Abstract
Previous studies have shown that physiological responses to cigarette smoke can be detected via bronchial airway epithelium gene expression profiling and that heterogeneity in this gene expression response to smoking is associated with lung cancer. In this study, we sought to determine the similarity of the effects of tobacco smoke throughout the respiratory tract by determining patterns of smoking-related gene expression in paired nasal and bronchial epithelial brushings collected from 14 healthy nonsmokers and 13 healthy current smokers. Using whole genome expression arrays, we identified 119 genes whose expression was affected by smoking similarly in both bronchial and nasal epithelium, including genes related to detoxification, oxidative stress, and wound healing. While the vast majority of smoking-related gene expression changes occur in both bronchial and nasal epithelium, we also identified 27 genes whose expression was affected by smoking more dramatically in bronchial epithelium than nasal epithelium. Both common and site-specific smoking-related gene expression profiles were validated using independent microarray datasets. Differential expression of select genes was also confirmed by RT-PCR. That smoking induces largely similar gene expression changes in both nasal and bronchial epithelium suggests that the consequences of cigarette smoke exposure can be measured in tissues throughout the respiratory tract. Our findings suggest that nasal epithelial gene expression may serve as a relatively noninvasive surrogate to measure physiological responses to cigarette smoke and/or other inhaled exposures in large-scale epidemiological studies.
Keywords: cigarette smoke, Affymetrix exon arrays, bronchial airway epithelium, nasal epithelium
although cigarette smoking is well recognized as the major cause of lung cancer and chronic obstructive pulmonary disease (5), only 10–20% of smokers develop these diseases (18). It is unclear what factors might contribute to differences in risk for tobacco-related disease among current or former smokers with similar smoking histories. The development of noninvasive methods that identify differences in the physiological response to tobacco smoke represents an approach for identifying factors that contribute to differences in risk of tobacco-related lung disease.
Based on the concept that cigarette smoke creates a “field of injury” throughout the respiratory tract, our hypothesis is that it is possible to characterize the physiological response to cigarette smoke in epithelial cells lining the respiratory tract using genome-wide gene expression profiling. We have previously defined the impact of tobacco smoke on bronchial airway epithelial cells by comparing gene expression patterns among healthy nonsmokers and smokers using samples collected from the main stem bronchus by bronchoscopy (19). Subsequently we have identified how the expression of these genes change as a result of smoking cessation (1), as well as a pattern of gene expression in these cytologically normal bronchial airway epithelial cells that can distinguish smokers with and without lung cancer and serve as an early diagnostic biomarker for disease (2, 20). Although gene expression differences in large airway epithelial cells obtained via bronchoscopy have successfully identified candidate biomarkers of smoking-related lung damage, the invasiveness of bronchoscopy (which involves continuous monitoring of the heart and lungs due to risks associated with conscious sedation and airway brushings) prevents it from being used in large-scale studies to assess variability in the physiological response to smoking or as a screening tool for assessing smoking-related lung cancer risk in asymptomatic individuals.
A potential alternative to measuring the response to tobacco smoke exposure in cells from the bronchial airways is to measure this response in oral or nasal mucosa, tissues that are also directly exposed to tobacco smoke. We have recently shown that genes affected by smoking in bronchial airways can be used to distinguish oral and nasal mucosa from smokers and nonsmokers using oral, nasal, and bronchial epithelium samples collected from different subjects, suggesting that aspects of the physiological response to tobacco smoke might be shared across different airway epithelial sites (21). In that study, the similarity between the bronchial and nasal gene expression response to smoking was most pronounced. That study, however, did not establish specific genes that are affected by smoking at all sites or genes that might be affected by smoking at some sites but not others. Furthermore, the previous study was only able to show a general relationship in airway patterns of smoking-related gene expression using Gene Set Enrichment Analysis (GSEA) (22) and was unable to identify specific gene expression similarities and differences between sites, because the bronchial and nasal samples were collected from different groups of volunteers and the nasal sample size was small. Therefore, while this early study is encouraging (as it suggests there is a degree of overall similarity in the response to smoking throughout the airway), without a detailed understanding of the homogeneity of the smoking effect throughout the airway, it is difficult to determine whether these sites might be largely equivalent for measuring the physiological consequences of smoking in the airway.
To address this issue, we have now sought to determine the relationship between smoking-related gene expression changes in nasal and bronchial epithelium from smokers and nonsmokers in which both tissues were collected from each of the study participants. Genome-wide gene expression profiling of these samples showed that the majority of the gene expression consequences of smoking are common to both nasal and bronchial epithelium. We also identified a small number of genes that show site-specific effects of smoking, with most of these being changed in bronchial epithelium but not nasal epithelium.
MATERIAL AND METHODS
Study population.
We recruited 14 healthy never smokers and 13 current smokers for the study at Boston Medical Center [see Supplemental Table S11 for comparison to cohorts in previously published datasets (19, 21)]. Nonsmokers with history of significant second-hand environmental cigarette exposure, respiratory symptoms, or regular use of inhaled medications were excluded. For each participant, a detailed smoking history was obtained including cumulative tobacco exposure (measured in pack-years), age when they began smoking and second-hand tobacco exposure. All individuals were screened with routine chest X-ray and spirometry and were excluded if they had evidence of pulmonary pathology. The study was approved by the Institutional Review Board of Boston Medical Center and all participants provided written informed consent.
