Abstract
Rodents homozygous for autosomal leptin receptor gene mutations not only become obese, insulin resistant, and hyperleptinemic but also develop a dysregulated immune system. Using marker-assisted breeding to introgress the Koletsky rat leptin receptor mutant (lepr−/lepr−), we developed a novel congenic BBDR.lepr−/lepr− rat line to study the development of obesity and type 2 diabetes (T2D) in the BioBreeding (BB) diabetes-resistant (DR) rat. While heterozygous lepr (−/+) or homozygous (+/+) BBDR rats remained lean and metabolically normal, at 3 wk of age all BBDR.lepr−/lepr− rats were obese without hyperglycemia. Between 45 and 70 days of age, male but not female obese rats developed T2D. We had previously developed congenic BBDR.Gimap5−/Gimap5− rats, which carry an autosomal frameshift mutation in the Gimap5 gene linked to lymphopenia and spontaneous development of type 1 diabetes (T1D) without sex differences. Because the autoimmune-mediated destruction of pancreatic islet β-cells may be affected not only by obesity but also by the absence of leptin receptor signaling, we next generated BBDR.lepr−/lepr−,Gimap5−/Gimap5− double congenic rats carrying the mutation for Gimap5 and T1D as well as the Lepr mutation for obesity and T2D. The hyperleptinemia rescued end-stage islets in BBDR.lepr−/lepr−,Gimap5−/Gimap5− congenic rats and induced an increase in islet size in both sexes, while T1D development was delayed and reduced only in females. These results demonstrate that obesity and T2D induced by introgression of the Koletsky leptin receptor mutation in the BBDR rat result in islet expansion associated with protection from T1D in female but not male BBDR.lepr−/lepr−,Gimap5−/Gimap5− congenic rats. BBDR.lepr−/lepr−,Gimap5−/Gimap5− congenic rats should prove valuable to study interactions between lack of leptin receptor signaling, obesity, and sex-specific T2D and T1D.
Keywords: obesity, diabetes, islet inflammation, lymphopenia
obesity and diabetes are two prevalent chronic diseases that have become a major public health concern in industrialized countries, affecting at least 16 million people in the United States alone (29, 43). While type 2 diabetes (T2D) was previously thought to be an adult-onset disease, the prevalence of T2D is increasing in an alarming manner in children and adolescents, likely because of the epidemic rise in obesity in old and young alike (38). Also, the incidence rate of type 1 diabetes (T1D) is increasing worldwide, and it is among the most common chronic childhood illnesses, affecting 18–20 per 100,000 children a year with an increasing incidence of 3–4% annually, currently afflicting 1.4 million Americans (36, 41). There is much discussion about the possible contribution of obesity in children to the current increase in the incidence rate of T1D (18, 49, 50). Associated with these epidemiologic changes are a reduction in high-risk HLA genotypes among new T1D patients and an increase in previously protective or neutral haplotypes (7, 39). These findings suggest that the increased incidence of T1D may well be related to the obesity epidemic among children and adolescents in the Western world.
The obesity epidemic may contribute to the rising trend of T1D not only in developed but also in developing countries. The mechanisms mediating this epidemic are not understood, but it is speculated that adipokines or cytokines may affect the islet autoimmune pathogenetic process. It cannot be excluded that processes related to different etiologies of obesity may accelerate while others decelerate the pathogenic process leading to autoimmune killing of β-cells.
The BioBreeding Diabetes-Prone (BBDP) rat is widely used to study the etiopathogenic processes of T1D (for a review see Ref. 32). There are many similarities between the BioBreeding (BB) rat and humans in terms of clinical presentation and disease progression including hyperglycemia, polyuria, polydipsia, ketoacidosis, and insulin dependence for life (33, 42). T1D develops in all (100%) rats, regardless of sex, between 50 and 70 days of age. As in humans, diabetes in the BB rat is dependent on the major histocompatibility complex (MHC), a semidominant factor localized on rat chromosome 20 (Iddm1), and the single MHC u haplotype. The RT1u/u genotype is implicated, which is an ortholog of human HLA-DQ (16, 25, 33). A primary difference between the BB rat and human T1D is a lymphopenia defect present at birth in the rat, which results in a dramatic decrease in the number of CD4+ and CD8+ T lymphocytes. The lymphopenia (lyp/Iddm2) locus was cloned by position (14, 24) and the lymphopenia phenotype attributed to a frameshift deletion in the Gimap5 gene of the novel GTPase-immune-associated protein (Gimap)-related gene family (6, 17, 20, 35). The deletion results in a null allele, as the truncation of a significant portion of the Gimap protein was associated with a complete loss of the protein (30). Gimap5 belongs to a gene family of GTP-binding proteins and is normally present in T and B lymphocytes (5, 12, 35). Dissection of the genetics of BB rat T1D has, as in humans (4), revealed non-MHC genetic factors for diabetes risk on rat chromosome 4 proximal to the Gimap genes (4, 10) and on rat chromosome 2 to include the ortholog of the human T1D PTPN22 factor (4) in addition to a number of other genetic factors (32).
In humans, T1D requires MHC haplotype association and autoimmune destruction of pancreatic β-cells, whereas T2D requires both peripheral insulin resistance and inadequate β-cell compensation. While the BB rat has made it possible to dissect the genetics of spontaneous rat T1D, inbred and genetically well-characterized obese rats with T2D are not immediately available. Here we describe the utility of marker-assisted breeding to introgress a known leptin receptor mutation onto our BB diabetes-resistant (BBDR or DR) rat line (1, 4) and to generate DR.lepr−/lepr−,Gimap5−/Gimap5− double congenic rats carrying the mutation for Gimap5 and T1D as well as the Lepr mutation for obesity and T2D. The aim of our study was to examine the pathogenesis of T1D and T2D in the congenic DR.lepr−/lepr− and DR.lepr−/lepr−,Gimap5−/Gimap5− rats.
The Koletsky rat possesses features of metabolic syndrome such us obesity, hyperlipidemia, hyperinsulinemia, insulin resistance, and proteinuric kidney disease (19). The obese, normotensive, and hyperphagic phenotype in the Lister and Albany (LA/N) rat is a congenic strain derived from the Koletsky rat (40). The LA/N rat has the same mutation of the leptin receptor inherited as a single homozygous recessive trait, originally designated f, shown to be allelic with the Zucker fatty trait (fa) but of distinct origin. The obesity mutation designated fak, where the “k” identifies Koletsky, is a nonsense mutation of the leptin receptor gene, resulting in a premature stop codon in the leptin receptor extracellular domain at amino acid +763 (45, 52). This suggests that in the fak rat none of the leptin receptor isoforms encodes a transmembrane domain necessary for signal transduction. In contrast, the Zucker fatty (fa) rat has a missense mutation at position +269 coding a different exon in the extracellular domain in the leptin receptor (3, 15, 45).
Leptin is an adipocyte-derived hormone/cytokine that links nutritional status with neuroendocrine and immune functions (21). It is mainly produced by adipocyte tissue in relation to body fat mass and, at lower levels, by stomach, skeletal muscle, and placenta (9). A very well-studied role of leptin is to regulate body weight by reduction of food intake via a satiety signal and to increase energy expenditure through stimulation of thermogenesis. The leptin receptor is a member of the class I cytokine receptor family (8, 46, 47) and is expressed in the hypothalamus in areas that regulate appetite and body weight. It is also expressed in endothelial cells, pancreatic β-cells, bone marrow precursors, monocytes/macrophages, and T and B cells, and an increasing number of studies implicate leptin signaling in immune function (8, 27, 28, 46, 47).
In the present study we used an approach based on marker-assisted breeding to accomplish the introgression of genetic factors (11, 24, 31). Marker-assisted breeding differs from previously reported strategies that developed obese BB rats by classical genetic methods resulting in removal of the Iddm2 gene and retention of the mutated Lepr (fa) gene from the Zucker fatty rat (48). We used our recently developed DR.Gimap5−/Gimap5− congenic rat (8) to introgress the Koletsky LA/N.lepr− mutation from chromosome 5 by marker assisted breeding. First, a congenic DR.+/+,lepr−/lepr− rat was developed to determine the effect of the lepr−/lepr− mutation on the congenic DR.+/+ rat. Second, this congenic rat was crossed with the DR.Gimap5−/Gimap5− congenic rat (1) to generate a double congenic rat carrying both the mutation for Gimap5 (lyp) and spontaneous T1D as well as the mutation for obliteration of leptin signaling leading to obesity and T2D.
MATERIALS AND METHODS
Introgression of Koletsky mutation onto congenic DR.+/+ rat.
The introgression of the Lepr gene from the Koletsky rat onto the BBDR rat was initiated in 2001 in collaboration with Dr. J. T. Hansen, Animal Genetic Resource, National Institutes of Health and completed in 2007 after 15 cycles of backcross breeding by marker-assisted breeding (31). The first eight breeding cycles were bred in the laboratory of Dr. Hansen and the next nine cycles at the University of Washington in Seattle.
Genotyping lepr− gene on chromosome 5.
Between 25 and 30 days of age, 2-mm tail biopsies were obtained and DNA was isolated according to the following protocol: Tail biopsies were digested overnight in a water bath at 55°C with 12 μl of a 20 mg/ml stock of proteinase K (Promega, Madison, WI) in 500 μl SET (1% SDS in 150 mmol/l NaCl, 5 mmol/l EDTA, and 50 mmol/l Tris, pH 8.0). The following morning the samples were centrifuged at 20,800 g for 15 min, and the supernatant was transferred to 500 μl of cold isopropanol. The samples were again centrifuged at 20,800 g for 5 min, the isopropanol was discarded, and the DNA pellet was washed twice in cold 70% EtOH. The pellet was air dried for 1 h and resuspended in 200 μl of TE (10 mmol/l Tris, 1 mmol/l EDTA, pH 7.4) at 55°C overnight. The samples were diluted to 25 ng/μl in double-distilled (dd)H2O, and 2 μl of this genomic DNA was used in the following 10-μl PCR reaction: 1 μl of 10× reaction buffer (Promega), 0.8 μl of MgCl2 (Promega), 0.2 μl of 10 mmol/l dNTPs (Promega), 0.5 μl of 1 μmol/l M13-labeled forward primer (Qiagen), 0.5 μl of 20 μmol/l reverse primer (Qiagen), 0.1 μl of Taq DNA polymerase (Promega), 1 μl of M13 700 (LiCor Biosciences), and 3.9 μl of ddH2O. All samples were amplified according to the following standard PCR protocol: 1) 95°C for 5 min, 2) 95°C for 20 s, 3) 60°C for 20 s, and 4) 72°C for 30 s, steps 2–4 repeated 30 times, and then 72°C for 3 min. PCR products were diluted to 25% with STOP solution (LiCor Biosciences) and analyzed with a NEN Global IR2 DNA Analyzer System (model 4200S-2) with a 6.5% gel matrix (LiCor Biosciences). Repetitive simple DNA sequences were found with the University of California–Santa Cruz (UCSC) rat genome browser at http://genome.ucsc.edu/index.html, and the primers were designed with Primer3 (Massachusetts Institute of Technology, Cambridge, MA).
