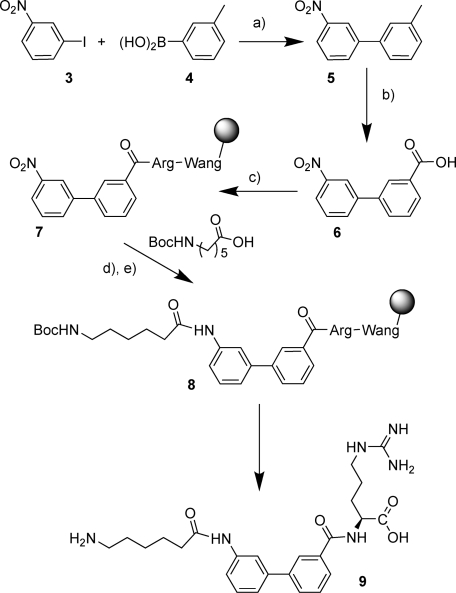
Simple linear peptides were synthesized by standard 9-fluorenylmethoxycarbonyl (FMOC) solid phase methodology as previously reported.(9) For the small molecules a highly flexible mixed solution phase/solid phase synthesis was devised that allowed the preparation of multiple analogues in a timely fashion. The lysine analogue and linking scaffold were assembled in solution phase and then coupled in one unit to the arginine-O-Wang resin using standard coupling conditions (Scheme 1). A representative example is shown in Scheme 2, which illustrates the range of chemical manipulations applied to different building blocks. Thus, Suzuki coupling of 3-nitroiodobenzene 3 with 3-methyphenylboronic acid 4 provided the biphenyl derivative 5 which was oxidized to the acid 6 using potassium permanganate. Coupling to preformed Wang-Arg resin with the reactive coupling agent bromo-tris-pyrrolidinophosphonium hexafluorophosphate (PyBrop) gave the nitro compound 7 linked to solid phase. Tin(II) chloride reduction followed by coupling with the Boc (tert-butyloxycarbonyl) protected acid gave the intermediate 8. The target compound 9 was then produced by deprotection with trifluoroacetic acid/triisopropylsilane (TFA/TIPS). For further testing larger amounts of compounds were required, and these were synthesized by solution phase chemistry as shown in Scheme 3 for 2. Reaction of the thiadiazolesulfonyl chloride 10 with methyl 3-aminothiophene-2-carboxylate 11 was achieved in pyridine at room temperature to give the sulfonamide 12. Hydrolysis to the acid 13 with lithium hydroxide and amide coupling of 13 with the 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl (Pbf) protected arginine methyl ester 14 gave 15. Finally deprotection with lithium hydroxide followed by TFA and water gave 2.
Scheme 1. Mixed Solid Phase−Solution Phase Synthetic Strategy.
Scheme 2. Typical Analogue Synthesis.
Reagents: (a) Pd2(dba)3, Pd(PPh3)4, KF, dioxane, reflux 24 h; (b) KMnO4, py, H2O, reflux 3 h; (c) HOBt, DCI, DMF, Wang-Arg-OH; (d)SnCl2·H2O, DMF; (e) PyBrOP, RCOOH, 2,6-lutidine, CH2Cl2; (f) TFA, TIPS, H2O.
Scheme 3. Solution Phase Methodology.
Reagents: (a) pyridine, 20 °C; (b) LiOH·H2O, 50 °C, THF, MeOH, H2O; (c) PyBrOP, DIPEA, DCM, 20 °C; (d) TFA/H2O 9:1.
Design of Small Molecules Mimicking the C-Terminal Amino Acid Residues of 1
Our strategy for designing small molecule inhibitors was initially to perform structure−activity relationship studies of short peptides in assays of VEGF-A binding to porcine aortic endothelial (PAE) cells expressing NRP1 in the absence of other VEGF receptors (PAE/NRP1) and thereby to identify the essential properties of peptides necessary for inhibition of binding. With this information, we then constructed a design template for the peptide−NRP1 interaction (Scheme 1, box) and undertook a systematic search for chemistry scaffolds that would retain the high affinity of the peptides in binding assays.
The C-terminal peptide Ac-RXDKPRR(9) of VEGF-A retains significant affinity for NRP1. Substitution of the nonessential penultimate arginine to alanine 16 and the synthesis of increasingly N terminal truncated peptides produced H-KPAR-OH, 17, a minimal length significantly active peptide (Table 1). Substitution with d-amino acids for the lysine or alanine 18, 19 lowered inhibitory activity. The penultimate position was tolerant of proline 20 and phenylalanine 21 in addition to alanine. The terminal arginine was essential for activity, as replacement with d-arginine 22, lysine 23, or glutamine 24 removed activity. The essential C-terminal arginine was retained with different chemical scaffolds being used to connect it to the lysine (equivalent to K162 in VEGF-A165) (Table 2). These scaffolds were selected on the basis of an approximate modeling fit of the full small molecule mimetic with the H-KPAR-OH peptide, and a representative subset is shown. Biphenyl scaffolds 9, 25, furanylenephenyl 26, and cycloalkylaryl scaffolds 27, 28, 29 gave modest inhibitions. However a thiophene-based scaffold 30 had an IC50 of 13 μM for inhibition of VEGF-A165 binding to NRP1. Follow-up studies showed that other five-membered ring heterocycles were less effective (data not shown). Reversal of the sulfonamide linkage 31 or replacement of the thiophene with a phenyl ring 32 removed activity, as did removal of the 4-aminophenylsulfonamide group 33. The best scaffold exemplified by 30 was progressed with an extensive structure−activity study; salient features are shown (Table 3). Thus, H-bond acceptor groups in the 2 or 3 position of the aromatic or aryl group were beneficial, 34 and 35, whereas methylation of the sulfonamide NH removed activity, 36 and 40. Other 4-substitutions were tolerated, 37 and 38, while a 2-nitro group, 39, showed the best activity of the simple substituents. A fluorinated analogue, 41, was less active. Insertion of a benzothiadiazole heteroaryl produced compound 2, with an IC50 of 8 μM in a cell based assay. The sulfur atom had a positive effect on potency because the O analogue 42 was less active. Other similarly active compounds were found, such as 43 and 44. Most alterations to the arginine, including the unnatural d-enantiomer 45 or 46 (Table 4) or substitution by lysine 47, resulted in a partial or complete loss of activity (Table 4). The methyl ester 48 had poor activity, while replacement of the alkyl side chain by phenyl, 49, also reduced activity. Extension of the alkyl chain to four carbons, 50, again reduced activity; however, introduction of a carbonyl group into the side chain of the arginine 51 was partially tolerated. Methylation of the guanidine as in 52 or the nitroguanidine 53 had poor activity. Further manipulations to the carboxylic acid group in the thiadiazole series, such as reduction to the alcohol 54 or conversion to an amino acid mimic 55 were deleterious. In contrast the hydroxamic acid group in 56 was at least partially tolerated . Thus, 2 was identified as a non-peptide antagonist of VEGF-A binding to NRP1.