Sample collection.
Bronchial airway epithelial cells were obtained from bronchial brushings of the right main stem bronchus taken during fiber optic bronchoscopy with an endoscopic cytobrush (Cellebrity Endocscopic Cytobrush; Boston Scientific, Boston, MA). The epithelial cell content of the bronchial brushings was >90% (19). The brushes were immediately placed in RNAprotect Cell Reagent (Qiagen, Valencia, CA) and stored at 4°C until RNA was isolated. RNA was extracted from the cells using the miRNeasy mini kit (Qiagen) according to manufacturer's protocol. Integrity of the RNA samples was assessed by Agilent BioAnalyzer, and purity of the RNA was confirmed using a NanoDrop spectrophotometer.
During the same clinic visit, we collected nasal epithelial cells by brushing the inferior turbinate as previously described (21). Briefly, the right nare was lavaged with 1 ml of 1% lidocaine. A nasal speculum (Bionox, Toledo, OH) then spread the nare while a standard cytology brush was inserted underneath the inferior nasal turbinate. The brush was rotated in place for 3 s, removed, and immediately placed in 1 ml RNA Later (Qiagen). After storage at 4°C, RNA was isolated via Qiagen RNeasy Mini Kits as per the manufacturer protocol. Unlike bronchoscopy, continuous cardiac and respiratory monitoring of subjects during and after the procedure was not required.
We obtained 4–5 ml of blood from a subset of study participants (8 current and 5 never smokers) for determination of plasma cotinine. Samples were centrifuged, and 2.2 ml of plasma was stored at −80°C and then shipped on dry ice to the University of California - San Francisco Division of Clinical Pharmacology and Experimental Therapeutics. Gas chromatography (quantitation limit = 10 ng/ml) or liquid chromatography-tandem mass spectrometry (quantitation limit = 0.02 ng/ml) was used to analyze samples for the presence of this nicotine metabolite from self-reported current and never smokers, respectively.
Microarray data acquisition and preprocessing.
We used 1 μg of RNA from the bronchial and nasal epithelium samples as starting material for the microarray studies. Ribosomal RNA was first removed using the RiboMinus Human/Mouse Transcriptome Isolation Kit (Invitrogen, Carlsbad, CA). This treated RNA was then converted to cDNA and subsequently processed, labeled, and hybridized onto Human Exon 1.0 ST GeneChips as previously described (25). Following hybridization, each array was washed and stained according to the standard Affymetrix protocol. The stained array was scanned using an Affymetrix GeneChip Scanner 3000, resulting in a raw data file for each array.
The ∼230,000 “core” exon probe sets on the Exon array, which map with a high degree of confidence to ∼17,800 empirically supported transcripts (RefSeq and full-length GenBank mRNAs), were used for transcript-level analysis. Transcript-level expression values were derived by quantile sketch normalization using the model-based iterPLIER algorithm as implemented in the ExACT software package (Affymetrix, Santa Clara, CA). A global median normalization method was then applied to the data sets. The gene annotations used for each probe set were from the annotation file obtained from Affymetrix (www.affymetrix.com). After preprocessing, one bronchial sample from one never smoker was identified as an outlier based on box plots of the relative log expression and Principal Components Analysis (PCA). This sample was excluded from further analysis.
Identification of smoking-related gene expression changes.
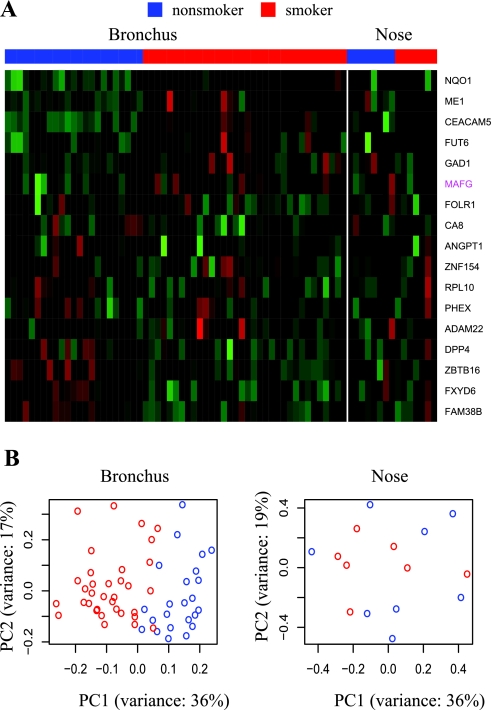
To identify genes that are differentially expressed in response to tobacco smoke exposure in both nasal and bronchial epithelial cells, as well as genes where tobacco smoke exposure has a site-specific effect, we used a mixed linear model that included smoking status and site (nasal vs. bronchial) as fixed effects, and random effects to take into account patient matching and batch effects. The regression equation is given below:
where geneijk is the log2 expression value for gene i, in patient j, and batch k; the parameter μ represents the log2 expression value of the referent group (bronchus of never smoker). The random term εijk represents the random error that was assumed to be normally distributed, while εpatientj and εbatchk represent the patient and batch random effects, respectively. The remaining regression terms parameterize the site-specific effects. Specifically, the dummy variables Xnose and Xstatus specify the site (Xnose = 1 for nose and 0 for bronchus) and the smoking status (Xstatus = 1 for current smokers and 0 for never smokers), the parameters βnose and βstatus represent the site and smoking status-specific main effects, and the term βnose.status represents the interaction between site and smoking status. These parameters can be interpreted as fold changes in log2 scale. For example, for a gene in which there is no interaction between site and smoking status (βnose.status = 0), the regression coefficient βstatus represents the log2 fold change between expression in smokers and nonsmokers, and the lack of interaction means that the change in expression is the same in bronchial and nasal epithelium. If the interaction between site and smoking status is significant (βnose.status ≠ 0), the changes in expression between sites is dependent on the smoking status, and βstatus represents the log2 fold change between smokers and nonsmokers in bronchial epithelium, while βstatus +βnose.status is the log2 fold change between smokers and nonsmokers in nasal epithelium.