Whole genome scan.
At the N11 generations, we genotyped 140 simple sequence length polymorphisms (SSLPs) with an average genome coverage of 10 cM to ensure that the genomic background of the new DR.lepr−/lepr− strain was fixed for BBDR with the exception of the recombinant fragment from Koletsky DNA that was introgressed. At the N11 generation, the number of SSLPs was reduced to 46, including only markers on chromosomes that were not fully fixed along the entire chromosome length (at a 10-cM resolution). All SSLPs were amplified, and genotypes were determined with fluorescent genotyping on an ABI3730XL capillary sequencer, as outlined in detail elsewhere (34). Briefly, each forward primer for the SSLP is synthesized with a 5′ tail containing the universal M13 primer sequence (5′-TGTAAAACGACGGCCAGT-SSLPf-3′). The PCR is a two-step reaction containing the primers specific for the SSLP and an additional fluorophore-labeled M13 primer. The initial amplification steps incorporate and amplify the SSLP-specific primers, thus incorporating the M13 tail. The latter rounds incorporate the labeled M13 dye-conjugate primer, allowing for detection on the ABI3730XL capillary sequencer and analysis with ABI GeneMapper software. Multiple fluorophores are used, in combination with different PCR product sizes, to allow multiplexing of four SSLPs in a single capillary.
Introgression of congenic DR.lepr−/lepr− rat with congenic DR.Gimap5−/Gimap5− rat.
In the second step, a brother-sister litter of DR.lepr−/lepr+ congenic rats was crossed with the DR.Gimap5−/Gimap5− congenic rat (1) to generate DR.lepr−/lepr+,Gimap5−/Gimap5+ rats, followed by an intercross of the F1 generation rats. In the F1 generation only DR.lepr−/lepr+,Gimap5−/Gimap5− males were crossed with DR.lepr−/lepr+,Gimap5−/Gimap5+ females, and from this cross we generated a double congenic rat carrying both the mutation for Gimap5 (lyp or gimap5−) and T1D as well as the mutation for Lepr and obesity and T2D.
Rat housing.
Rats were housed in a specific pathogen-free facility at the University of Washington, Seattle at controlled temperature (21–25°C) and relative humidity (40–50%) and maintained on a 12:12-h light-dark cycle with 24-h access to regular food (5053 Harlan Teklad, Madison, WI) and water. All protocols were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Washington, Seattle. The University of Washington Rodent Health Monitoring Program was used to track infectious agents via a quarterly sentinel monitoring system, and sentinels were negative for agents tested. Excluded infectious agents are listed at http://depts.washington.edu/compmed/rodenthealth/index.html#excluded.
Blood cells counts and flow cytometry of lymphocyte phenotypes.
Animals were anesthetized with intraperitoneal pentobarbital sodium and exsanguinated. Peripheral blood samples were collected into heparinized tubes at 45 days for the analyses of white blood cells (K/μl), red blood cells (RBC, M/μl), hemoglobin (g/dl), hematocrit (%), mean corpuscular volume (MCV, fl), mean corpuscular hemoglobin (MCH, pg), mean corpuscular hemoglobin concentration (%), polymorphonuclear cells (%), lymphocytes (%), and monocytes (%). Rat thymocytes, spleen, and mesenteric lymph node (MLN) were minced in phosphate-buffered saline (PBS) containing 10% fetal calf serum (FCS) and filtered with a 40-μm cell strainer. The single-cell suspensions from spleen were washed with lysis buffer (BD Pharm Lyse, BD Biosciences, San Diego, CA) for the elimination of RBC.
Peripheral blood mononuclear cells were isolated by Ficoll-Hypaque gradient centrifugation. Approximately 1 × 106 cells were stained with fluorescence-labeled antibodies in 100 μl of staining buffer (PBS containing 4% BSA) for 30 min on ice in 96-well U-bottom plates. Cells were washed twice in staining buffer before being fixed for 20 min on ice in PBS containing 1% paraformaldehyde (PFA), washed, suspended in 300 μl of PBS, and analyzed by flow cytometry. Two- and three-color flow cytometric analysis were performed on a FACS-440 (Becton Dickinson Immunocytometry Systems, Sunnyvale, CA) equipped with a helium-neon and argon laser. Acquisition was performed with the Consort 30 program, and analysis was done with Lysis 1 (Becton Dickinson). The flow cytometer was calibrated with QC3 (FITC/PE/Cy-Chrome) beads. Forward scatter and side scatter were used for electronic exclusion of contaminating dead cells. The panel of monoclonal antibodies prelabeled with FITC, PE, and Cy-chrome and PerCP monoclonal antibodies were purchased from Pharmingen (San Diego, CA) to be used for analysis of T cell subsets: anti-CD3 (G4.18), anti-CD4 (OX35), anti-CD8 (OX8), anti-CD90 (Thy-1), anti α-β T cell receptor (R73), anti-CD25 (OX39), anti-CD45 (OX1), anti-CD45RC (OX22), and anti-CD45RA (OX33).
Phenotypic examinations.
Starting at 35–40 days of age, all rats were weighed daily (Sartorious, Edgewood, NY) and blood glucose was tested (Ascensia Contour, Bayer, Leverkusen, Germany) until 180 days of age. Diabetes was diagnosed when blood glucose exceeded 200 mg/dl for 2 consecutive days. Animals were anesthetized with pentobarbital sodium intraperitoneally, weighed, measured (body length from anus to nose), and then exsanguinated. Blood samples were collected at 180 days of age into heparinized tubes for white blood cell counts and then centrifuged to obtain the supernatant-plasma to analyze cytokine and adipokine profiles and to measure the concentration of insulin, leptin, cholesterol, glucagon, and triglycerides. Insulin, leptin, and glucagon were measured by ELISA (Linco). Colorimetric assay kits were used to assay total cholesterol (Diagnostic Chemicals Limited) and triglycerides (Roche Diagnostics). Adipokine and cytokine profiles [IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p70), IL-13, IL-17, IL-18, eotaxin, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), GRO/KC, IFN-γ, leptin, monocyte chemoattractant protein-1 (MCP-1), macrophage inflammatory protein-1α (MIP-1α), IFN-inducible protein-10 (IP-10), regulated on activation, normal T expressed and secreted (RANTES), TNF-α, and VEGF] were determined with the BeadLyte cytokine assay kit (Millipore, Billerica, MA). Samples were diluted 1:2.5 with RPMI + 10% FCS and loaded onto a Millipore Multiscreen BV 96-well filter plate. Serial dilutions of cytokine standards were prepared in parallel and added to the plate. BeadLyte 24-Plex Cytokine beads were vortexed for 30 s, and 25 μl was added to each well with plasma samples. Samples were then incubated on a plate shaker at 600 rpm in the dark at room temperature for 2 h. The plate was applied to a Millipore Multiscreen Vacuum Manifold and washed twice with 50 μl of assay buffer (PBS, pH 7.4, 1% BSA, 0.05% Tween 20, 0.05% sodium azide), and each well was resuspended with 75 μl of assay buffer. Twenty-five microliters of biotinylated Anti-Human Multi-Cytokine Reporter was added to each well. The plate was incubated at room temperature for 1.5 h on a plate shaker at 600 rpm in the dark. Streptavidin-PE was diluted 1:12.5 in assay buffer, and then 25 μl was added directly to each well. The plate was incubated on a plate shaker at 600 rpm in the dark at room temperature for 30 min. Twenty-five microliters of stop solution [0.2% (vol/vol) formaldehyde in PBS, pH 7.4] was added to each well and incubated at room temperature for 5 min. The plate was then applied to the vacuum manifold, and each well was resuspended in 125 μl of assay buffer and shaken for 1 min. The assay plate was transferred to the Bio-Plex Luminex 100 XYP instrument for analysis. Cytokine concentrations were calculated with Bio-Plex Manager 4.1 software with a five-parameter curve-fitting algorithm applied for standard curve calculations. Body mass index (BMI, g/cm2) was determined by dividing body weight (g) by the square of body length (cm2). The abdominal fat pads, consisting of mesenteric, retroperitoneal, and gonadal fat pads, were removed and weighed. The adiposity index was calculated from each fat pad and body weight (% of fat pad weight/body weight).
Histological assessment of pancreatic islets and islet immunostaining.
Multiple organs and tissues were removed and fixed in 4% (wt/vol) PFA in PBS. The specimens were washed three times in 50% EtOH for 1 h each and preserved in 70% EtOH before being embedded in paraffin, and 5-μm sections were cut and stained with hematoxylin and eosin (H & E). Histological sections were examined by a pathologist blind to experimental groups. All tissues were given a morphological diagnosis, and the pancreatic islet changes were categorized and scored.