Table 1. Establishing Small Peptide Structure−Activity Relationshipsa.
| inhibition of 125I-VEGF binding to PAE/NRP1 cells |
|||
|---|---|---|---|
| compd | structure | at 100 μM | IC50 (μM) |
| 16 | Ac-RXbDKPAR-OH | 71 ± 6.8% | 18 ± 0.11 |
| 17 | H-KPAR-OH | 83 ± 1.2% | 30 ± 0.04 |
| 18 | H-kPAR-OH | 51 ± 7.9% | |
| 19 | H-KPaR-OH | 36 ± 5.1% | |
| 20 | H-KPPR-OH | 96 ± 1.2% | 14 ± 0.01 |
| 21 | H-KPFR-OH | 63 ± 3.3% | |
| 22 | H-KPAr-OH | 22 ± 9.5% | |
| 23 | H-KPAK-OH | 22 ± 1.2% | |
| 24 | H-KPAQ-OH | 19 ± 7.6% | |
IC50 values were measured on all compounds showing inhibition of >70%. PAE/NRP1 cells in 24-well plates were incubated with different concentrations (0.1−100 μM) of compounds followed by addition of 0.1 nM 125I-VEGF-A165. Nonspecific binding was determined in the presence of 100-fold excess unlabeled VEGF. Results were obtained from three independent experiments, each performed using triplicate determinations at each concentration of compound, and are presented as the mean ± SEM. R2 values ranged from 0.9874 to 0.9996.
X is 2-aminobutyric acid.
Table 2. Search for Active Core Scaffoldsa.
 |
IC50 values were measured on all compounds showing inhibition of >70%. PAE/NRP1 cells in 24-well plates were incubated with different concentrations (0.1−100 μM) of compounds followed by addition of 0.1 nM 125I-VEGF-A165. Nonspecific binding was determined in the presence of 100-fold excess unlabeled VEGF. Results were obtained from three independent experiments, each performed using triplicate determinations at each concentration of compound, and are presented as the mean ± SEM. R2 values ranged from 0.9874 to 0.9996.
Table 3. Structure−Activity Relationships for Phenyl Sulfonamidesa.
 |
IC50 values were measured on all compounds showing inhibition of >70%. PAE/NRP1 cells in 24-well plates were incubated with different concentrations (0.1−100 μM) of compounds followed by addition of 0.1 nM 125I-VEGF-A165. Nonspecific binding was determined in the presence of 100-fold excess unlabeled VEGF. Results were obtained from three independent experiments, each performed using triplicate determinations at each concentration of compound, and are presented as the mean ± SEM. R2 values ranged from 0.9874 to 0.9996.
Table 4. Structure−Activity Relationships for Arginine Mimeticsa.
 |
IC50 values were measured on all compounds showing inhibition of >70%. PAE/NRP1 cells in 24-well plates were incubated with different concentrations (0.1−100 μM) of compounds followed by addition of 0.1 nM 125I-VEGF-A165. Nonspecific binding was determined in the presence of 100-fold excess unlabeled VEGF. Results were obtained from three independent experiments, each performed using triplicate determinations at each concentration of compound, and are presented as the mean ± SEM. R2 values ranged from 0.9874 to 0.9996.
NMR and X-ray Structural Data on 2 Confirm Binding to the Cleft Formed by the NRP1 Loops and Indicate a Key Hydrogen Bond in the Structure Stabilizing the Bound Conformation
Definitive evidence for the binding site of 2 was provided by monitoring the NRP1 b1 domain 2D 15N,1H-heteronuclear single quantum coherence (HSQC) NMR spectrum in the presence of increasing concentrations of the ligand. 15N,1H HSQC spectra are universally used to monitor the local “chemical environment” of NH groups, and in a ligand titration, the exchange regime exhibited by the perturbed cross-peaks can be related to the ligand binding kinetics such that slow−intermediate exchange behavior is typically observed for ligands with a slow off-rate consistent with tight binding. Consistent with the measured IC50 for NRP1 b1 binding, the NMR titration yielded intermediate-to-slow exchange characteristics for the majority of perturbed resonances. Both the distribution of residues that exhibited ligand-dependent chemical shift perturbations (Supporting Information Figures S2 and S3) and the directional pattern of those chemical shift changes (Supporting Information Figure S4) are highly similar to those obtained with 1, Ac-DKPRR-OH, and tuftsin. These data point to the conservation of interactions between the backbone and side chain elements of the terminal arginyl moiety of 2 with the binding pocket on NRP1 b1.