Figure 1 gives a schematic summary of the analysis. The lme function in R package nlme was used to first identify genes with a significant site-status interaction effect [false discovery rate (FDR) < 0.05]. We then characterized these genes for which tobacco-smoke exposure has a site-specific effect by examining the fold change between smokers and nonsmokers in nasal and bronchial epithelium. Here, we used |fd| B to represent the absolute log2 fold-change in bronchus between smokers and nonsmokers, which is measured by the regression coefficient βstatus. |fd| N was used to represent the absolute log2 fold-change in nose between smokers and nonsmokers, which is measured by βstatus +βnose.status. If |fd| B > 0.5, but |fd| N < 0.3, we considered tobacco smoke exposure to more dramatically affect expression of that gene in the bronchus. If |fd| B < 0.3, but |fd| N > 0.5, we considered smoke exposure to more dramatically change gene expression in the nose. We considered those genes with a significant site-status interaction effect where both |fd| B > 0.5 and |fd| N > 0.5 to be affected by smoking exposure differently in the two sites. The fold change threshold to define significant feature genes after FDR correction was set to 0.5 based on our finding that of 2,904 negative control probe sets (derived from intronic regions), only one had an apparent fold difference > 0.5 between smokers and nonsmokers.
Fig. 1.
Methodology for identifying genes that are commonly and site-specifically differentially expressed in response to tobacco smoke exposure. For each gene, the relationship between gene expression in log2 scale, site, status, and the interaction between site and status was examined with a mixed linear regression model. |fd| B represents the absolute log2 magnitude of the difference between current and never smoker in bronchial samples (fdB = βstatus), and fdN is the magnitude of the difference in nasal samples (fdN = βstatus +βnose.status). FDR, false discovery rate.
Genes with a significant site-status interaction effect were then removed from further analysis. For the remaining genes, we used the same lme function to fit the regression model without the site-status interaction term and identify genes with a significant status effect (FDR < 0.05). Here, the regression coefficient βstatus represents the log2 fold change between smokers and nonsmokers when the site effect is fixed. As genes with a significant site-specific smoking effect had already been removed in the previous step, those that had an absolute βstatus > 0.5 were considered to be affected by tobacco exposure at both sites.
To further characterize the smoking-related genes, hierarchical clustering of all never and current smokers using the differentially expressed genes was performed. Hierarchical clustering of the genes and samples was performed on z-score normalized data using a Pearson correlation (uncentered) similarity metric and average linkage clustering.
To identify gene ontology (GO) molecular function categories (6), KEGG pathways (13), and GenMAPP pathways (15) that are overrepresented within the genes differentially expressed between current vs. never smokers, Expression Analysis Systematic Explorer (EASE) was used to functionally classify these genes using the distribution of the 17,881 annotated genes on the Affymetrix Human Exon arrays among these annotation terms as the background distribution. Fisher's exact test was used to assess enrichment of differentially expressed genes in annotation terms.
In silico validation using other gene expression data sets.
For the genes that are affected by smoking status independently of collection site in both bronchial and nasal epithelium, we searched for corresponding probe sets on the Affymetrix U133A array. PCA was performed using the gene expression values for these probe sets in previously published U133A datasets from our group where we had profiled bronchial and nasal epithelium samples from current and never smokers who were not participants in the current study. This analysis included bronchus samples from 23 never smokers and 34 smokers, and nasal epithelium samples from 8 never smokers and 7 current smokers from different groups of volunteers. Additionally, we mapped U133A probe sets that we previously identified to be differentially expressed with smoking in bronchial epithelium (19) to probe sets on the Human Exon arrays. The expression level of these probe sets as measured in the current study was used for PCA.
The relationship between smoking-related gene expression differences observed in the current study and those observed in earlier studies was also explored using GSEA (22). For determining whether the genes we identified as differentially expressed in the current dataset were also observed to be affected by smoking in previous studies using the U133A array (19, 21), we ranked the U133A probe sets according to the previously observed signal-to-noise ratio for the effect of smoking. We next used GSEA to examine the distribution of genes whose expression was identified using the Exon array dataset as being affected by smoking independently of collection site within these ranked lists. We also examined the distribution of genes whose expression is more dramatically altered by smoking in the bronchus. Conversely, we ranked genes by the absolute value of the smoking status coefficient from the mixed-linear models described above and used GSEA to examine the distribution of genes we previously identified as being perturbed by smoking in bronchial epithelium (19). The significance (P value) of the distribution of gene sets within the ranked list was determined by gene set permutation (22) and corrected for multiple hypothesis testing (FDR q-value).