After initial histological evaluation, pancreatic islet size, degeneration, and inflammation were scored for severity as follows. Histological islet sizes, which can be influenced by sectioning, were graded within the study first by initial blind read of all animals to categorize as small, normal, or enlarged. Then the wild-type controls were identified for the pathologist, and the remaining animals were graded in relation to wild-type islet size. The size severity scores ranged from −1, where islets were either obliterated by inflammation or smaller than normal; 0 for normal-sized islets; 1 for islets 1.5–2× normal; 2 for islets that were 2.5–3× normal with few entrapped exocrine acinar cells; and 3 for islets >3× normal with numerous entrapped acinar cells. Enlarged islets had an irregular shape, encroachment, and occasional entrapment of acinar exocrine cells. Islet cell degeneration was characterized by swollen cells with vacuolated to clear cytoplasm and condensed nuclei. Degeneration was scored as follows: 0, no degeneration in islet; 1, <10% of cells within islets; 2, 11–25%; 3, >26%. Inflammation was characterized by perivascular, periductular, and intraislet mixed inflammation composed of lymphocytes and macrophages that were occasionally hemosiderin laden, eosinophils, and neutrophils. Inflammatory changes were scored as follows: 0, none; 1, minimal, <10 cells/islet; 2, mild multifocal, few foci; 3, moderate, coalescing foci within islet; 4, marked with obliteration of islet architecture.
Pancreatic tissue was also subjected to dual immunofluorescence staining using glucagon- and insulin-specific antibodies for the identification of islet α- and β-cells, respectively. Briefly, pancreatic tissues were fixed in 10% phosphate-buffered formalin at 4°C and then processed (Sakura Tissue-Tek VIP 5) and embedded (Sakura Tissue-Tek TEC 5 Tissue Embedding Console System) in paraffin (Fisher Scientific, Fair Lawn, NJ). Three- to four-micrometer sections were prepared on a standard microtome (MICROM HM 355-S) and mounted on Fisher PLUS slides. Before staining, slides were dried (MOPEC medite TDO 60) at 65°C for at least 1 h and then deparaffinized and hydrated down to distilled water (Sakura Tissue-Tek DRS). Sections were heat treated at 96°C in target antigen retrieval solution (DakoCytomation), rinsed in water and then 50 mM Tris buffer (pH 7.6), and blocked for 1 h at room temperature with 5% donkey serum (Sigma-Aldrich, St. Louis, MO). For dual immunofluorescence, sections were incubated overnight at room temperature with a combined 1/10 dilution of goat polyclonal anti-glucagon (Santa Cruz) and a 1/2,000 dilution of mouse monoclonal anti-insulin (Sigma-Aldrich) in PBS with sodium azide. Sections were rinsed and incubated with a 1/40 dilution of Texas Red-conjugated donkey anti-goat IgG (Jackson ImmunoResearch Laboratories) and FITC-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories) secondary antibodies in Tris buffer for 1 h. After staining, slides were washed with PBS and dried by centrifugation; coverslips were mounted with Vectashield mounting medium (Vector Laboratories) and imaged and analyzed on a Nikon E600 upright microscope system (Nikon USA, Melville, NY) equipped with infinity optics, a Princeton Instruments MicroMAX 5 MHz cooled charge-coupled device camera, and MetaMorph version 6.3r3 software (Universal Imaging).
Statistical analysis.
Data are shown as means ± SE. Statistical differences between DR.lepr−/lepr−, DR.Gimap5−/Gimap5−, DR.lepr−/lepr−,Gimap5−/Gimap5−, and DR.+/+ congenic rats were determined by t-test using unequal variances. All two-tail statistical tests were judged statistically significant at P < 0.05. Survival time is presented in days when 25% of a line develops diabetes. A comparison of the equality of survival functions was tested with the log-rank test. Statistical analyses were performed with Stata 8 (StataCorp 2003, Stata Statistical Software release 8, StataCorp, College Station, TX).
RESULTS
Establishment of congenic BBDR.lepr+/lepr− line.
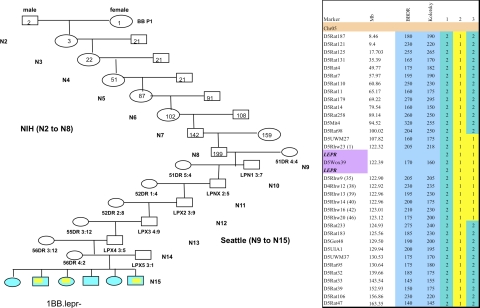
The introgression of the Lepr− gene from the Koletsky rat onto the diabetes-resistant BBDR rat was completed in 2007 after 15 cycles of backcrossing by marker-assisted breeding. A female BBDR.+/+ rat provided the genetic background of the first intercross with a male heterozygous DR.lepr+/lepr− to obtain the F1 hybrids carrying the Lepr− allele. The heterozygous F1 DR.lepr+/lepr− females were backcrossed to male BBDR.+/+, and the N2 generation was obtained (Fig. 1, left). Subsequently, 15 backcross matings were performed between the progeny and the recipient BBDR rats for the unique congenic DR.lepr−/lepr− rat (Figs. 1 and 2). All rats were screened for recombination events between the D5UWM27 and D5Rhw20 markers representing an introgression of ∼20 Mb from Koletsky rat DNA onto the DR background (Fig. 1, right).
Fig. 1.
Breeding strategy (left) and genotyping of chromosome 5 (right). Left: the breeding approach of total N15 backcrosses that led to the generation of the new congenic line. The first intercross and from N2 to N8 backcrosses were generated in the laboratory of Dr. J. T. Hansen at the National Institutes of Health (NIH). From N9 to N15 backcrosses were produced in the Robert H. Williams Laboratory, University of Washington, of Å. Lernmark. At each backcross the DNA was sent to Seattle for genotyping assessment. Numbers and names in and below each box represent rat ID. Right: simple sequence length polymorphism (SSLP) marker map of chromosome 5. Marker annotations are shown together with the University of California-Santa Cruz (UCSC) map marker location and the size of the marker in the 3 parental strain BioBreeding diabetes-resistant (BBDR) and Koletsky rats. Purple, localization of the Lepr gene expanded from 122.320.075 bp to 122.503.437 bp with the marker D5Wox39 inside the DNA fragment that comprised the gene; blue, markers polymorphic between the parental strains. Animals in lanes 1–3 represent the parental and new Lepr−/− congenic line. The animal in lane 1 is diabetes resistant (DR), lane 2 Koletsky (Kol), lane 3 BBDR.Lepr− (D5Rat98-D5Rat233)Rhw. The numerical color code is green (2) DR/DR and yellow (1) Kol/Kol. Colors in left panel correspond with colors in lanes 1 and 3 in right panel.
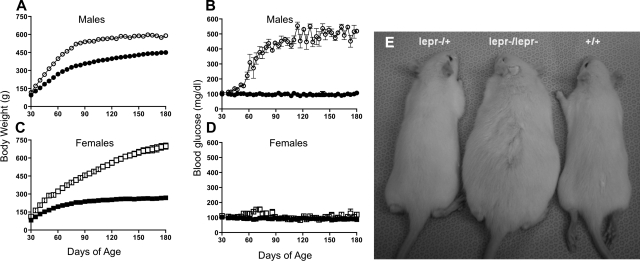
Fig. 2.
Time course of changes in body weight and blood glucose of male and female congenic rats. Rat body weights (A and C) and blood glucose (B and D) of DR.lepr−/lepr− [n = 3 (○) and n = 4 (□)] and DR.+/+ [n = 4 (● and ■)]. All rats were monitored daily from 30 to 180 days of age. Data are means ± SE. Most bars are not visible because of small SE. P < 0.05 vs. age-matched wild-type control group, which persisted up to 180 days of age (ANOVA). Data for DR.lepr−/lepr+ were the same as for DR.+/+. E: representative image of the 3 genotypes at 45 days of age.
The successful generation of the double congenic DR.lepr−/lepr−,Gimap5−/Gimap5− line was initiated after the single congenic lines for Lepr− and Gimap5− were established. The intercrossing of the single congenic lines generated DR.lepr−/lepr+,Gimap5−/Gimap5+ animals, and then from the F1 intercross selected DR.lepr−/lepr+,Gimap5−/Gimap5− males were used to cross with the DR.lepr−/lepr+,Gimap5−/Gimap5+ females to produce the double congenic rats.
Sex difference in obesity and hyperglycemia in DR.lepr−/lepr− rats.
Obesity differences were first discernible starting around 30 days of age (Fig. 2, A and C), with a robust phenotype visible at 45 days of age between homozygous DR.lepr−/lepr− and homozygous DR.+/+ rats (Fig. 2E). Sex differences were readily apparent. DR.lepr−/lepr− males gained weight until they developed hyperglycemia between 60 and 90 days of age (Fig. 2, A and B). At that time body weight remained constant, although DR.lepr−/lepr− males were still ∼29% heavier than DR.+/+ control rats at 180 days (P < 0.001) and blood glucose levels were increased fourfold (P < 0.001). In contrast, DR.lepr−/lepr− females continued to gain weight and were >160% heavier than DR.+/+ female control rats at 180 days (P < 0.001) but showed no differences in blood glucose levels (P = not significant; Fig. 2, C and D).
Visceral adiposity.
Phenotypic differences in abdominal, mesenteric, retroperitoneal, and gonadal fat pad weights and indexes and BMI at 180 days of age (Table 1) paralleled weight differences between DR.lepr−/lepr− and DR.+/+ rats as described above (Fig. 2). With the exception of body length in males, the leptin receptor mutation was associated with increases in all of these measures in the DR.lepr−/lepr− rats (P < 0.001 for all except body length in females, which was P < 0.01).
Table 1.
Phenotypic analyses of abdominal fat of lean and obese male and female DR.lepr−/lepr− rats at 180 days of age
| Males |
Females |
|||||
|---|---|---|---|---|---|---|
| DR.+/+ | DR.lepr−/lepr− | P value | DR.+/+ | DR.lepr−/lepr− | P value | |
| No. of animals | 5 | 3 | 3 | 4 | ||
| Body wt, g | 451.4 ± 15 | 589 ± 10.1 | * | 263 ± 13.6 | 699 ± 49 | * |
| Body length, cm | 25.3 ± 0.5 | 25 ± 0.50 | ns | 21.3 ± 0.30 | 24 ± 0.8 | * |
| BMI, g/cm2 | 7.2 ± 0.2 | 9.7 ± 0.20 | * | 5.7 ± 0.20 | 12 ± 0.5 | * |
| Mesenteric fat pad, g | 3.6 ± 0.3 | 14 ± 1.70 | * | 2.2 ± 0.60 | 15 ± 1.8 | * |
| Retroperitoneal fat pad, g | 8 ± 1.2 | 31 ± 5.50 | * | 2.3 ± 0.20 | 32 ± 1.4 | * |
| Gonadal fat pad, g | 10 ± 2.2 | 25 ± 0.60 | * | 4.1 ± 0.50 | 24 ± 1.1 | * |
| Mesenteric fat index | 0.8 ± 0.0 | 2.4 ± 0.30 | * | 0.8 ± 0.20 | 2.1 ± 0.1 | * |
| Retroperitoneal fat index | 1.7 ± 0.2 | 5.3 ± 0.80 | * | 0.9 ± 0.20 | 4.6 ± 0.3 | * |
| Gonadal fat index | 2.1 ± 0.4 | 4.3 ± 0.20 | * | 1.5 ± 0.20 | 3.4 ± 0.2 | * |
Values represent means ± SE. Body mass index (BMI) is defined as body weight (g) divided by square of body length (cm). Fat pad index is defined as fat pad weight divided by body weight. P values were determined by Student's t-test assuming unequal variances.