The precise binding mode and ligand conformation of 2 were established by X-ray crystallography of human NRP1 b1. The crystal structure, containing a dimer in the asymmetric unit (interchain Cα root-mean-square deviation of <0.5 Å), was determined at 2.9 Å resolution, with electron density readily observable for 2 in the binding pockets of both noncrystallographic symmetry related molecules (Figure 5a). The carboxylate group of 2 superimposes almost perfectly with the carboxylate group of tuftsin in the rat NRP1 b1b2 crystal structure (Supporting Information Figure S5a).(7) The guanidine groups of the two ligands also overlay. The structure also reveals an intramolecular shared hydrogen bond (Figure 5b) between the carbonyl amide NH and the sulfonamide nitrogen, providing a stable central conformation of the molecule. We postulate that this bond is critical for biological activity. In chain A the benzothiadiazole ring adopts a conformation on the same side to the guanidine chain with respect to the central planar portion of the molecule containing the thiophene ring. In chain B the benzothiadiazole ring is on the opposite side. In both arrangements the benzothiadiazole ring is involved in stacking interactions with a side chain residue. In chain A this interaction is provided by the M276B side chain of a symmetry related molecule (not shown). In chain B the benzthiadiazole ring stacks against the N300B side chain (Supporting Information Figure S5b). The part of the pocket binding the arginine seems to be an almost perfect “arginine receptor”, and few changes to the l-arginine were tolerated.
Figure 5.
X-ray analysis of 2. (a) FOM weighted difference map of 2-bound NRP1 b1 crystal structure calculated from the model refined without atomic coordinates for 2. The calculated electron density (green mesh) clearly shows the presence of the ligand in two different conformations in the binding sites A and B. Final stick models for 2 in chain A (left) and chain B (right) are overlaid over the density. Colors are green, red, blue, and yellow for carbon, oxygen, nitrogen, and sulfur, respectively. (b) Detail of conformation of chain A bound 2. The position of the (putative) shared hydrogen is indicated by a magenta sphere and arrow.
Binding and Functional Biological Activity of 2 in Endothelial and Lung Carcinoma Cells
Compound 2 selectively inhibited radiolabeled 125I-VEGF-A binding to PAE/NRP1, but not VEGFR2-expressing cells, with an IC50 of 8 μM (Table 3) and inhibited bt-VEGF-A binding to purified NRP1 b1 domain in a cell-free assay with an IC50 of 3 μM (Figure 6a). Compound 2 also inhibited VEGF-A binding to lung carcinoma A549 and prostate carcinoma DU145 cells, which express NRP1, but not VEGFR1 and VEGFR2, with similar potency (Supporting Information Figure S6). Binding of VEGF-A to human umbilical vein endothelial cells (HUVECs), which express VEGFR2, VEGFR1, and NRP1, was also inhibited by 2 with an IC50 of 23 μM, the reduced potency reflecting the specificity of 2 for NRP1 and its inability to compete with VEGF-A binding to other receptors (Figure S6). The potency of compound 2 in inhibiting VEGF-A binding to NRP1 was thus very similar to that of the bicyclic peptide compound 1, which inhibits 125I-VEGF-A binding to PAE/NRP1 and tumor cells with IC50 values of 2−3 μM and inhibits 125I-VEGF-A binding to HUVECs with an IC50 of 20 μM.(9)
Figure 6.
Binding and functional activity of 2. (a) 2 displaces biotinylated VEGF from NRP1 b1 in a cell-free assay. Plates precoated with purified recombinant NRP1 b1 domain were incubated for 2 h at room temperature with 0.25 nM bt-VEGF-A165 in the presence of the indicated concentrations of 2, and specific VEGF binding was determined. Values presented are the means ± SEM obtained from three independent experiments each performed in triplicate. R2 = 0.9965. (b) 2 inhibits VEGF-stimulated VEGFR2 phosphorylation. Confluent HUVECs were pretreated for 30 min with the indicated concentrations of 2 or with an equivalent volume of solvent (DMSO) and were then treated with or without 25 ng/mL VEGF-A165 for 5 min at 37 °C. Total VEGFR2 and VEGFR2 phosphorylated at Tyr1175 were then determined in treated cell lysates using a specific ELISA. Values presented are the means ± SEM obtained from three independent experiments each performed in duplicate: (∗) p < 0.05 versus no 2; (∗∗) p < 0.01 versus no 2; (∗∗∗) p < 0.001 versus no 2. (c) 2 decreases chemotactic responses to VEGF. HUVECs were pretreated for 30 min with the indicated concentrations of 2 or with an equivalent volume of solvent (DMSO) and placed into top inserts. The chemotaxis of cells toward either VEGF at 25 ng/mL or medium control (C) in bottom wells was determined after 4 h of incubation. Values presented are the means ± SEM obtained from three independent experiments each performed in duplicate: (∗) p < 0.05 for 100 μM versus 0 μM 2 pretreated cells toward VEGF. (d) Sensitization of carcinoma cells to a chemotherapeutic agent by 2. A549 cells were incubated in serum-free medium containing paclitaxel at the indicated concentrations in the absence or presence of 100 μM 2. Cell viability was measured after 48 h of treatment. Values presented are the means ± SEM obtained from three independent experiments each performed in triplicate: (∗∗) p < 0.01 for the chemotherapeutic drug alone versus drug plus 2.