Validation of candidate genes by real-time RT-PCR.
Real-time PCR was used to confirm the differential expression of a select number of genes. Primer sequences for candidate genes (CYP1B1, CYP1A1, AKR1B10, TMEM45A, SEC14L3, MAFG, CCL28, and DNER) and a control gene (GAPDH) were designed with Primer Express software (Applied Biosystems) and are listed in Supplemental Table S2. RNA samples (500 ng of residual sample from the microarray experiment) were treated with TURBO DNA-free (Ambion) for 30 min, as per the manufacturer's protocol, to remove contaminating genomic DNA. Total RNA was reverse transcribed using random hexamers (Applied Biosystems) and Superscript II reverse transcriptase (Invitrogen). The resulting cDNA product was added to SYBR Green PCR master mix (Applied Biosystems). Forty cycles of amplification, data acquisition, and data analysis were carried out using an ABI Prism 7000 Sequence Detector (Applied Biosystems). Threshold determinations were automatically performed by the instrument for each reaction. All real-time PCR experiments were carried out in triplicate on each sample.
Additional information.
All statistical analyses described above were performed with R 2.8.0 (available at http://r-project.org) and Bioconductor (4). All microarray data from this study have been deposited in GEO under accession #GSE16008.
RESULTS
Smoking induces a common transcriptional response across nasal and bronchial epithelium.
There were no significant differences in age, race, or sex between current and never smokers for the 27 healthy volunteers recruited into this study (Table 1). Cotinine analysis confirmed the self-reported smoking status of all 13 subjects tested. Cotinine levels ranged from 81.1–397.5 ng/ml (mean = 222.1; SD = 124.5) among current smokers and nondetectable to 0.07 ng/ml among never smokers. Smokers reported currently smoking 5–23 cigarettes per day with lifetime exposure histories ranging from 0.5 to 30 yr.
Table 1.
Demographics of study participants
| Current Smokers (n = 13) | Never Smokers (n = 14) | P Value | |
|---|---|---|---|
| Age, yr* | 35.4 ± 9.9 | 31.6 ± 10.8 | 0.357 |
| Race | 6 AFA, 4 HIS, 3 CAU | 5 AFA, 2 HIS, 7 CAU | 0.372 |
| Sex | 7 M, 6 F | 4 M, 10 F | 0.252 |
| Pack years* | 10.77 ± 9.3 | ||
| Cigarettes per day* | 12.87 ± 5.16 |
P values for sex and race were calculated by Fisher exact test. P value for age was calculated by a Student's t-test.
Values represent means ± SD. AFA, African American; HIS, Hispanic; CAU, Caucasian; M, male; F, female.
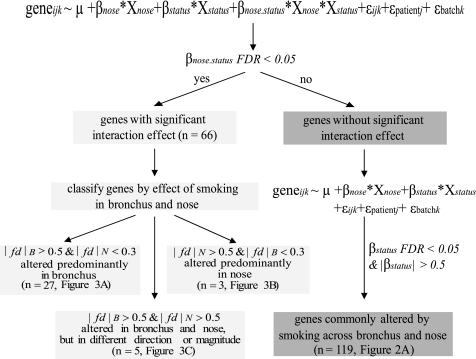
After correcting for multiple comparisons, we identified 255 genes as significantly differentially expressed in response to smoking independently of site (FDR <0.05), using the strategy outlined in Fig. 1. Of these, 119 genes remained after a filter was applied to retain only genes for which the magnitude of the mixed-model smoking-status coefficient was >0.5 (after adjusting for the effects of site, patient, and batch). This corresponds to a covariate-corrected fold change between current and never smokers of 1.4 (Fig. 2A). Differential expression of candidate genes from this list was confirmed by real-time PCR (Fig. 2B).
Fig. 2.
A: similarity between bronchial and nasal gene expression. Gene expression estimates for the 119 genes (see Supplemental Table S4) that vary between smokers and nonsmokers were z-score normalized within each tissue and organized using hierarchical clustering (green indicates lower than average expression, red indicates higher than average expression). B: real-time PCR validation of select genes (colored pink in A) in 3 smokers and 3 nonsmokers. The y-axis represents relative log2 expression. Error bars indicate SE. Differences in expression were evaluated using Student's t-test.
Site-specific changes in genes expression across the respiratory tract in smokers.