Significant (<0.01); ns, not significant.
Blood cell counts.
Early blood cell counts at 45 days of age showed that DR.lepr−/lepr− rats had higher MCV, MCH, and percentage of polymorphonuclear cell values compared with DR.+/+ rats (P < 0.05 for all; Table 2). However, DR.lepr−/lepr− rats also had lower levels of RBC and percentage of lymphocytes (P < 0.05 for both). The only significant differences remaining by 180 days of age were the higher percentage of polymorphonuclear cells and lower percentage of lymphocytes in DR.lepr−/lepr− rats (P = 0.01 for both).
Table 2.
Blood cell counts at different ages for DR.+/+ and DR.lepr−/lepr− rats
| 45 Days |
180 Days |
|||||
|---|---|---|---|---|---|---|
| DR.+/+ | DR.lepr−/lepr− | P value | DR.+/+ | DR.lepr−/lepr− | P value | |
| WBC, K/μl | 5.9 ± 0.85 | 4.5 ± 0.51 | ns | 6.96 ± 0.4 | 6.68 ± 1.4 | ns |
| RBC, M/μl | 7.01 ± 0.12 | 5.97 ± 0.03 | * | 7.66 ± 0.3 | 8.32 ± 0.4 | ns |
| HGB, g/dl | 15.1 ± 0.28 | 13.77 ± 0.11 | ns | 15.04 ± 0.5 | 15.8 ± 0.76 | ns |
| HCT,% | 51.4 ± 0.99 | 47.52 ± 0.32 | ns | 50 ± 1.9 | 53.3 ± 3.19 | ns |
| MCV, fl | 73.3 ± 0.56 | 79.83 ± 0.31 | * | 65.6 ± 1 | 63.8 ± 1.07 | ns |
| MCH, pg | 21.5 ± 0.1 | 23.05 ± 0.08 | * | 19.68 ± 0.3 | 19.1 ± 0.19 | ns |
| MCHC, % | 29.3 ± 0.16 | 28.85 ± 0.3 | ns | 30.14 ± 0.4 | 29.9 ± 0.55 | ns |
| Polys, % | 8.43 ± 1.19 | 15.5 ± 3 | * | 7.75 ± 1.2 | 13.5 ± 0.29 | * |
| Lymph, % | 88.9 ± 1.79 | 81.17 ± 2.48 | * | 89 ± 1.5 | 82 ± 1.22 | * |
| Monos, % | 2.43 ± 0.68 | 3.33 ± 0.67 | ns | 3 ± 0.4 | 3.5 ± 1.19 | ns |
Data represent means ± SE for n = 6 rats/group at 45 days of age and 5 rats/group at 180 days of age. Data are combined for male and female rats. WBC, white blood cells; RBC, red blood cells; HGB, hemoglobin; HCT, hematocrit; MCV, mean corpuscular volume; MCH, mean corpuscular hemoglobin; MCHC, mean corpuscular hemoglobin concentration; Polys, polymorphonuclear cells; Lymph, lymphocytes; Monos, monocytes. P values were determined by Student's t-test.
P < 0.05.
Serum cytokines and adipokines.
Concentration of serum cytokines and adipokines differed between DR.+/+ and DR.lepr−/lepr− rats (Table 3). Eotaxin levels were elevated twofold in female DR.lepr−/lepr− rats and fourfold in male DR.lepr−/lepr− rats compared with sex-matched control rats (P = 0.06). The cytokines IL-13 and IFN-γ were significantly elevated in male DR.lepr−/lepr− rats (P = 0.03 and 0.05, respectively), whereas the adipokine MCP-1 was increased more than fourfold in female DR.lepr−/lepr− rats (P < 0.001) compared with sex-matched control rats. Serum levels of IL-1α, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12 (p70), IL-13, IL-17, IL-18, G-CSF, GM-CSF, MIP-1α, IP-10, TNF-α, and VEGF were not significantly different between male or female DR.+/+ and DR.lepr−/lepr− rats (data not shown).
Table 3.
Serum cytokines and adipokines at 180 days of age in DR.lepr−/lepr− line
| Females |
Males |
|||||
|---|---|---|---|---|---|---|
| DR.+/+ | DR.lepr−/lepr− | P value | DR.+/+ | DR.lepr−/lepr− | P value | |
| Eotaxin | 69.45 ± 25.9 | 136.3 ± 95.0 | ns | 45.55 ± 5.0 | 196.5 ± 53.1 | 0.06 |
| GRO/KC | 718.1 ± 45.5 | 2,830 ± 787.7 | 0.07 | 949.2 ± 61.1 | 1,009 ± 192.4 | ns |
| IL-13 | 1,005 ± 432.9 | 1,941 ± 1,358 | ns | 551.1 ± 113.4 | 2,174 ± 450.0 | * |
| IFN-γ | 418.0 ± 180.7 | 926.0 ± 655.9 | ns | 254.5 ± 59.1 | 1,103 ± 261.8 | * |
| RANTES | 9,245 ± 359.9 | 13,470 ± 1,513 | 0.06 | 15,450 ± 2,244 | 10,050 ± 2,020 | ns |
| MCP-1 | 173.3 ± 30.7 | 720.9 ± 75.8 | † | 337.6 ± 44.6 | 296.4 ± 39.3 | ns |
Data (in pg/ml) represent means ± SE for n = 4 rats/group for each genotype in male and female rats. RANTES, regulated on activation, normal T expressed and secreted; MCP-1, monocyte chemoattractant protein-1. Significance values of the means were determined by Student's t-test.
P < 0.05,
P < 0.001.
Phenotypes of thymus, spleen, MLN, and peripheral blood immune cells.
Obese DR.lepr−/lepr− rats had a higher proportion of thymus cells that were CD4−CD8− (double negative, DN) and single positive (SP) CD4+ and a lower proportion that were CD4+CD8+ (double positive, DP), CD4−CD8+, α-β TCR (R73+), and CD90+ compared with lean rats (Table 4; P < 0.05 with the exception of DN). The lean DR.+/+ rat had a higher proportion of spleen cells that were R73+, R73+CD8+, R73+CD4−CD8−, CD45+CD45RA+, CD45+CD45RC+, and CD45+ and a lower proportion that were R73+CD4+ and R73+CD4+CD8+ compared with the obese rats (P < 0.05). In the MLN and peripheral blood the R73+CD4+CD8+ DP cell subset was higher in obese compared with lean rats, while the R73+ T cell was lower (P < 0.05). The proportion of CD4+ cells that expressed CD25+ did not differ between lean and obese rats.
Table 4.
Thymus, spleen, mesenteric lymph nodes, and peripheral blood immune cell phenotype of DR.lepr−/lepr− line
| DR.+/+ | DR.lepr−lepr− | P | |
|---|---|---|---|
| Thymus | |||
| DN | 5.35 ± 1.1 | 8.06 ± 2.8 | ns |
| DP | 82.04 ± 0.8 | 78.18 ± 2.4 | * |
| CD4+ | 7.89 ± 0.7 | 10.03 ± 0.5 | * |
| CD8+ | 4.73 ± 0.6 | 3.73 ± 0.3 | * |
| R73+ | 94.55 ± 0.9 | 89.50 ± 2.6 | * |
| CD4+CD25+ | 0.89 ± 0.2 | 0.79 ± 0.1 | ns |
| CD90+ | 97.31 ± 1.1 | 93.25 ± 2.8 | * |
| CD45+CD45RA+ | 0.63 ± 0.2 | 0.49 ± 0.1 | ns |
| CD45+CD45RC+ | 0.72 ± 0.1 | 0.87 ± 0.2 | ns |
| CD45+ | 97.08 ± 1.1 | 93.99 ± 3.4 | ns |
| Spleen | |||
| R73+ | 25.08 ± 1.5 | 20.25 ± 1.7 | * |
| R73+CD4+ | 63.41 ± 1.7 | 67.99 ± 1.4 | * |
| R73+CD8+ | 27.59 ± 1.1 | 21.03 ± 1.2 | * |
| R73+CD4+CD8+ | 4.18 ± 0.3 | 5.98 ± 0.6 | * |
| R73+CD4−CD8− | 3.98 ± 0.4 | 2.45 ± 0.2 | * |
| CD4+CD25+ | 3.14 ± 0.3 | 3.21 ± 0.4 | ns |
| CD90+ | 29.55 ± 2.4 | 24.93 ± 1.5 | * |
| CD45+CD45RA+ | 42.23 ± 2.5 | 32.32 ± 1.1 | * |
| CD45+CD45RC+ | 30.85 ± 2.6 | 19.15 ± 1.1 | * |
| CD45+ | 92.25 ± 1.6 | 84.86 ± 2.7 | * |
| MLN | |||
| R73+ | 75.30 ± 3.9 | 77.20 ± 7.5 | ns |
| R73+CD4+ | 77.98 ± 6.3 | 76.07 ± 3.0 | ns |
| R73+CD8+ | 14.66 ± 4.5 | 14.39 ± 3.7 | ns |
| R73+CD4+CD8+ | 2.26 ± 0.4 | 3.39 ± 0.8 | * |
| R73+CD4−CD8− | 1.71 ± 0.2 | 1.66 ± 0.6 | ns |
| CD4+CD25+ | 6.03 ± 1.1 | 6.66 ± 0.7 | ns |
| CD45+CD45RA+ | 18.29 ± 3.5 | 17.31 ± 2.5 | ns |
| CD45+CD45RC+ | 20.60 ± 3.3 | 19.91 ± 2.4 | ns |
| CD45+ | 98.05 ± 1.6 | 96.93 ± 2.5 | ns |
| Peripheral blood | |||
| R73+ | 31.08 ± 4.8 | 19.83 ± 6.7 | ns |
| R73+CD4+ | 80.25 ± 2.3 | 78.52 ± 1.9 | ns |
| R73+CD8+ | 16.31 ± 3.2 | 14.60 ± 3.9 | ns |
| R73+CD4+CD8+ | 1.75 ± 0.3 | 3.09 ± 0.4 | * |
| R73+CD4−CD8− | 0.93 ± 0.4 | 0.69 ± 0.0 | ns |
| CD4+CD25+ | 0.95 ± 0.3 | 1.20 ± 0.1 | ns |
| CD45+CD45RA+ | 12.93 ± 1.4 | 13.13 ± 4.3 | ns |
| CD45+CD45RC+ | 19.25 ± 2.7 | 19.10 ± 5.1 | ns |
| CD45+ | 33.83 ± 7.1 | 38.40 ± 7.1 | ns |
Data are from male and female rats and represent means ± SD for n = 4/group for each genotype. Values are the proportion (%) of the total gated cells as determined by immunofluorescence. MLN, mesenteric lymph node. Differences among DR.+/+ control and DR.lepr−/lepr− rats were determined by Student's t-test.