The effects of 2 on VEGF-A signaling and function were examined in human endothelial cells. Compound 2 reduced VEGF-A-induced VEGFR2 tyrosine phosphorylation in HUVECs in a dose-dependent fashion, with a maximum inhibition of 34% at 100 μM (Figure 6b and Supporting Information Figure S7). Compound 2 also significantly reduced VEGF-A induced migration of HUVECs (Figure 6c). These results agree well with our previous findings showing partial inhibition of VEGF-A-induced VEGFR2 tyrosine phosphorylation by the bicyclic peptide NRP1 antagonist 1 and partial inhibition of both VEGFR2 activation and VEGF-induced migration by an inhibitory NRP1 antibody.3,9 Overall, compound 2 caused a partial inhibition of VEGF receptor activity and biological function, consistent with the current model for the role of NRP1 in VEGF function, in which NRP1 is required for optimal signaling and certain biological functions downstream of VEGFR2, particularly migration.(1)
The functional effect of 2 was also examined in A549 lung carcinoma cells, which express NRP1 but no detectable VEGFR2. Treatment of A549 cells with 2 caused a significant reduction in cell viability over a 48 h incubation (Supporting Information Figure S7). Pretreatment of A549 cells with 2 significantly enhanced the cytotoxic effects of two widely used chemotherapeutic drugs, paclitaxel (Figure 6d) and 5-fluorouracil (Supporting Information Figure S8) by approximately 4- and 10-fold, respectively. NRP1 down-regulation by siRNA has previously been shown to increase the chemosensitivity of pancreatic cancer cells,(10) while cytotoxic synergy has been noted for VEGF pathway inhibition with a combination of the multikinase inhibitor sorafenib and the proteasome inhibitor bortezomib.(11)
Conclusion
Compound 2 exhibited a biological profile consistent with inhibition of VEGF-A binding to NRP1 demonstrating an impairment, but not complete inhibition, of cell migration and VEGFR2 phosphorylation. The viability of carcinoma cells was also decreased by 2, and it enhanced the potency of cytotoxics. NRP1 blockers have the potential to target both angiogenesis and tumor growth and metastasis and may provide a valuable alternative targeted cancer therapy, particularly relevant in the light of concern over recent findings showing that anti-VEGF agents may stimulate metastasis.(12) Compound 2 and its analogues make excellent tools for the study of NRP1 biology and provide a point of departure for development of more potent anti-NRP1 drugs in the future.
Experimental Section
All commercially available solvents and reagents were used without further treatment as received unless otherwise noted. NMR spectra were measured with a Bruker DRX 400 or 600 MHz spectrometer; chemical shifts are expressed in ppm relative to TMS as an internal standard and coupling constants (J) in Hz. Mass spectra were obtained using a Waters ZQ2000 single quadrupole mass spectrometer with electrospray ionization (ESI). High resolution mass spectra were acquired on a Waters LCT time-of-flight mass spectrometer with electrospray ionization (ESI). Analysis of compounds by reverse-phase LCMS was carried out on an analytical C18 column (Phenomenex Gemini, 50 mm × 3.0 mm, 5 μm) and an AB gradient of 5−95% for B at a flow rate of 1 mL/min, where eluent A was 0.1% formic acid/water and eluent B was 0.1% formic acid/acetonitrile. Silica gel (60 Å, 40−63 μm, Fisher) was used for flash column chromatography. Routine analytical thin layer chromatography was performed on precoated plates (60F254, Machery-Nagel). All compounds tested were at least 95% pure by LCMS and NMR.
Chemistry
Linear Peptide Synthesis
Linear peptides were synthesized by Fmoc solid-phase synthesis using Wang or 2-chlorotrityl linkers in accordance with our previously reported methods.(9) The resins and the amino acid derivatives, Fmoc-Abu-OH, Fmoc(Me)-Ala-OH, Fmoc-Ala-OH, Fmoc-Arg(Pbf)-OH, Fmoc-Asp(OtBu)-OH, Fmoc-Lys(Boc)-OH, Fmoc-Phe-OH, and Fmoc-Pro-OH were purchased from Calbiochem Novabiochem (Nottingham, U.K.) and Bachem AG (Bubendorf, Switzerland). All solvents used were of HPLC-grade quality.
Molecular Modeling
Molecules were analyzed using SYBYL 7.0 (Tripos Inc., St. Louis, MO). The Biopolymer tools in SYBYL were used to build some of the peptides, to check structures for errors, to correct atom types, and to add/remove hydrogens as necessary. Structures were minimized using the Tripos force field with a steepest descent gradient of 100 iterations followed by a conjugate gradient of 0.01 kcal/mol or a maximum of 10000K iterations as termination criteria. During minimizations, a 12 Å nonbonded cutoff was applied and when solvent was not present and ionic strength was set to 4.00. The collected structural data were analyzed with the graphic tools of SYBYL, version 7.0. All calculations were performed on the dual Hewlett-Packard workstation XW6000, two 2.8 GHz CPUs running REDHAT Enterprise Linux WS4. The possible binding sites of NRP1 b1 domain were searched using the SYBYL SITEID module (SYBYL, version 7.0; Tripos Inc., St. Louis, MO).
Large Scale Synthesis of 2
3-(Benzo[1,2,5]thiadiazole-4-sulfonylamino)thiophene-2-carboxylic Acid Methyl Ester (12)
To a stirred solution of the arylsulfonyl chloride (10) (7.68 g, 32.7 mmol, 1.2 equiv) in pyridine (30 mL) under nitrogen (balloon) at 45 °C was added dropwise over 1 h a solution of methyl 3-aminothiophene-2-carboxylate (11) (4.28 g, 27.3 mmol, 1 equiv) in pyridine (10 mL). The resulting suspension was stirred for 20 h at 20 °C. The reaction was monitored using TLC and was deemed complete after approximately 2 h. Water (25 mL) was added and the resulting mixture stirred for 15 min. The resulting pale-yellow precipitate was isolated by suction filtration and was washed with water (3 × 10 mL) followed by isohexane (3 × 10 mL), then dried (in vacuo) to afford the desired product. Yield: 9.30 g, 96%. Pale-yellow solid. TLC Rf = 0.2 (ethyl acetate/isohexane, 1:2). LCMS: tR = 3.09 min; purity >95%. MS: m/z 354.1 [M − 1]−.
3-(Benzo[1,2,5]thiadiazole-4-sulfonylamino)thiophene-2-carboxylic Acid (13)
The ester 12 (9.28 g, 26.1 mmol, 1 equiv) was stirred with lithium hydroxide monohydrate (5.48 g, 130.5 mmol, 5 equiv) in a tetrahydrofuran/methanol/water mixture (5:3:2; 20 mL) at 50 °C for 4 h. The reaction mixture was monitored using TLC; when deemed complete, the solvents were removed in vacuo. The product was then directly isolated by precipitation using 2 M hydrochloric acid (100 mL) followed by suction filtration. The precipitate was washed with water (3 × 50 mL) and then with isohexane (3 × 50 mL) and dried in vacuo at 40 °C for 14 h to afford the desired product. Yield: 8.75 g, 25.7 mmol, 98%. Beige solid. LCMS: tR = 2.74 min; purity >95%. MS m/z 340 [M − 1]−. 1H NMR (400 MHz, DMSO-d6): δ 10.43 (1H, br, s), 8.44−8.38 (2H, m), 7.86 (1H, d, J = 8.9, 7.1 Hz), 7.75 (1H, d, J = 5.5 Hz), 7.25 (1H, d, J = 5.5 Hz).