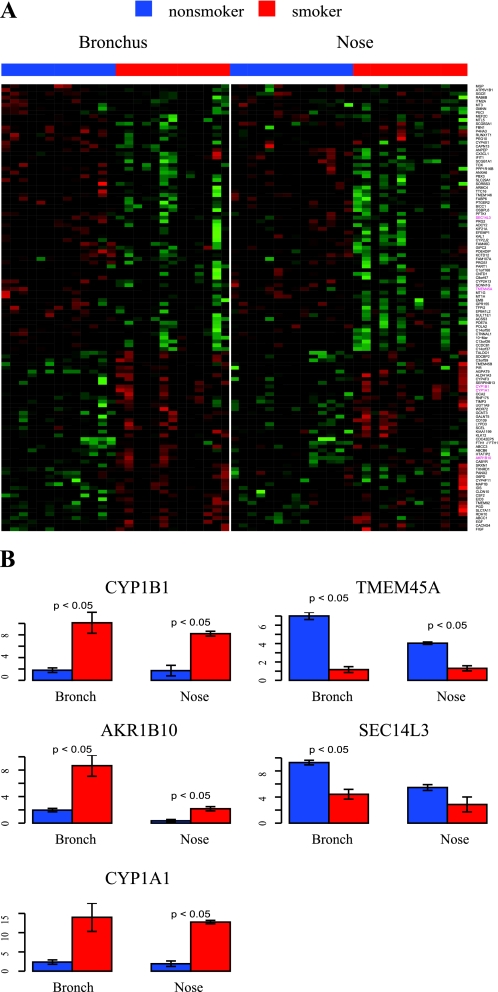
We identified 66 genes with expression patterns that have a significant interaction term for the effect of smoking status and site in the mixed linear model (FDR < 0.05). Using the procedure in Fig. 1, we characterized 27 genes as being differentially expressed as a result of smoking predominantly in bronchial epithelium (Fig. 3A). Conversely, three genes (CCL28, ALPL, and GBP3) were categorized as having expression levels that are altered by smoking predominantly in nasal epithelium (the expression of gene CCL28 is shown in Fig. 3B). The expression of another five genes (ALDH3A1, GPX2, AKR1C2, TLL1, DNER) was found to be affected by smoking in both sites (|effect of smoking| > 0.5 in both sites), but these five genes differed between bronchial and nasal epithelium with regard to either the magnitude (ALDH3A1, GPX2, AKR1C2) or the direction (TLL1, DNER) of the effect of smoking on gene expression (the expression estimates of DNER are shown in Fig. 3C). Differential expression of a subset of these genes was validated by RT-PCR (S2).
Fig. 3.
Site-specific changes in gene expression induced by smoking. A: heat map of 27 genes differentially expressed as a result of smoking more dramatically in bronchial epithelium. B: CCL28 is differentially expressed between smokers and nonsmokers in nose, but not in bronchus. C: DNER is upregulated by smoking in bronchus but is downregulated in nose.
In silico validation of airway-wide and site-specific gene expression changes in response to smoking.
Previous studies from our group characterized the effect of cigarette smoking on bronchial epithelium gene expression (19) and demonstrated, at a global level, similarities in the gene expression response of nasal epithelium (21) using gene expression data collected with Affymetrix U133A microarrays from unmatched samples. We used these previous datasets to validate the genes we identified here as being differentially expressed in response to smoking in both bronchial and nasal epithelium, or in only one of these sites.
We mapped 85 of the 119 genes that are affected by smoking similarly in both bronchial and nasal epithelium to probe sets on the U133A array. We used the expression levels of these 85 probe sets from a U133A dataset consisting of bronchial epithelium samples from 23 never smokers and 34 smokers, and nasal epithelium samples from 8 never smokers and 7 current smokers to perform a PCA. While the effect of smoking on the expression of these genes is less pronounced in nasal epithelium samples, the PCA indicates that, as a set, the expression of these genes is able to distinguish both bronchial and nasal epithelial samples by smoking status in this dataset (Supplemental Fig. S1A). To further validate this relationship, we performed two GSEA of the U133A dataset. In the first analysis we ranked the U133A probe sets according to the absolute signal-to-noise ratio for the effect of smoking observed in the U133A bronchial samples (19). In the second analysis, we ranked the probe sets by this metric for the effect of smoking in the U133A nasal samples (21). By GSEA of these ranked lists, we found that the genes we identified as being affected by smoking similarly in both bronchial and nasal epithelium are among the genes most strongly affected by smoking in the U133A bronchial (P < 0.001) and U133A nasal datasets (P < 0.001). We also did the converse experiment. For 97 genes that we had found to be differentially expressed by smoking in bronchial airway samples (19), we were able to find corresponding Exon array probe sets for 71 genes. We found that expression of probe sets corresponding to these previously identified genes in the Exon array dataset is able to distinguish both bronchial and nasal epithelial samples by smoking status using PCA (Supplemental Fig. S1B). Using GSEA, we also found that these probe sets were among the genes most affected by smoking in bronchial (P < 0.001) and nasal epithelium (P = 0.002).