P < 0.05.
Serum phenotypes of DR.+/+, DR.lepr−/lepr−, and DR.lepr−/lepr−,Gimap5−/Gimap5− rats.
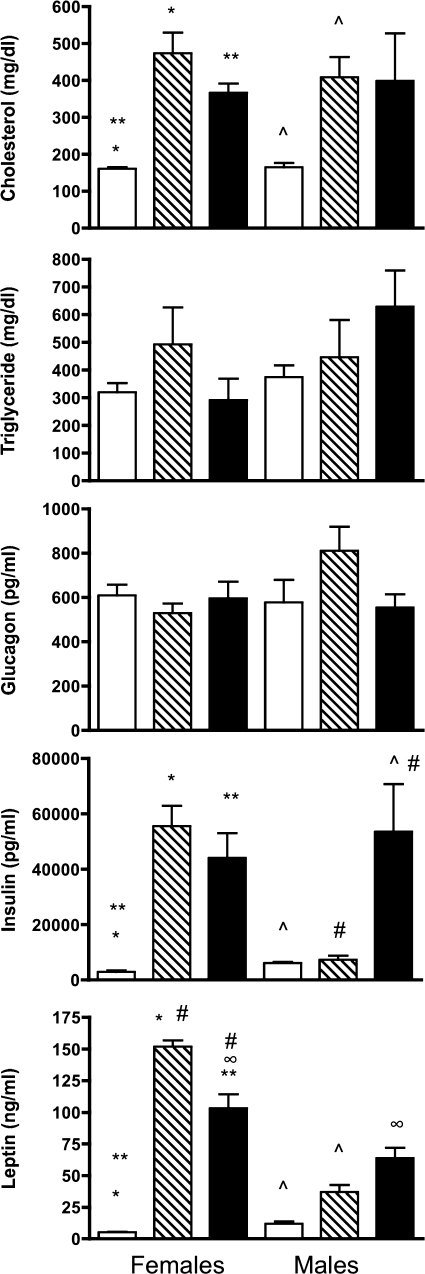
The levels of cholesterol, triglycerides, glucagon, insulin, and leptin were assayed in serum from DR.+/+, DR.lepr−/lepr−, and DR.lepr−/lepr−,Gimap5−/Gimap5− male and female rats at 180 days of age (Fig. 3). Cholesterol levels were elevated in both male and female DR.lepr−/lepr− and DR.lepr−/lepr−,Gimap5−/Gimap5− female rats compared with DR.+/+ control rats (P < 0.05). Triglyceride and glucagon levels did not differ between any of the lines in either males or females. Insulin levels were significantly elevated in female DR.lepr−/lepr− and in male and female double congenic DR.lepr−/lepr−,Gimap5−/Gimap5− rats compared with control rats (P < 0.05). However, insulin levels in male DR.lepr−/lepr− rats did not differ from levels observed in male control rats. As anticipated, leptin levels were significantly elevated in both female and male leptin receptor-deficient rats. The highest leptin levels, 151.8 ± 4.9 ng/ml, were observed in female DR.lepr−/lepr− rats, which contrasted with 5.1 ± 0.3 ng/ml observed in female control rats. The differences were not as large in males. Leptin concentration in male DR.lepr−/lepr− rats was 36.7 ± 5.5 ng/ml compared with 11.9 ± 1.6 ng/ml in male control DR.+/+ rats. Leptin levels for both male and female DR.lepr−/lepr−,Gimap5−/Gimap5− rats were also elevated compared with control rats (P < 0.05 for all) (Fig. 3). Leptin levels in female DR.lepr−/lepr− rats were about twofold elevated over their male counterparts, which may be related to the development of diabetes in male but not female DR.lepr−/lepr− rats.
Fig. 3.
Values for cholesterol, triglycerides, glucagon, insulin, and leptin in serum of female and male DR.+/+, DR.lepr−/lepr−, and DR.lepr−/lepr−,Gimap5−/Gimap5− rats. Serum samples were evaluated at 180 days of age for the +/+ (open bars) and lepr−/lepr− (hatched bars) rats and at 150 days of age for lepr−/lepr−,Gimap5−/Gimap5− (filled bars) double congenic animals. Bars that share common symbols in the same chart are significantly different (P < 0.05) (Student's t-test). Leptin levels in DR.lepr−/lepr−,Gimap5−/Gimap5−: female > male (P = 0.051). Data are means ± SE (n = 3–5/group).
Weights and blood glucose in DR.lepr−/lepr−,Gimap5−/Gimap5− female rats.
There were no differences in body weight between DR.lepr−/lepr− and DR.lepr−/lepr−,Gimap5−/Gimap5− nondiabetic rats until 150 days of age (data not shown). As expected, as hyperglycemia developed in the female DR.lepr−/lepr−,Gimap5−/Gimap5− rats they were unable to maintain the same body weight growth curve as their nondiabetic double congenic littermates. At 150 days of age the five diabetic rats weighed less (443 ± 73 vs. 609 ± 52 g) and had higher blood glucose levels (430 ± 92 vs. 117 ± 42 mg/dl) than nondiabetic rats (P < 0.05 and P < 0.001, respectively).
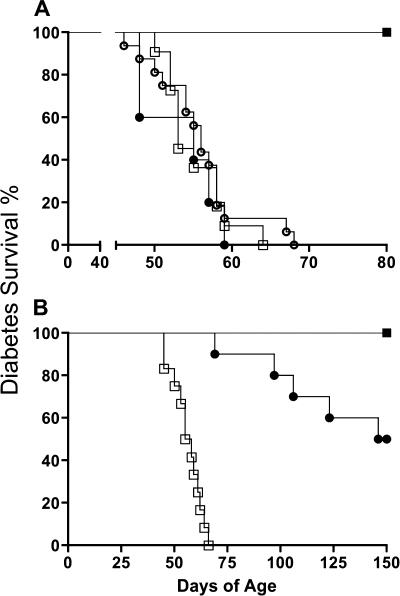
Survival estimates and time to diabetes of DR.Gimap5−/Gimap5−, DR.lepr−/lepr−, and DR.lepr−/lepr−,Gimap5−/Gimap5− rats.
Kaplan-Meier survival curves and the log-rank test for equality of survivor functions were used to compare time to diabetes onset in male and female DR.Gimap5−/Gimap5−, DR.lepr−/lepr−, and DR.lepr−/lepr−,Gimap5−/Gimap5− rats (Fig. 4). As we have previously described (10), 25% of male and female DR.Gimap5−/Gimap5− rats develop diabetes around 52 days of age (P = 0.18; Fig. 4). The survivor functions for the three male DR.Gimap5−/Gimap5−, DR.lepr−/lepr−, and DR.lepr−/lepr−,Gimap5−/Gimap5− rat lines did not differ (P = 0.69); 100% of all three lines develop diabetes. In contrast to males, female DR.lepr−/lepr− rats did not develop diabetes, showing that leptin receptor deficiency induces diabetes selectively in males (P < 0.001). Survivor functions for male and female double congenic DR.lepr−/lepr−,Gimap5−/Gimap5− rats also differed (P < 0.001); half of the male rats were diabetic by 55 days of age, while no females developed diabetes by that age.
Fig. 4.
Kaplan-Meier survival estimates in male (A) and female (B) rats. A: days of age of diabetes onset in DR.+/+ [n = 9 (■)], DR.lepr−/lepr− [n = 16 (○)], DR.Gimap5−/Gimap5− [n = 11 (□)], and DR.lepr−/lepr−,Gimap5−/Gimap5− [n = 5 (●)] male rats. B: time of diabetes onset in DR.+/+ and DR.lepr−/lepr− [n = 6 for each genotype (■)], DR.Gimap5−/Gimap5− [n = 12 (□)], and DR.lepr−/lepr−,Gimap5−/Gimap5− [n = 10 (●)] female rats.
Histological assessment of pancreatic islets and islet immunostaining.