(S)-2-{[3-(Benzo[1,2,5]thiadiazole-4-sulfonylamino)thiophene-2-carbonyl]amino}-5-guanidino(Pbf)pentanoic Acid Methyl Ester (15)
The acid (13) (5.0 g, 14.7 mmol, 1 equiv) and bromo-tris-pyrrolidinophosphonium hexafluorophosphate (PyBroP, 8.25 g, 17.7 mmol, 1.2 equiv) were suspended in dichloromethane (50 mL), under nitrogen (balloon), and stirred for 2 min at 20 °C. N,N-Diisopropylethylamine (23 mL, 132.3 mmol, 9 equiv) was added in one portion and the orange solution stirred at room temperature for 10 min. The protected amino acid (H-Arg(Pbf)-OMe; 8.40 g, 17.7 mmol, 1.2 equiv) was added as a single portion, and the reaction mixture was stirred at 20 °C for 4 h. The solvent was removed in vacuo and the residue taken up in ethyl acetate (approximately 200 mL). The solution was washed with 1 M hydrochloric acid (10 × 50 mL) and then evaporated directly to give a viscous brown gum that was maintained under vacuum for 17 h. The resulting brown glassy solid was carried forward without further purification. Yield: 11.2 g, 14.7 mmol, 100%. Brown glassy solid. TLC Rf = 0.44 (methanol/ethyl acetate (10:90)). LCMS tR = 1.81 min; purity >85%.
(S)-2-{[3-(Benzo[1,2,5]thiadiazole-4-sulfonylamino)thiophene-2-carbonyl]amino}-5-guanidino(Pbf)pentanoic Acid
The crude ester 15 (11.2 g, 14.7 mmol) and lithium hydroxide hydrate (3.08 g, 73.5 mmol, 5 equiv) were stirred in tetrahydrofuran/water (2:1, 300 mL) for 30 min at 20 °C. After this time, LCMS indicated complete reaction, The organic solvent was removed in vacuo and hydrochloric acid (1 M, aqueous solution, 20 mL) added to precipitate the product. The resulting solid was collected by filtration and washed with 1 M hydrochloric acid (100 mL), water (2 × 100 mL), and isohexane (3 × 50 mL) and then carried forward directly. Yield: quantitative recovery assumed. Yellow solid. Rf = (ethyl acetate) baseline. LCMS tR = 3.17 min. 1H NMR (400 MHz, DMSO-d6): δ 11.36 (1H, br s), 8.42−8.34 (3H, m), 7.84 (1H, dd, J = 8.8 and 7.0 Hz), 7.66 (1H, d, J = 5.3 Hz), 7.19 (1H, d, J = 5.3 Hz), 4.23−4.20 (1H, m), 3.06−3.01 (2H, m), 2.92 (2H, s), 2.46 (3H, s), 2.41 (3H, s), 1.95 (3H, s), 1.82−1.73 (1H, m), 1.69−1.60 (1H, m), 1.40−1.35 (8H, m).
(S)-2-(3-(Benzo[c][1,2,5]thiadiazole-4-sulfonamido)thiophene-2-carboxamido)-5-guanidinopentanoic Acid (2)
The acid (11.0 g, 14.7 mmol, 1 equiv) was stirred with trifluoroacetic acid/water (9:1, 150 mL) for 65 h at 25 °C. After this time the acidic solution was added to an ice/water mixture, causing the immediate precipitation of a pale-green solid that was collected by filtration and washed copiously with ether before being suspended in further ether and subjected to ultrasonic irradiation for ∼5 min. The resulting fine powder was collected and dried under vacuum at ∼40 °C. Yield: 5.16 g, 8.44 mmol, 57%. Pale-green solid. LCMS tR = 4.77 min; purity >95%. MS m/z 498 [M + 1]+. 1H NMR (400 MHz, (CD3)2SO/D2O): δ 8.26−8.23 (2H, m), 7.78 (1H, dd, J = 8.6 and 7.0 Hz), 7.45 (1H, d, J = 5.3 Hz), 7.17 (1H, d, J = 5.3 Hz), 4.26 (1H, dd, J = 9.6 and 4.5 Hz), 3.16−3.13 (2H, m), 1.88−1.81 (1H, m), 1.77−1.70 (1H, m), 1.60−1.54 (2H, m). 13C NMR (400 MHz, (CD3)2SO): δ 173.0, 163.2, 156.8, 154.7, 148.1, 131.9, 130.1, 128.7, 127.0, 119.8, 113.7, 52.0, 40.3, 27.3, 25.4. LCMS: tR = 4.48 min; purity >96%. MS m/z 498 [M + 1]+. HRMS (m/z): [M]+ calcd for C17H20N7O5S3, 498.0688; found, 498.0697.
Structural Biology Methods
Protein NMR Methods
NMR spectroscopy was performed at either 500 or 600 MHz 1H frequency at 25 °C. 15N or 13C,15N-isotope labeled NRP1 b1 samples were prepared in 50 mM phosphate buffered saline (100 mM sodium chloride), 1.0 mM dithiothreitol, 1.0 mM ethylenediaminetetraacetic acid, pH 6.0. Backbone resonance assignments were obtained by analysis of 3D HNCA, HN(CO)CA, HNCACB, HNCO, HN(CA)CO spectra. Ligand titrations were monitored by 2D 15N,1H HSQC spectroscopy of 0.3 mM NRP1 b1 to which aliquots of buffered ligand (200 mM) were added until saturation was achieved;(13) the maximal protein concentration dilution was <10%. Control experiments were recorded to assess the sensitivity of the NRP1 b1 cross-peak positions to small changes in protein concentration and sample pH. NMR spectra were processed with nmrPipe(14) and analyzed in CCPN analysis.(15) Cross-peak chemical shift perturbations were declared significant when the compound change in 1H and 15N resonance position Δδ = {[(ΔδN/6.5)2 + ΔδH2]}1/2 > 0.15 where ΔδN and ΔδH are measured in ppm. Essentially complete backbone NMR assignments were obtained by standard triple resonance NMR spectroscopy of a uniformly double 13C,15N-isotope labeled sample of NRP1 b1. The pattern of secondary 13C chemical shifts obtained is consistent with the known globular structure of NRP1 b1 from X-ray diffraction studies, indicating that the overall structure in solution is unperturbed (data not shown).