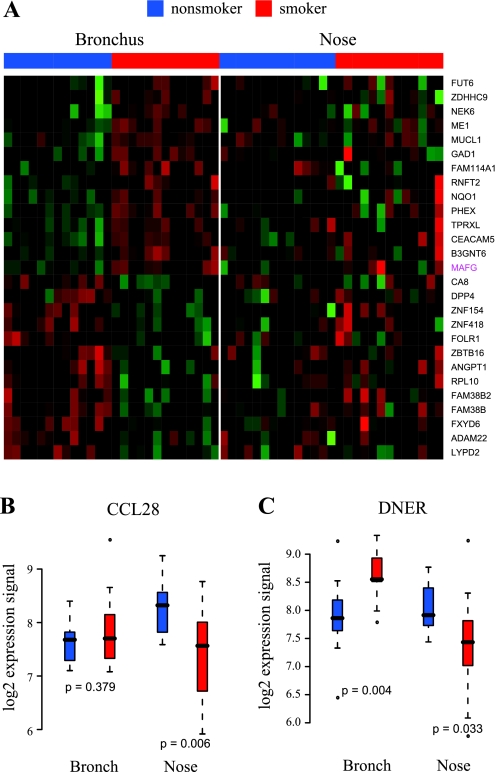
Of the 27 genes characterized as being strongly affected by smoking in bronchial but not nasal epithelium, we identified probe sets for measuring 17 on the U133A platform by gene symbol. We examined their expression in our U133A dataset of bronchial and nasal epithelium samples from smokers and nonsmokers. We did this first by hierarchical clustering (Fig. 4A). Consistent with the Exon array expression profiles, expression of these genes appears to vary between never and current smokers in bronchial samples but not in samples from nasal epithelium. PCA of these expression profiles similarly indicates that they are able to distinguish bronchial but not nasal epithelial samples by smoking status (Fig. 4B). Perhaps not surprisingly, GSEA of the U133A dataset also showed that the genes we identified as having bronchial-epithelium-specific smoking-related expression profiles are amongst the genes most affected by smoking in bronchial epithelium (P = 0) in the U133A dataset but are not among the genes that are most affected by smoking in nasal epithelium (P = 0.56).
Fig. 4.
In silico validation of genes more dramatically altered by smoking in bronchus (see Fig. 3A), using an independent U133A data. Among 27 genes, 17 genes with corresponding U133A probe sets were identified. A: heat map of the expression profiles of these 17 genes in previously published U133A datasets consisting of bronchial and nasal samples from current and never smokers. B: principal component analysis (PCA) of 17 genes in U133A bronchus and nose data. Each point represents a subject (red: smokers, blue: nonsmokers).
GO and pathway analysis.
The Database for Annotation, Visualization, and Integrated Discovery (DAVID) and EASE (8) were used to identify GO molecular function categories (6), human KEGG pathways (13), and GenMAPP pathways (15) that are overrepresented among the 119 genes with bronchial and nasal smoking-related expression profiles (Supplemental Table S3). Genes involved in oxidoreductase activity, xenobiotic metabolism, oxygen binding, tryptophan metabolism, the pentose phosphate pathway, and structural components of the extracellular matrix were significantly enriched among 119 genes (FDR < 0.05). The 27 genes with expression profiles that are affected by smoking much more so in bronchial than nasal epithelium were not significantly enriched for any functional category (FDR > 0.05).
DISCUSSION
Working from the hypothesis that cigarette smoke creates a field of injury throughout the respiratory tract and the desire to develop noninvasive biomarkers of physiological response to cigarette smoke exposure, we have investigated the relationship between the bronchial and nasal gene expression response to smoking. The majority of transcripts that are affected by smoking are affected similarly at both sites. The largely common response to cigarette exposure in epithelium collected from different airway sites likely reflects a conserved cellular response to cigarette smoke components, though the similarities in the tobacco smoke response are likely amplified by the similar cellular architecture and physiological functions shared by the epithelial cells lining the bronchus and nares: these airway compartments are both lined with ciliated pseudostratified columnar epithelial cells, and both sites participate in the detoxification of airborne compounds and the antioxidant response to redox stress (9). Taken together with our earlier work showing that upon smoking cessation the majority of genes that are differentially expressed in smokers revert rapidly toward the baseline levels observed in nonsmokers, these results suggest that the bulk of the gene expression differences in smokers–and therefore, the bulk of the physiological response to cigarette smoke–is an airway-wide and acute response to tobacco smoke.
The genes whose expression is affected by smoking at both sites are enriched for genes encoding proteins with oxidoreductase activity including the cytochrome P450 genes (e.g., CYP1A1, CYP1B1), aldo-keto reductases (AKR1B10), aldehyde dehydrogenase (ALDH1A3), and thioredoxin reductase (TXNRD1), with all of these genes being expressed more highly in smokers. CYP1A1 is involved in the metabolism of benzopyrene, a lung carcinogen, and component of tobacco smoke (7, 23). We have previously shown that the oxidoreductase genes induced by smoking are among the most rapidly reversible genes upon smoking cessation (1). The induction of these genes in both bronchial and nasal epithelium from current smokers further supports the idea of a core protective or detoxifying response to tobacco exposure that both sites share. Additionally, we found several genes with enzyme inhibitor activity including SERPINB13, TIMP3, and SCGB1A1 to be changed in both bronchial and nasal epithelium in smokers. SERPINB13, which is induced by smoking at both sites, has been linked to differentiation and apoptosis of human keratinocytes and protection of epithelial cells from cellular stress (24). TIMP3, also induced at both sites, inhibits angiogenesis by inhibiting VEGF binding to VEGF receptor-2 (14). SCGB1A1, which we found to be repressed by smoking at both sites, has been shown to have potent anti-inflammatory properties (17). Additionally, components of the pentose phosphate pathway, which serves to generate NADPH, an important defense against oxidative stress caused by cigarette smoke and other environmental exposures (10), were altered by smoking. All of the above pathways have been found to be enriched among genes with smoking-related expression profiles in other datasets (1).