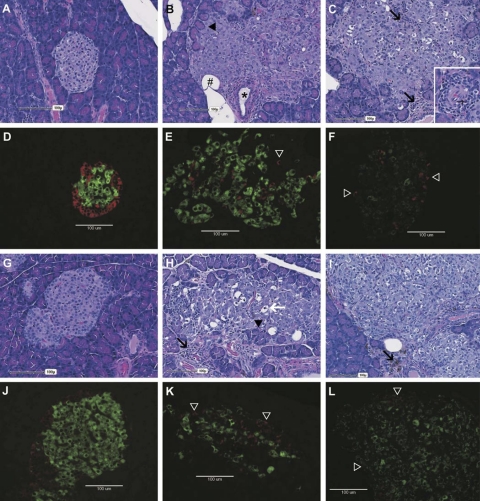
Pancreatic sections from female (Fig. 5, A–F) and male (Fig. 5, G–L) rats at various stages of diabetes were stained for comparison. Histological sections were stained with H & E, and an immunofluorescent staining protocol was used to identify insulin- and glucagon-producing cells. Nondiabetic DR.lepr−/lepr− rat islets (Fig. 5, A and C) and diabetic DR.lepr−/lepr−,Gimsp5−/Gimap5− rat islets (Fig. 5, G and I) were histologically enlarged and irregular (likely hypertrophied) with variable encroachment and entrapment of acinar exocrine cells, inflammation, and prominent vasculature compared with wild-type rats (Fig. 5, D and J). Nondiabetic DR.lepr−/lepr− rat islets (Fig. 5, D and F) and diabetic DR.lepr−/lepr−,Gimsp5−/Gimap5− rat islets (Fig. 5, J and L) were immunostained for insulin and glucagon to confirm that the enlarged islets were due to expansion of islet cells and dispersion of α- and β-cells with marked disruptions in islet architecture. The insulin/FITC intensity per square millimeter (data not shown) indicated lower insulin intensity in the DR.lepr−/lepr− female and male rat islets (P = 0.06 for both) compared with DR.+/+ controls, respectively (Fig. 5, E vs. D and K vs. J).
Fig. 5.
Representative hematoxylin and eosin (H & E)-stained histological and insulin and glucagon immunofluorescent sections from female (A–F) and male (G–L) rats. Wild-type DR.+/+ (left); DR.lepr−/lepr− (center), and DR.lepr−/lepr−,Gimap5−/Gimap5− (right) rats are shown. Age and no. of days with diabetes: DR.+/+ control rat at 180 days of age (A, D, G, and J); nondiabetic female DR.lepr−/lepr− rat at 180 days of age (B and E); 113-day diabetic DR.lepr−/lepr− male rat at 180 days of age (H and K); 27-day diabetic DR.lepr−/lepr−,Gimap5−/Gimap5− rat at 150 days of age (C and F), and 36-day diabetic DR.lepr−/lepr−,Gimap5−/Gimap5− rats at 81 days of age (I and L). Wild-type islets are clearly demarcated from exocrine acinar cells with central insulin-producing β-cells (green) surrounded by a ring of glucagon-producing α-cells (red) in females (A and D) and males (G and J). In contrast, DR.lepr−/lepr− islets in females (B and E) and males (H and K) are enlarged with an irregular shape, encroachment, and occasional entrapment of acinar exocrine cells (black arrowheads). Immunofluorescent sections stained for insulin and glucagon confirm that the enlarged islets noted on H & E-stained sections are composed of abundant β-cells and disrupted α-cells (E and K, open arrowheads) with multifocal black regions within the islets likely representing entrapped acinar cells (B and H, black arrowheads), lipocytes (B, #), ducts (B, *) or regions of inflammatory (H, black arrows) or degenerate islet cells characterized by enlarged cells with condensed to pyknotic nuclei, occasional karyorrhexis, and vacuolated cytoplasm (H, white arrow). Note the relative increase in degenerate cells within the male DR.lepr−/lepr− islet compared with the female DR.lepr−/lepr− islet. Enlarged islets were also noted in DR.lepr−/lepr−,Gimap5−/Gimap5− double congenic rats (C and F, female; I and L, male), immunostaining again confirming that enlarged islets contained abundant β-cells and disrupted α-cells (F and L, open arrowheads). Note the perivascular, periductular, and intraislet mixed inflammation (C, inset and I, black arrow) composed of lymphocytes and macrophages, occasionally hemosiderin laden (I, arrow), eosinophils (C, inset and black arrows), and neutrophils.
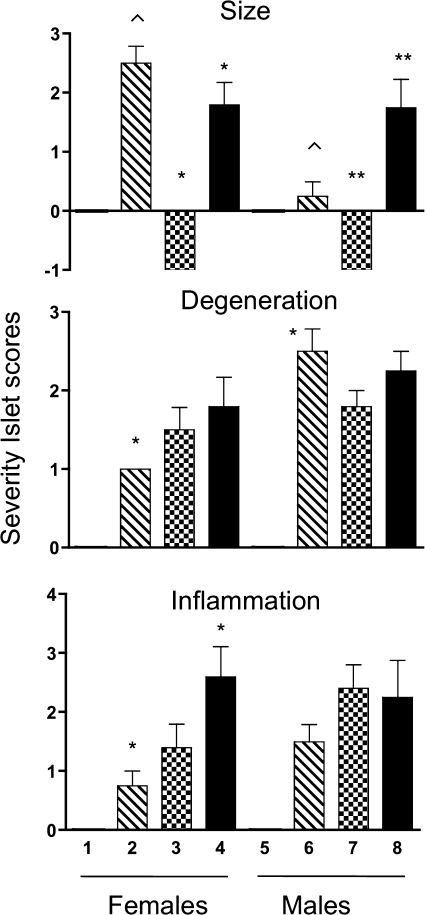
Assessment of pancreatic islet size, degeneration, and inflammation.
Pancreatic islets from all genotypes at different stages of diabetes were assessed for size, degeneration, and inflammation. Compared with DR.+/+ rats, which were assigned a score of 0 (Fig. 6), islet size was increased in female DR.lepr−/lepr− rats (P < 0.05) but not males. However, both male and female DR.lepr−/lepr−,Gimap5−/Gimap5− rats had increased islet size compared with DR.Gimap5−/Gimap5− rats (P < 0.05 for both). In contrast, islet size was reduced within 1 day after the onset of diabetes in all DR.Gimap5−/Gimap5− rats. Only male DR.lepr−/lepr− rats showed a high score in islet degeneration compared with female rats from the same genotype (P < 0.05) (Fig. 6). There was an increase in islet inflammation in DR.lepr−/lepr−,Gimap5−/Gimap5− female rats compared with DR.lepr−/lepr− (P < 0.05) and DR.Gimap5−/Gimap5− female rats. Compared with controls, all three genotypes showed islet degeneration and islet inflammation.
Fig. 6.
Score assessment of pancreatic islet size (top), degeneration (middle), and inflammation (bottom). Values for female (1–4) and male (5–8) rats. Open bars, DR.+/+ (1 and 5); hatched bars, DR.lepr−/lepr− (2 and 6); checkerboard bars, DR.Gimap5−/Gimap5− (3 and 7); filled bars, DR.lepr−/lepr−,Gimap5−/Gimap5− (4 and 8). Ages for scores are as follows: 1, 2, 5, and 6, 180 days of age; 3 and 7, a day after diabetes onset; 4, 150 days of age; 8, between 15 and 27 days after diabetes onset. Groups marked by common symbols in the same chart are significantly different (P < 0.05). Data are means ± SE (n = 3–5/group).
Comparison of diabetes, obesity, sex phenotype, and islet histology.
The data in Table 5 display a summary of the phenotypic differences for the BBDR congenic lines with respect to obesity, diabetes, islet size, and α- and β-cell staining. T2D affected both DR.lepr−/lepr− and DR.lepr−/lepr−,Gimap5−/Gimap5− male but only half of female DR.lepr−/lepr−,Gimap5−/Gimap5− rats. In both female DR.lepr−/lepr− and DR.lepr−/lepr−,Gimap5−/Gimap5− rats β-cell insulin staining was reduced and α-cell glucagon staining was fragmented.
Table 5.
Summary of BBDR congenic lines
| Gimap5−/Gimap5− | lepr−/lepr− | lepr−/lepr−,Gimap5−/Gimap5−- | |
|---|---|---|---|
| Obesity | No | Yes | Yes |
| Sex | N/A | M = F | M = F |
| Diabetes | T1D | T2D | T2D |
| Sex* | M = F(100%) | M(100%) F(0%) | M(100%) F(50%) |
| Islet anatomy | |||
| Islet size | End stage | Enlarged | Enlarged |
| β-Cell staining | |||
| Male | Absent | Reduced | Reduced |
| Female | Absent | Reduced | Reduced |
| α-Cell mantle | |||
| Male | Disrupted | Fragmented | Fragmented |
| Female | Disrupted | Fragmented | Fragmented |
M, males; F, females; N/A, not applicable.
Percentage of animals that develop diabetes by sex.
DISCUSSION
The present study demonstrates that by introgressing the defective leptin receptor gene from the Koletsky rat onto the BB DR rat we effectively generated early age at onset obesity followed by a sex-specific diabetes occurring only in males. Hence, in the DR.lepr−/lepr− rats the lack of leptin signaling renders diabetes protection only in female rats. Preservation of the leptin receptor mutation was ensured by initial genotyping of the Koletsky founder animals and was confirmed by genotyping the generations bred during the 15 backcrosses. The rats are kept in heterozygous breeding because all DR.lepr−/lepr− male develop obesity, diabetes, and hypogonadism. An advantage of heterozygous breeding is that all pups are born to nonobese and nondiabetic parents.
The pathogenic process in the DR.lepr−/lepr− rat can be summarized as follows. First, onset is age dependent: no rats develop diabetes before 40 days of age, and in our present DR.lepr−/lepr− rats, 100% of males develop diabetes before 80 days. Second, the onset is sex specific: no female DR.lepr−/lepr− rats developed diabetes (Figs. 2 and 4). Third, the mode of inheritance is recessive in the cross between DR.lepr−/+ rats: 25% of offspring develop obesity. Fourth, the onset of diabetes is rapid in the males: the animals progress from normoglycemia to hyperglycemia within a week. At 180 days of age the visceral adiposity index is the same in male and female DR.lepr−/lepr− rats, but female rats have higher body weight and leptin levels than male DR.lepr−/lepr− rats (Table 1 and Fig. 3). It can be of interest to know whether the excess of body weight in females was due to higher content of subcutaneous adipose tissue, since it is known that the higher leptin expression levels in this fat depot contribute to blood leptin concentration and whereas visceral adipose tissue is a risk factor for the metabolic syndrome and diabetes, the subcutaneous adipose tissue seems to protect against ectopic deposition of excess lipids in liver, muscle, pancreas, adversely affected in obesity and diabetes. Histological analysis of the DR.lepr−/lepr− rat pancreas indicates the presence of enlarged islets with multifocal vacuolization compared with DR.+/+ rats (Fig. 5), suggesting cellular degeneration. It has been previously reported in the BioBreeding Zucker diabetes-prone (BBZDP)/Wor rat β-cell that hyperplasia and diabetes develop because of a combination of insulin resistance and autoimmune insulitis (48). Our breeding strategy to yield the present congenic line of DR.lepr−/lepr− rats, in which the nonsense mutation of the Lepr (fak) gene was fixed on the DR rat, has resulted in an animal with extreme obesity and diabetes. In contrast, the (BBZDR)/Wor rat was generated by a classical genetic method by which the Iddm2 gene was removed and the missense mutation of the Lepr (fa) gene from the Zucker fatty rat was retained (48).