Protein Generation, Purification, and Crystallization with 2
Recombinant human NRP1-b1 domain was expressed in E. coli strain Rosetta (DE3) (Novagen) as a fusion protein containing a tobacco etch virus protease cleavable N-terminal hexahistidine tag. The protein was purified by combination of immobilized metal affinity chromatography, size exclusion chromatography (Superdex75), and ion exchange chromatography on a SP FF Sepharose column. Purified NRP1-b1 protein in 20 mM Tris-chloride (pH 7.9) and 50 mM sodium chloride buffer was incubated with a 10-fold molar excess of 2 for 1 h at 4 °C. The protein was concentrated to 10 mg/mL and crystallized in a 1:1 volume mixture with 100 mM MES (pH 6.0), 10% PEG 8K, and 200 mM zinc acetate at 20 °C employing hanging drop vapor-diffusion method. Crystals appeared within 3 days, with thick needle morphology and a yellow hue, from compound 2. Crystals belonged to space group P21 with cell dimensions of a = 40.96 Å, b = 89.60 Å, c = 41.60 Å, α = 90.00°, β = 97.72°, γ = 90.00° and contained two NRP1-b1 molecules in the asymmetric unit.
X-ray Crystallography
A data set at 2.9 Å resolution was obtained at the London School of Pharmacy by using a R-AXISIV image plate detector system and Osmic mirrors. Data were merged and indexed using the program d*TREK(16) in the CrystalClear(17) software suite. Molecular replacement was performed using PHASER(18) with apo NRP1-b1 domain (PDB code 1KEX). Iterative rounds of building and refinement were carried out in COOT(19) and REFMAC5.(20) TLS groups were generated using the TLSMD Web server.(21) The dictionary file for 2 was generated using the PRODRG Web server.(22) The structure was refined to final R/Rfree values of 21.6/28.2% with reasonable geometry (Table 5).
Table 5. Data Collection and Refinement Statistics (Molecular Replacement).
| parameter | |
|---|---|
| space group | P21 |
| cell dimension | |
| a, b, c (Å) | 41.0, 89.6, 41.6 |
| α, β, γ (deg) | 90, 97.7, 90 |
| resolution (Å) | 30.3−2.9 (3.0−2.9)a |
| Rsym or Rmerge | 0.079 (0.191)a |
| I/σ(I) | 8.5 (4.0)a |
| completeness (%) | 88.0 (85.4)a |
| redundancy | 2.4 (2.3)a |
| Refinement | |
| resolution (Å) | 30.3−2.9 |
| no. reflections | 5852 |
| Rwork/Rfree | 21.6/28.2 |
| no. atoms | |
| protein | 2455 |
| ligand/ion | 70 |
| water | 8 |
| rms deviation | |
| bond length (Å) | 0.012 |
| bond angle (deg) | 1.42 |
Highest-resolution shell is shown in parentheses.
Plasmids, Adenoviruses, and Mutagenesis
The full-length open reading frame of human NRP1 (Origene) was cloned into the pENTR directional TOPO vector. Subsequently plasmid expression constructs (pcDNA3.2/V5-DEST) and adenoviral vectors encoding NRP1 constructs (pAd/CMV/V5-DEST) were generated by LR recombination using the Gateway system (Invitrogen). Site-directed mutagenesis was performed with the QuikChange XL site-directed mutagenesis kit (Stratagene). All constructs were verified by DNA sequencing.
Reagents
Recombinant human VEGF (VEGF-A165) was obtained from R & D Systems (Abingdon, U.K.). Collagen type I, Dulbecco’s modified Eagle’s medium (DMEM)/25 mM HEPES, 5-fluorouracil, and paclitaxel were purchased from Sigma-Aldrich (Dorset, U.K.). All other reagents used were of the purest grade available.
Cell Culture and Transfection
Human umbilical vein endothelial cells (HUVECs) were purchased from TCS CellWorks (Buckingham, U.K.) and cultured in endothelial cell basal medium (EBM) supplemented with 10% fetal bovine serum (FBS), 10 ng/mL human epidermal growth factor, 12 μg/mL bovine brain extract, 50 μg/mL gentamicin sulfate, and 50 ng/mL amphotericin-8. Porcine aortic endothelial cells expressing NRP1 (PAE/NRP1) were provided by Dr. Shay Soker. The cells were grown in Ham’s F12 medium containing 10% FBS and 25 μg/mL hygromycin B. Human tumor cell lines including carcinoma lung A549 and prostate DU145 cells were provided by Quintiles Limited (Edinburgh, U.K.) and grown in RPMI 1640 medium (Invitrogen, Paisley, U.K.) containing 10% and l-glutamine. HUVECs or DU145 cells were seeded at the density of 2 × 104 cells per well of 96-well plates in 0.1 mL growth medium and transfected with adenovirus vectors containing the full-length open-reading frame of human NRP1. The adenovirus NRP1-transfected cells grew for 2 days prior to a binding assay. COS7 cells were obtained from ECACC and maintained in DMEM supplemented with 10% FBS (Invitrogen). COS7 cells were grown in 24-well plates to 80% confluency and transfected with NRP1 plasmid constructs using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Immunoblotting
Cells were extracted by lysis buffer (64 mM Tris-HCl, pH 6.8, 0.2 mM Na3VO4, 2% sodium dodecyl sulfate (SDS), 10% glycerol, protease inhibitors for serine, cysteine, and metalloproteases). The whole cell lysate samples were separated by SDS−PAGE and transferred to Immobilon membranes (Millipore). The membranes were immunoblotted with specific primary antibodies. Immunoreactive bands were visualized by chemiluminescence using horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence reagent (Amersham Biosciences, Bucks, U.K.).