In addition to identifying specific genes that are affected by smoking throughout the airway, we have also identified a smaller number of genes that are more dramatically altered by smoking in bronchial epithelium relative to nasal epithelium. MAFG, a transcription factor that interacts with Nrf1 and Nrf2 to regulate transcription of detoxification enzymes and components of the oxidative stress response (12), was among these genes. We previously found MAFG to be more highly expressed in bronchial epithelium from smokers relative to nonsmokers (19). We have shown that this induction of MAFG is regulated by mir218, a miRNA that is repressed in smokers (16). MAFG binding sites are overrepresented among the genes that are more dramatically affected by smoking in bronchial epithelium than in nasal epithelium (P = 0.05). This suggests that bronchial epithelium-specific regulation upstream of MAFG may account for some of the bronchial-specific gene expression responses to tobacco smoke. Whether mir218 or some other mechanism contributes to this bronchial-specific regulation of MAFG and the consequences of this site-specific response remains to be determined. In particular, it will be important to distinguish between the possibility that site-specific genes include those for which expression levels are sensitive to the concentration of tobacco-smoke components (reflecting the higher concentration of cigarette smoke components to which bronchial airway cells are exposed) and mechanisms that result from the repertoire of physiological responses to tobacco smoke components varying between the sites. Additionally, it will be of great interest to determine whether or not the mechanisms that contribute to these site-specific differences in gene expression also contribute to the higher cancer incidence in the bronchial airway compared with the nose.
We only identified three genes with a nasal-specific smoking-responsive expression profile. One of these genes is MEC (mucosal epithelial chemokine, CCL28), a member of the CC family of small cytokines, which are involved in immunoregulatory and inflammatory processes. MEC/CCL28 is abundantly expressed by epithelia in the bronchi, colon, and salivary gland (11). In our study, CCL28 was expressed at relatively high levels in both bronchial and nasal epithelium in never smokers, but its levels were decreased in nasal epithelium from smokers.
Interestingly, we also identified a small number of genes that appear to be affected by smoking at both sites, but are either changed in the same direction, but to a different extent in bronchial and nasal epithelium or are induced by smoking at one site and repressed by smoking at the other. Aldehyde dehydrogenase (ALDH3A1), GPX2 glutathione peroxidase 2, and AKR1C2 aldo-keto reductase were induced by smoking at different magnitudes at each site (each of the three genes was induced fourfold in the bronchus, but only induced 1.5-fold in the nose), suggesting a stronger induction of the detoxifying response to tobacco exposure in bronchus. DNER, a delta and notch-like epidermal growth factor-related receptor, was induced by smoking in bronchus and repressed in nose. The Cancer Genome Anatomy Project (CGAP) detected increased expression of DNER in lung cancer samples, relative to their normal controls by using serial analysis of gene expression (3), suggesting a potential link to the higher rate of cancer development in the bronchial airway compared with the nose.
In summary, our study has demonstrated that the gene expression response to smoking in nasal and bronchial epithelium is largely similar. This supports the hypothesis that gene expression profiling of the nasal epithelium may be an effective strategy for studying many of the physiological responses to tobacco smoke exposure. The existence of subtle differences in the gene expression response to smoking between the airway sites suggests that the validity of nasal profiling will need to be established for specific epidemiologic and clinical applications. Such efforts are warranted given the relatively noninvasive procedures by which nasal epithelium can be collected, and the fact that nasal gene expression profiling may facilitate large-scale studies that would be impossible with bronchial airway sampling. For example, longitudinal sampling of nasal epithelium may provide an opportunity to explore the acute and chronic effects of tobacco exposure and other inhaled toxins. We have previously shown that gene expression differences distinguish bronchial airway samples from healthy smokers and those with lung cancer (20) and are currently exploring the possibility of gene expression changes indicative of other tobacco-related diseases being detectable in these specimens. If these gene expression differences extend to the epithelial cells lining the nares, nasal epithelium gene expression could serve as the basis for screening tools to identify smokers at highest risk for tobacco-related lung disease.
GRANTS
This work was supported by National Institute of Environmental Health Sciences Grant UO1ES-016035 (Genes, Environment and Health Initiative).