Serum cytokine and adipokine levels in DR.lepr−/lepr− rats.
The serum cytokine and adipokine levels in DR.lepr−/lepr− rats suggest that the leptin receptor mutation is associated with the induction of cytokines coupled with adipocyte expansion that is the result of increased cytokine or adipokine secretion, or both. Eotaxin, IL-13, and INF-γ are increased in male rats. Consistent with the possibility that these cytokines contribute to diabetes development is the observation that obese male DR.lepr−/lepr− rats do not show a significant increase in serum insulin. Although leptin levels are increased, as expected, in both female and male DR.lepr−/lepr− rats, leptin levels in female rats were more than twofold elevated over their male counterparts. In contrast, the serum levels of GRO/KC, RANTES, VEGF, and MCP are increased in females but not in male DR.lepr−/lepr− rats. These data suggest that certain cytokines and adipokines may contribute to diabetes resistance in female DR.lepr−/lepr− rats. These serum cytokine and adipokine data suggest that the lack of the leptin receptor induces pathogenetic processes that affect T2D development in male but not in female rats. Additional evidence of innate immune activation in DR.lepr−/lepr− rats is reflected by granulocyte cell counts, which were twice those of DR+/+ rats. The increased granulocyte levels parallel the eosinophilia associated with islet inflammation in the development of T2D in DR.lepr−/lepr− rats (Fig. 5) (13, 26).
Leptin receptor and immunity.
DR.lepr−/lepr− rats express a nonfunctional form of leptin receptor due to a nonsense mutation in the leptin receptor gene (40). As a consequence, the extracellular domain of the leptin receptor is absent and thus obese DR.lepr−/lepr− rats lack the ability to respond to leptin via the long isoform of the leptin receptor. Recent evidence indicates that leptin has a significant role in immune cell function (2, 22, 23, 28, 37, 44) and is expressed on several immune cell types, including macrophages, dendritic cells, and T and B lymphocytes (27). Therefore the absence of a functional leptin receptor signaling via the long isoform b likely accounts, at least in part, for the impaired production of inflammatory mediators in the splenocytes of the JCR:LA-cp rat (51). The present finding that the leptin receptor mutation in DR.lepr−/lepr− rats alters the phenotype of normal immune cells particularly in the thymus and spleen compared with DR.+/+ rats suggests that the leptin receptor may function as a key gene in the development of T cells in the thymus.
DR.lepr−/lepr−,Gimap5−/Gimap5− double congenic rats.
We generated DR.lepr−/lepr−,Gimap5−/Gimap5− double congenic rats carrying the mutation for Gimap5 that confers lymphopenia and T1D as well as the Koletsky leptin receptor mutation for obesity and T2D. In double congenic DR.lepr−/lepr−,Gimap5−/Gimap5− rats we showed that males all developed diabetes but that in females onset was delayed. Therefore, the introgression of the leptin receptor mutation onto the DR.Gimap5−/Gimap5− rat is inducing protection of T1D pathogenesis in the females but not in the males. These findings strongly suggest that sex-specific mechanisms are important to the development of DR.Gimap5−/Gimap5− rat T1D similar to the human disease. In humans more males than females develop diabetes after 10–12 yr of age. We showed that hyperleptinemia rescues end-stage islets in DR.lepr−/lepr−,Gimap5−/Gimap5− congenic rats and induces an increase in islet size in both sexes, while there is a modulated T1D development only in the females. This may be related to the significantly higher levels of leptin observed in female over male DR.lepr−/lepr−,Gimap5−/Gimap5− rats. The insulin levels in DR.+/+, DR.lepr−/lepr−, and DR.lepr−/lepr−,Gimap5−/Gimap5− rats seem to correlate positively with pancreatic islet size and negatively with islet degeneration and inflammation (Fig. 6). However, glucagon levels are not affected, even though in immunofluorescent sections stained for glucagon lower levels can be observed. Importantly, female double congenic rats do not seem to require insulin treatment, unlike DR.Gimap5−/Gimap5− lymphopenic rats. It is likely that these differences in serum insulin can be explained by the abnormal pancreatic islets observed in male DR.lepr−/lepr− rats (Table 5 and Fig. 6). It is therefore possible that the leptin receptor mutation modulates the immune system in its acute attack on the islet β-cells in the lymphopenic rat.
Conclusion.
This is the first study to describe a DR.lepr−/lepr−,Gimap5−/Gimap5− double congenic rat that is a genetic rodent model of obesity and T2D. In this model the impaired immune response caused by the Gimap5− lymphopenia phenotype delays the onset of diabetes in DR.lepr−/lepr−,Gimap5−/Gimap5− females by modulating the immune-mediated destruction of pancreatic islets. These data suggest that leptin receptor deficiency protects DR.lepr−/lepr−,Gimap5−/Gimap5− rats from spontaneous T1D, indicating a potential role for the leptin receptor in development of their characteristic autoimmune diabetes. DR.lepr−/lepr−,Gimap5−/Gimap5− congenic rats may provide a unique model to study interactions between T1D and obesity.*****
GRANTS
This work was supported by a Junior Faculty Award (1-05-JF-32) (D. H. Moralejo) and a Mentor Based Postdoctoral Fellowship Award (Å. Lernmark) from the American Diabetes Association, the National Institutes of Health (AI-42380), the Virus Molecular Biology and Cell Core (W. Osborne and B. Van Yserloo) of the University of Washington Diabetes and Endocrinology Research Center (DK-17047), as well as by the Robert H. Williams Endowment at the University of Washington.
DISCLOSURES
There are no conflicts of interest.
REFERENCES
- 1.Bieg S, Koike G, Jiang J, Klaff L, Pettersson A, MacMurray AJ, Jacob HJ, Lander ES, Lernmark A. Genetic isolation of iddm 1 on chromosome 4 in the BioBreeding (BB) rat. Mamm Genome 9:324–326, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Busso N, So A, Chobaz-Peclat V, Morard C, Martinez-Soria E, Talabot-Ayer D, Gabay C. Leptin signaling deficiency impairs humoral and cellular immune responses and attenuates experimental arthritis. J Immunol 168:875–882, 2002 [DOI] [PubMed] [Google Scholar]
- 3.Chua SC, Jr, Chung WK, Wu-Peng XS, Zhang Y, Liu SM, Tartaglia L, Leibel RL. Phenotypes of mouse diabetes and rat fatty due to mutations in the OB (leptin) receptor. Science 271:994–996, 1996 [DOI] [PubMed] [Google Scholar]
- 4.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, Warram JH, Todd JA. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet 40:1399–1401, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalberg U, Markholst H, Hornum L. Both Gimap5 and the diabetogenic BBDP allele of Gimap5 induce apoptosis in T cells. Int Immunol 19:447–453, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Dion C, Carter C, Hepburn L, Coadwell WJ, Morgan G, Graham M, Pugh N, Anderson G, Butcher GW, Miller JR. Expression of the Ian family of putative GTPases during T cell development and description of an Ian with three sets of GTP/GDP-binding motifs. Int Immunol 17:1257–1268, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Fourlanos S, Varney MD, Tait BD, Morahan G, Honeyman MC, Colman PG, Harrison LC. The rising incidence of type 1 diabetes is accounted for by cases with lower-risk human leukocyte antigen genotypes. Diabetes Care 31:1546–1549, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friedman JM. Leptin, leptin receptors, and the control of body weight. Nutr Rev 56:s38–s46, 1998 [DOI] [PubMed] [Google Scholar]
- 9.Friedman JM, Halaas JL. Leptin and the regulation of body weight in mammals. Nature 395:763–770, 1998 [DOI] [PubMed] [Google Scholar]
- 10.Fuller JM, Bogdani M, Tupling TD, Jensen RA, Pefley R, Manavi S, Cort L, Blankenhorn EP, Mordes JP, Lernmark A, Kwitek AE. Genetic dissection reveals diabetes loci proximal to the gimap5 lymphopenia gene. Physiol Genomics 38:89–97, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuller JM, Kwitek AE, Hawkins TJ, Moralejo DH, Lu W, Tupling TD, Macmurray AJ, Borchardt G, Hasinoff M, Lernmark A. Introgression of F344 rat genomic DNA on BB rat chromosome 4 generates diabetes-resistant lymphopenic BB rats. Diabetes 55:3351–3357, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Hellquist A, Zucchelli M, Kivinen K, Saarialho-Kere U, Koskenmies S, Widen E, Julkunen H, Wong A, Karjalainen-Lindsberg ML, Skoog T, Vendelin J, Cunninghame-Graham DS, Vyse TJ, Kere J, Lindgren CM. The human GIMAP5 gene has a common polyadenylation polymorphism increasing risk to systemic lupus erythematosus. J Med Genet 44:314–321, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hessner MJ, Wang X, Meyer L, Geoffrey R, Jia S, Fuller J, Lernmark A, Ghosh S. Involvement of eotaxin, eosinophils, and pancreatic predisposition in development of type 1 diabetes mellitus in the BioBreeding rat. J Immunol 173:6993–7002, 2004 [DOI] [PubMed] [Google Scholar]