Cell-Based Ligand Binding
Radiolabeled binding displacement experiments were performed in cells grown to confluence in 24-well plates and using the indicated concentrations of compounds followed by addition of 0.1 nM 125I-VEGF-A165 (1200−1800 Ci/mmol, GE Healthcare, U.K.) as described previously (Jia et al., 2006).(9) Nonspecific binding was determined in the presence of 100-fold excess unlabeled VEGF.
For biotinylated (bt)-VEGF-A165 binding, confluent adenovirus NRP1-transfected HUVECs or DU145 cells in 96-well plates were washed twice with phosphate-buffered saline (PBS). The various concentrations of compounds diluted in binding medium (Dulbecco’s modified Eagle’s medium, 25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), pH 7.3, containing 0.1% bovine serum albumin (BSA)) were added, followed by addition of 2 nM bt-VEGF-A165. After 2 h of incubation at room temperature, the medium was aspirated and washed three times with PBS. The bound bt-VEGF-A165 to NRP1 was detected by streptavidin−horseradish peroxidase conjugates and the enzyme substrate and measured using a Tecan Genios plate reader at 450 nm absorbance with a reference wavelength at 595 nm. Nonspecific binding was determined in the presence of 100-fold excess unlabeled VEGF-A165. Half maximal inhibitory concentrations (IC50) for selected compounds were obtained from competition curves obtained from 125I-VEGF-A165 and bt-VEGF-A165 binding assays in which different concentrations (0.1−100 μM) of compound were tested, using Prism, version 4.0, software and the equation
where Top and Bottom are the responses, respectively, at the top and bottom of the curve, Y is the inhibitory response at a given compound concentration X, and the Hillslope describes the steepness of the curve. R2 values measuring goodness of fit of the competition curves were also determined using the Prism software.
Cell-Free bt-VEGF-A165 Binding
The 96-well plates were precoated with NRP1-b1 protein at 3 μg/mL overnight at 4 °C. On the following day, the plates were treated with blocking buffer (PBS containing 1% BSA) and washed three times with wash buffer (PBS containing 0.1% Tween-20). The various concentrations of compounds diluted in PBS containing 1% DMSO were added, followed by addition of 0.25 nM bt-VEGF-A165. After 2 h of incubation at room temperature, the plates were washed three times with wash buffer. The bt-VEGF-A165 bound to NRP1-b1 was detected by streptavidin−horseradish peroxidase conjugates and the enzyme substrate and measured using a Tecan Genios plate reader at 450 nm absorbance with a reference wavelength at 595 nm. Nonspecific binding was determined in the absence of NRP1-b1 coated wells of the plates.
Measurement of VEGFR2 Phosphorylation
Following treatment of HUVECs, phosphorylation at Tyr1175 of VEGFR2 and total VEGFR2 in cell lysates were detected by PathScan phopho-VEGFR2 and total VEGFR2 enzyme linked immunosorbant assay (ELISA) kits (Cell Signalling) according to the manufacturer’s instructions.
Cell Migration
Cell migration was measured in chemotaxis 24-transwell plates with collagen I-coated inserts incorporating polyethylene terephthalate track-etched membranes with 8 μm pores (Becton Dickinson Biosciences). VEGF in EBM/0.1% BSA was placed in the bottom wells of the plates. Cells were trypsinized, washed, and resuspended in EBM/0.1% BSA to give a final cell concentration of 3 × 105/mL. Then 1.5 × 105 cells with or without 2 treatment as indicated were loaded into each top inserts. The chemotaxis transwell plates were incubated at 37 °C for 4 h. After the incubation, nonmigrated cells on the top side of the transwell membranes were removed and migrated cells on the under side of the transwell membranes were stained with the REASTAIN Quick-Diff kit (Reagena, Ltd., Toivala, Finland). The stained cells from each well were counted in four fields at ×100 magnification using an eyepiece indexed graticule (100 grids).
Cell Viability
Cell viability was determined by measurement of conversion of the tetrazolium salt XTT to form formazan dye. Carcinoma cells were seeded at a density of 4 × 103 cells per well of 96-well plates in 100 μL of serum-free medium containing various concentrations of 5-FU or paclitaxel as indicated in the absence or presence of 2 at 100 μM. After 44 h of incubation, XTT labeling reagent mixture (Roche Diagnostics, East Sussex, U.K.) was added to the cultures and they were incubated for a further 4 h. The formazan product was then measured at 490 nm with a reference wavelength at 595 nm.
Acknowledgments
Research in the PCD group is supported by the MRC (file reference U117574559). Research in the IZ group is supported by the British Heart Foundation (grant RG/06/003). We thank Claudio Dagostin (compound synthesis), Emma Dugdale (analysis and purification), Stewart Kirton (computational chemistry), and Kerry Jenkins and Trevor Perrior (crystal structure interpretation of 2). We also thank Gary Parkinson (London School of Pharmacy) for assistance with data collection, Claire Newton for project management, and John Martin for his unwavering support.
Supporting Information Available
Full experimental details of the synthesis of the compounds mentioned in the text and additional figures. This material is available free of charge via the Internet at http://pubs.acs.org.
Atomic coordinates and structure factors for the reported crystal structure have been deposited in the Protein Data Bank under accession code 3I97.