DISCLOSURES
None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Beane J, Sebastiani P, Liu G, Brody JS, Lenburg ME, Spira A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol 8:R201, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beane J, Sebastiani P, Whitfield TH, Steiling K, Dumas YM, Lenburg ME, Spira A. A prediction model for lung cancer diagnosis that integrates genomic and clinical features. Cancer Prev Res (Phila Pa) 1: 56–64, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boon K, Osorio EC, Greenhut SF, Schaefer CF, Shoemaker J, Polyak K, Morin PJ, Buetow KH, Strausberg RL, De Souza SJ, Riggins GJ. An anatomy of normal and malignant gene expression. Proc Natl Acad Sci USA 99:11287–11292, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. CA Cancer J Clin 51: 15–36, 2001 [DOI] [PubMed] [Google Scholar]
- 6.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, Richter J, Rubin GM, Blake JA, Bult C, Dolan M, Drabkin H, Eppig JT, Hill DP, Ni L, Ringwald M, Balakrishnan R, Cherry JM, Christie KR, Costanzo MC, Dwight SS, Engel S, Fisk DG, Hirschman JE, Hong EL, Nash RS, Sethuraman A, Theesfeld CL, Botstein D, Dolinski K, Feierbach B, Berardini T, Mundodi S, Rhee SY, Apweiler R, Barrell D, Camon E, Dimmer E, Lee V, Chisholm R, Gaudet P, Kibbe W, Kishore R, Schwarz EM, Sternberg P, Gwinn M, Hannick L, Wortman J, Berriman M, Wood V, de la Cruz N, Tonellato P, Jaiswal P, Seigfried T, White R. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res 32:D258–D261, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91:1194–1210, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol 4:R70, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoshino Y, Mio T, Nagai S, Miki H, Ito I, Izumi T. Cytotoxic effects of cigarette smoke extract on an alveolar type II cell-derived cell line. Am J Physiol Lung Cell Mol Physiol 281: L509–L516, 2001 [DOI] [PubMed] [Google Scholar]
- 10.Kruger NJ, von Schaewen A. The oxidative pentose phosphate pathway: structure and organisation. Curr Opin Plant Biol 6:236–246, 2003 [DOI] [PubMed] [Google Scholar]
- 11.Kunkel EJ, Butcher EC. Chemokines and the tissue-specific migration of lymphocytes. Immunity 16:1–4, 2002 [DOI] [PubMed] [Google Scholar]
- 12.Myhrstad MC, Husberg C, Murphy P, Nordstrom O, Blomhoff R, Moskaug JO, Kolsto AB. TCF11/Nrf1 overexpression increases the intracellular glutathione level and can transactivate the gamma-glutamylcysteine synthetase (GCS) heavy subunit promoter. Biochim Biophys Acta 1517:212–219, 2001 [DOI] [PubMed] [Google Scholar]
- 13.Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res 36:W423–W426, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, Bond M, Baker A, Anand-Apte B. A novel function for tissue inhibitor of metalloproteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med 9:407–415, 2003 [DOI] [PubMed] [Google Scholar]
- 15.Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics 8:217, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schembri F, Sridhar S, Perdomo C, Gustafson AM, Zhang X, Ergun A, Lu J, Liu G, Zhang X, Bowers J, Vaziri C, Ott K, Sensinger K, Collins JJ, Brody JS, Getts R, Lenburg ME, Spira A. MicroRNAs as modulators of smoking-induced gene expression changes in human airway epithelium. Proc Natl Acad Sci USA 106: 2319–2324, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sengler C, Heinzmann A, Jerkic SP, Haider A, Sommerfeld C, Niggemann B, Lau S, Forster J, Schuster A, Kamin W, Bauer C, Laing I, LeSouef P, Wahn U, Deichmann K, Nickel R. Clara cell protein 16 (CC16) gene polymorphism influences the degree of airway responsiveness in asthmatic children. J Allergy Clin Immunol 111:515–519, 2003 [DOI] [PubMed] [Google Scholar]
- 18.Shields PG. Molecular epidemiology of lung cancer. Ann Oncol 10, Suppl 5: S7–S11, 1999 [DOI] [PubMed] [Google Scholar]
- 19.Spira A, Beane J, Shah V, Liu G, Schembri F, Yang X, Palma J, Brody JS. Effects of cigarette smoke on the human airway epithelial cell transcriptome. Proc Natl Acad Sci USA 101:10143–10148, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spira A, Beane JE, Shah V, Steiling K, Liu G, Schembri F, Gilman S, Dumas YM, Calner P, Sebastiani P, Sridhar S, Beamis J, Lamb C, Anderson T, Gerry N, Keane J, Lenburg ME, Brody JS. Airway epithelial gene expression in the diagnostic evaluation of smokers with suspect lung cancer. Nat Med 13:361–366, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Sridhar S, Schembri F, Zeskind J, Shah V, Gustafson AM, Steiling K, Liu G, Dumas YM, Zhang X, Brody JS, Lenburg ME, Spira A. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genomics 9:259, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102: 15545–15550, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vineis P, Veglia F, Benhamou S, Butkiewicz D, Cascorbi I, Clapper ML, Dolzan V, Haugen A, Hirvonen A, Ingelman-Sundberg M, Kihara M, Kiyohara C, Kremers P, Le ML, Ohshima S, Pastorelli R, Rannug A, Romkes M, Schoket B, Shields P, Strange RC, Stucker I, Sugimura H, Garte S, Gaspari L, Taioli E. CYP1A1 T3801 C polymorphism and lung cancer: a pooled analysis of 2451 cases and 3358 controls. Int J Cancer 104:650–657, 2003 [DOI] [PubMed] [Google Scholar]
- 24.Welss T, Sun J, Irving JA, Blum R, Smith AI, Whisstock JC, Pike RN, von Mikecz A, Ruzicka T, Bird PI, Abts HF. Hurpin is a selective inhibitor of lysosomal cathepsin L and protects keratinocytes from ultraviolet-induced apoptosis. Biochemistry 42:7381–7389, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Zhang X, Liu G, Lenburg ME, Spira A. Comparison of smoking-induced gene expression on Affymetrix Exon and 3′-based expression arrays. Genome Inform 18:247–257, 2007. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.