Abstract
Heterogeneous stock (HS) animals provide the ability to map quantitative trait loci at high resolution [<5 Megabase (Mb)] in a relatively short time period. In the current study, we hypothesized that the HS rat colony would be useful for fine-mapping a region on rat chromosome 1 that has previously been implicated in glucose regulation. We administered a glucose tolerance test to 515 HS rats and genotyped these animals with 69 microsatellite markers, spaced an average distance of <1 Mb apart, on a 67 Mb region of rat chromosome 1. Using regression modeling of inferred haplotypes based on a hidden Markov model reconstruction and mixed model analysis in which we accounted for the complex family structure of the HS, we identified one sharp peak within this region. Using positional bootstrapping, we determined the most likely location of this locus is from 205.04 to 207.48 Mb. This work demonstrates the utility of HS rats for fine-mapping complex traits and emphasizes the importance of taking into account family structure when using highly recombinant populations.
Keywords: type 2 diabetes, quantitative trait loci, mapping, mixed model analysis
heterogeneous stocks (HS) are a powerful tool for rapidly fine-mapping loci involved in complex traits (41). These animals are originally derived from eight inbred founder strains and then bred for 40–50 generations in a pattern that aims to minimize inbreeding (1, 16). The resulting colony represents a random mosaic of the founders, with the distance between recombination approaching 2 cM, enabling rapid fine-mapping of quantitative trait loci (QTLs) (31). An HS rat stock was developed in 1984 using the following eight inbred founder strains: ACI/N, BN/N, BUF/N, F344/N, M520/N, MR/N, WKY/N, WN/N (16). Multiple phenotypes have been fine-mapped using the HS mouse (41). To date, however, the HS rat colony has only been used to fine-map a single locus for fear-related behavior (18).
We hypothesized that HS rats would be useful for fine-mapping a region on rat chromosome 1 that has previously been identified for several metabolic traits, including glucose tolerance, in several linkage studies in the rat (3, 11, 12, 20, 32, 37, 45, 46). Human linkage and genome-wide association studies for type 2 diabetes have also identified the homologous human region (6, 19, 25). While many of the previous rat studies have used animal models of diabetes such as the Goto-Kakizaki (GK) or Otsuka Long-Evans Tokushima Fatty rat (11, 12, 20, 32, 46), this locus has also been mapped for diabetes or glucose tolerance using founders of the HS colony: Wistar Kyoto (WKY) and August Copenhagen Irish (ACI) (37, 45). These studies indicate that diabetes susceptibility alleles will segregate within the HS rat colony. While the WKY rat has previously been found to exhibit hyperglycemia and glucose intolerance (17, 21), no information is currently available regarding glucose regulation in the ACI rat.
We demonstrate here that both WKY and ACI strains exhibit glucose intolerance, and that the variation found between HS founders is represented within the HS colony. Using the HS rat colony, we have successfully identified a 2.44 Mb QTL within a 67 Mb region on rat chromosome 1 for glucose tolerance. This locus has not been identified in previous studies, indicating that a novel gene or genes involved in glucose regulation resides within this QTL. In addition, these data support both the complexity of this 67 Mb region, as well as the utility of the HS rat colony for rapidly fine-mapping loci involved in diabetic traits.
METHODS
Animals
Founding inbred substrains.
The inbred founders that make up the HS rat colony (ACI/N, BN/SsN, BUF/N, F344/N, M520/N, MR/N, WKY/N, and WN/N) are no longer available from outside vendors. Therefore, all testing was done on the following substrains of the original NIH founders: ACI/Eur, BN/NHsdMcwi, BUF/NHsd, F344/NHsd, WKY/NHsd. These substrains were chosen based on similarities in origin to the original founders as determined by data obtained from the Rat Genome Database: http://rgd.mcw.edu/strains/. ACI/Eur and BN/NHsdMcwi were obtained from colonies maintained at the Medical College of Wisconsin. BUF/NHsd, F344/NHsd, and WKY/NHsd were ordered from Harlan Sprague-Dawley (Indianapolis, IN). All experiments were conducted only in males. Three of the strains (M520/N, MR/N and WN/N) can no longer be obtained from outside vendors, so these strains were not tested. All protocols were approved by the Institutional Animal Care and Use Committee at the Medical College of Wisconsin.