- 14.Hornum L, Romer J, Markholst H. The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes 51:1972–1979, 2002 [DOI] [PubMed] [Google Scholar]
- 15.Iida M, Murakami T, Ishida K, Mizuno A, Kuwajima M, Shima K. Phenotype-linked amino acid alteration in leptin receptor cDNA from Zucker fatty (fa/fa) rat. Biochem Biophys Res Commun 222:19–26, 1996 [DOI] [PubMed] [Google Scholar]
- 16.Kawano K, Hirashima T, Mori S, Natori T. OLETF (Otsuka Long-Evans Tokushima Fatty) rat: a new NIDDM rat strain. Diabetes Res Clin Pract 24, Suppl: S317–S320, 1994 [DOI] [PubMed] [Google Scholar]
- 17.Keita M, Leblanc C, Andrews D, Ramanathan S. GIMAP5 regulates mitochondrial integrity from a distinct subcellular compartment. Biochem Biophys Res Commun 361:481–486, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Knip M, Reunanen A, Virtanen SM, Nuutinen M, Viikari J, Akerblom HK. Does the secular increase in body mass in children contribute to the increasing incidence of type 1 diabetes? Pediatr Diabetes 9:46–49, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Koletsky S. Obese spontaneously hypertensive rats—a model for study of atherosclerosis. Exp Mol Pathol 19:53–60, 1973 [DOI] [PubMed] [Google Scholar]
- 20.Kupfer R, Lang J, Williams-Skipp C, Nelson M, Bellgrau D, Scheinman RI. Loss of a gimap/ian gene leads to activation of NF-kappaB through a MAPK-dependent pathway. Mol Immunol 44:479–487, 2007 [DOI] [PubMed] [Google Scholar]
- 21.La Cava A, Alviggi C, Matarese G. Unraveling the multiple roles of leptin in inflammation and autoimmunity. J Mol Med 82:4–11, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Loffreda S, Yang SQ, Lin HZ, Karp CL, Brengman ML, Wang DJ, Klein AS, Bulkley GB, Bao C, Noble PW, Lane MD, Diehl AM. Leptin regulates proinflammatory immune responses. FASEB J 12:57–65, 1998 [PubMed] [Google Scholar]
- 23.Lord GM, Matarese G, Howard JK, Baker RJ, Bloom SR, Lechler RI. Leptin modulates the T-cell immune response and reverses starvation-induced immunosuppression. Nature 394:897–901, 1998 [DOI] [PubMed] [Google Scholar]
- 24.MacMurray AJ, Moralejo DH, Kwitek AE, Rutledge EA, Van Yserloo B, Gohlke P, Speros SJ, Snyder B, Schaefer J, Bieg S, Jiang J, Ettinger RA, Fuller J, Daniels TL, Pettersson A, Orlebeke K, Birren B, Jacob HJ, Lander ES, Lernmark A. Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Res 12:1029–1039, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markholst H, Eastman S, Wilson D, Andreasen BE, Lernmark A. Diabetes segregates as a single locus in crosses between inbred BB rats prone or resistant to diabetes. J Exp Med 174:297–300, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maruta K, Treichel U, Kolb-Bachofen V, Kolb H. Prospective analysis of eosinophilia in spontaneously diabetic BB rats: correlation with islet inflammation but not with diabetes development. Diabetes Res 11:173–176, 1989 [PubMed] [Google Scholar]
- 27.Matarese G, Di Giacomo A, Sanna V, Lord GM, Howard JK, Di Tuoro A, Bloom SR, Lechler RI, Zappacosta S, Fontana S. Requirement for leptin in the induction and progression of autoimmune encephalomyelitis. J Immunol 166:5909–5916, 2001 [DOI] [PubMed] [Google Scholar]
- 28.Mattioli B, Straface E, Quaranta MG, Giordani L, Viora M. Leptin promotes differentiation and survival of human dendritic cells and licenses them for Th1 priming. J Immunol 174:6820–6828, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Meetoo D, McGovern P, Safadi R. An epidemiological overview of diabetes across the world. Br J Nurs 16:1002–1007, 2007 [DOI] [PubMed] [Google Scholar]
- 30.Michalkiewicz M, Michalkiewicz T, Ettinger RA, Rutledge EA, Fuller JM, Moralejo DH, Van Yserloo B, MacMurray AJ, Kwitek AE, Jacob HJ, Lander ES, Lernmark A. Transgenic rescue demonstrates involvement of the Ian5 gene in T cell development in the rat. Physiol Genomics 19:228–232, 2004 [DOI] [PubMed] [Google Scholar]
- 31.Moralejo DH, Park HA, Speros SJ, MacMurray AJ, Kwitek AE, Jacob HJ, Lander ES, Lernmark A. Genetic dissection of lymphopenia from autoimmunity by introgression of mutated Ian5 gene onto the F344 rat. J Autoimmun 21:315–324, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mordes JP, Bortell R, Blankenhorn EP, Rossini AA, Greiner DL. Rat models of type 1 diabetes: genetics, environment, and autoimmunity. ILAR J 45:278–291, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Mordes JP, Desemone J, Rossini AA. The BB rat. Diabetes Metab Rev 3:725–750, 1987 [DOI] [PubMed] [Google Scholar]
- 34.Moreno C, Dumas P, Kaldunski ML, Tonellato PJ, Greene AS, Roman RJ, Cheng Q, Wang Z, Jacob HJ, Cowley AW., Jr Genomic map of cardiovascular phenotypes of hypertension in female Dahl S rats. Physiol Genomics 15:243–257, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Nitta T, Takahama Y. The lymphocyte guard-IANs: regulation of lymphocyte survival by IAN/GIMAP family proteins. Trends Immunol 28:58–65, 2007 [DOI] [PubMed] [Google Scholar]
- 36.Onkamo P, Vaananen S, Karvonen M, Tuomilehto J. Worldwide increase in incidence of Type I diabetes—the analysis of the data on published incidence trends. Diabetologia 42:1395–1403, 1999 [DOI] [PubMed] [Google Scholar]
- 37.Papathanassoglou E, El-Haschimi K, Li XC, Matarese G, Strom T, Mantzoros C. Leptin receptor expression and signaling in lymphocytes: kinetics during lymphocyte activation, role in lymphocyte survival, and response to high fat diet in mice. J Immunol 176:7745–7752, 2006 [DOI] [PubMed] [Google Scholar]
- 38.Pihoker C, Scott CR, Lensing SY, Cradock MM, Smith J. Non-insulin-dependent diabetes mellitus in African-American youths of Arkansas. Clin Pediatr (Phila) 37:97–102, 1998 [DOI] [PubMed] [Google Scholar]
- 39.Resic-Lindehammer S, Larsson K, Ortqvist E, Carlsson A, Cederwall E, Cilio CM, Ivarsson SA, Jonsson BA, Larsson HE, Lynch K, Neiderud J, Nilsson A, Sjoblad S, Lernmark A, Aili M, Baath LE, Carlsson E, Edenwall H, Forsander G, Granstro BW, Gustavsson I, Hanas R, Hellenberg L, Hellgren H, Holmberg E, Hornell H, Johansson C, Jonsell G, Kockum K, Lindblad B, Lindh A, Ludvigsson J, Myrdal U, Segnestam K, Skogsberg L, Stromberg L, Stahle U, Thalme B, Tullus K, Tuvemo T, Wallensteen M, Westphal O, Aman J. Temporal trends of HLA genotype frequencies of type 1 diabetes patients in Sweden from 1986 to 2005 suggest altered risk. Acta Diabetol 45:231–235, 2008 [DOI] [PubMed] [Google Scholar]
- 40.Russell JC, Amy RM. Plasma lipids and other factors in the LA/N corpulent rat in the presence of chronic exercise and food restriction. Can J Physiol Pharmacol 64:750–756, 1986 [DOI] [PubMed] [Google Scholar]
- 41.Schranz DB, Lernmark A. Immunology in diabetes: an update. Diabetes Metab Rev 14:3–29, 1998 [DOI] [PubMed] [Google Scholar]
- 42.Scott J. The spontaneously diabetic BB rat: sites of the defects leading to autoimmunity and diabetes mellitus. Curr Top Microbiol Immunol 156:1–14, 1990 [DOI] [PubMed] [Google Scholar]
- 43.Siegel K, Narayan KV. The Unite for Diabetes campaign: overcoming constraints to find a global policy solution. Global Health 4:3, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siegmund B, Sennello JA, Jones-Carson J, Gamboni-Robertson F, Lehr HA, Batra A, Fedke I, Zeitz M, Fantuzzi G. Leptin receptor expression on T lymphocytes modulates chronic intestinal inflammation in mice. Gut 53: 965–972, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takaya K, Ogawa Y, Hiraoka J, Hosoda K, Yamori Y, Nakao K, Koletsky RJ. Nonsense mutation of leptin receptor in the obese spontaneously hypertensive Koletsky rat. Nat Genet 14:130–131, 1996 [DOI] [PubMed] [Google Scholar]
- 46.Tartaglia LA. The leptin receptor. J Biol Chem 272:6093–6096, 1997 [DOI] [PubMed] [Google Scholar]
- 47.Tartaglia LA, Dembski M, Weng X, Deng N, Culpepper J, Devos R, Richards GJ, Campfield LA, Clark FT, Deeds J, Muir C, Sanker S, Moriarty A, Moore KJ, Smutko JS, Mays GG, Wool EA, Monroe CA, Tepper RI. Identification and expression cloning of a leptin receptor, OB-R. Cell 83:1263–1271, 1995 [DOI] [PubMed] [Google Scholar]
- 48.Tirabassi RS, Flanagan JF, Wu T, Kislauskis EH, Birckbichler PJ, Guberski DL. The BBZDR/Wor rat model for investigating the complications of Type 2 diabetes mellitus. ILAR J 45:292–302, 2004 [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Lobstein T. Worldwide trends in childhood overweight and obesity. Int J Pediatr Obes 1:11–25, 2006 [DOI] [PubMed] [Google Scholar]
- 50.Wilkin TJ. Diabetes: 1 and 2, or one and the same? Progress with the accelerator hypothesis. Pediatr Diabetes 9:23–32, 2008 [DOI] [PubMed] [Google Scholar]
- 51.Wu-Peng XS, Chua SC, Jr, Okada N, Liu SM, Nicolson M, Leibel RL. Phenotype of the obese Koletsky (f) rat due to Tyr763Stop mutation in the extracellular domain of the leptin receptor (Lepr): evidence for deficient plasma-to-CSF transport of leptin in both the Zucker and Koletsky obese rat. Diabetes 46:513–518, 1997 [DOI] [PubMed] [Google Scholar]
- 52.Yamashita T, Murakami T, Iida M, Kuwajima M, Shima K. Leptin receptor of Zucker fatty rat performs reduced signal transduction. Diabetes 46:1077–1080, 1997. [DOI] [PubMed] [Google Scholar]