Footnotes
Abbreviations: G, glycine; P, proline; A, alanine; V, valine; L, leucine; I, isoleucine; M, methionine; C, cysteine; F, phenylalanine; Y, tyrosine; W, tryptophan; H, histidine; K, lysine; R, arginine; Q, glutamine; N, asparagine; E, glutamic acid; D, aspartic acid; S, serine; T, threonine; bt, biotinylated; Boc, tert-butyloxycarbonyl; BSA, bovine serum albumin; DMEM, Dulbecco’s modified Eagle’s medium; DMSO, dimethyl sulfoxide; EBM, endothelial cell basal medium; ELISA, enzyme-linked immunosorbent assay; FBS, fetal bovine serum; FMOC, 9-fluorenylmethyl)oxy]carbonyl; 5-FU, 5-fluorouracil; HEPES, N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid; HSQC, heteronuclear single quantum coherence; HUVECs, human umbilical vein endothelial cells; NRP1, neuropilin 1; NRP2, neuropilin 2; Pbf, 2,2,4,6,7-pentamethyldihydrobenzofuran-5-sulfonyl; PBS, phosphate buffered saline; PyBrOP, bromo-tris-pyrrolidinophosphonium hexafluorophosphate; PAE, porcine aortic endothelial; SDS, sodium dodecyl sulfate; SEMA3A, semaphorin 3A; TIPS, triisopropylsilane; TFA, trifluoroacetic acid; Tris, tris(hydroxymethyl)aminomethane; VEGF, vascular endothelial growth factor; VEGFR2, vascular endothelial growth factor receptor 2.
Supplementary Material
References
- Pellet-Many C.; Frankel P.; Jia H.; Zachary I. Neuropilins: structure, function and role in disease. Biochem. J. 2008, 411, 211–226. [DOI] [PubMed] [Google Scholar]
- Kruger R. P.; Aurandt J.; Guan K. L. Semaphorins command cells to move. Nat. Rev. Mol. Cell Biol. 2005, 6, 789–800. [DOI] [PubMed] [Google Scholar]
- Pan Q.; Chanthery Y.; Liang W. C.; Stawicki S.; Mak J.; Rathore N.; Tong R. K.; Kowalski J.; Yee S. F.; Pacheco G.; Ross S.; Cheng Z.; Le C. J.; Plowman G.; Peale F.; Koch A. W.; Wu Y.; Bagri A.; Tessier-Lavigne M.; Watts R. J. Blocking neuropilin-1 function has an additive effect with anti-VEGF to inhibit tumor growth. Cancer Cell 2007, 11, 53–67. [DOI] [PubMed] [Google Scholar]
- Wells J. A.; McClendon C. L. Reaching for high-hanging fruit in drug discovery at protein−protein interfaces. Nature 2007, 450, 1001–1009. [DOI] [PubMed] [Google Scholar]
- Hajduk P. J.; Huth J. R.; Tse C. Predicting protein druggability. Drug Discovery Today 2005, 10, 1675–1682. [DOI] [PubMed] [Google Scholar]
- Lee C. C.; Kreusch A.; McMullan D.; Ng K.; Spraggon G. Crystal structure of the human neuropilin-1 b1 domain. Structure 2003, 11, 99–108. [DOI] [PubMed] [Google Scholar]
- Vander Kooi C. W.; Jusino M. A.; Perman B.; Neau D. B.; Bellamy H. D.; Leahy D. J. Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 6152–6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Single letter amino acid nomenclature is used for amino acids in mutations.
- Jia H.; Bagherzadeh A.; Hartzoulakis B.; Jarvis A.; Lohr M.; Shaikh S.; Aqil R.; Cheng L.; Tickner M.; Esposito D.; Harris R.; Driscoll P. C.; Selwood D. L.; Zachary I. C. Characterization of a bicyclic peptide neuropilin-1 (NP-1) antagonist (EG3287) reveals importance of vascular endothelial growth factor exon 8 for NP-1 binding and role of NP-1 in KDR signaling. J. Biol. Chem. 2006, 281, 13493–13502. [DOI] [PubMed] [Google Scholar]
- Wey J. S.; Gray M. J.; Fan F.; Belcheva A.; McCarty M. F.; Stoeltzing O.; Somcio R.; Liu W.; Evans D. B.; Klagsbrun M.; Gallick G. E.; Ellis L. M. Overexpression of neuropilin-1 promotes constitutive MAPK signalling and chemoresistance in pancreatic cancer cells. Br. J. Cancer 2005, 93, 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.; Friday B. B.; Lai J. P.; Yang L.; Sarkaria J.; Kay N. E.; Carter C. A.; Roberts L. R.; Kaufmann S. H.; Adjei A. A. Cytotoxic synergy between the multikinase inhibitor sorafenib and the proteasome inhibitor bortezomib in vitro: induction of apoptosis through Akt and c-Jun NH2-terminal kinase pathways. Mol. Cancer Ther. 2006, 5, 2378–2387. [DOI] [PubMed] [Google Scholar]
- Schneider B. P.; Sledge G. W. Jr. Anti-VEGF therapy as adjuvant therapy: clouds on the horizon?. Breast Cancer Res. 2009, 11, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J.; Fairbrother W. J.; Palmer A. G.; Rance M.; Skelton N. J.. Protein NMR Spectroscopy: Principles and Practice, 2nd ed.; Academic Press: Boston, MA, 2007. [Google Scholar]
- Delaglio F.; Grzesiek S.; Vuister G. W.; Zhu G.; Pfeifer J.; Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Vranken W. F.; Boucher W.; Stevens T. J.; Fogh R. H.; Pajon A.; Llinas M.; Ulrich E. L.; Markley J. L.; Ionides J.; Laue E. D. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 2005, 59, 687–696. [DOI] [PubMed] [Google Scholar]
- Pflugrath J. W. The finer things in X-ray diffraction data collection. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999, 55, 1718–1725. [DOI] [PubMed] [Google Scholar]
- CrystalClear: An Integrated Program for the Collection and Processing of Area Detector Data; Rigaku Corporation, 1997. [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Painter J.; Merritt E. A. TLSMD Web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 2006, 39, 109–111. [Google Scholar]
- Schuttelkopf A. W.; van Aalten D. M. F. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 1355–1363. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.