Heterogeneous stock colony.
We obtained 25 heterogeneous stock breeding pairs from Dr. Eva Redei's colony at Northwestern University (Chicago, IL). Dr. Redei obtained her colony from Dr. Carl Hansen at the National Institutes of Health (NIH) in 2003. At the time we received the animals, they had been through 55 generations of breeding (50 at the NIH and 5 at Northwestern University). We increased the number of breeding pairs to 46 and have maintained these animals using a rotational breeding strategy to minimize inbreeding (1, 23). The HS animals at the Medical College of Wisconsin have been named NMcwi:HS and have the following Rat Genome Database identification number 2314009.
Experiments were conducted in 515 NMcwi:HS male progeny from the first four generations of breeders set up at the Medical College of Wisconsin.
Phenotyping Protocol
In both founder substrains and the heterogeneous stock animals, we conducted an intraperitoneal glucose tolerance test (IPGTT) at 16 wk of age. Animals were fasted overnight for 16 ± 1 h. In the morning, a basal sample of blood was taken from the tail and subsequent samples were collected 15, 30, 60, and 90 min after a 1 g/kg body wt glucose injection. We used the Ascensia Elite system for reading blood glucose values (Bayer, Elkhart, IN). Experiments were conducted over a period of 2 yr in two different buildings (due to a move of the animal facility after phenotypes in less than half of the animals were complete).
Genotyping
Tail samples from the original eight inbred founders were obtained from the NIH. Tail samples for the inbred substrain founders and the HS animals were collected at death. We extracted DNA from the tail tissue using an ethanol extraction procedure.
Original founders, founder substrains, and 515 HS were genotyped using 69 SSLP markers (see Supplementary Table S11) within a 67 Mb region on rat chromosome 1 (from 200 Mb to 267 Mb). Coordinates of markers are based on rat genome assembly version 3.4. Markers were spaced an average of just under 1 Mb apart. PCR was conducted as previously described (30) and analyzed on the ABI 3730.
Statistical Analysis
A repeated measures ANOVA was used to determine if there were statistical differences between inbred founder substrains for glucose during the IPGTT. Tukey-Kramer was the post hoc test used.
Statistical Genetic Analysis
We fine-mapped QTLs in the region by first inferring the underlying haplotype structure of each rat and then using those inferred haplotype states for an association analysis. Haplotype descent along each HS rat genome was inferred using the haplotype reconstruction method HAPPY (http://www.well.ox.ac.uk/happy/). This method, which has been used extensively to map loci in HS mice (31, 39, 41), uses a hidden Markov model to calculate the probability of descent from each founder at each genotyped locus. The method then estimates the expected proportion of genetic material originating from each founder for each interval between adjacent markers. The vector of haplotype proportions gi(m) for marker interval m of rat i is then used in place of the raw genotype to characterize variation at the locus. Initial studies using the HAPPY reconstruction method to fine-map in heterogeneous stock mice (31, 39) modeled gi(m) as the predictor in a simple regression model. However, it was recognized that the family structure of the HS, a consequence of the multiple generations needed to create a highly recombinant population suitable for fine-mapping, resulted in a complex correlation structure that led to confounded associations and an excess of false positives when analyzed using that simple model (40, 41). When whole genome information spanning the genome is available for each animal, these confounded associations can be effectively controlled by modeling multiple QTL associations simultaneously, by variance component modeling or by a combination of the two (40). When only a region of the genome is genotyped, however, multiple QTL modeling is not applicable. In their fine-mapping of a fear-related locus on a single chromosome, Johannesson et al. (18) overcame this excess of potential false positives by adopting a stricter significance threshold in which family structure is incorporated into the threshold procedure. Here we developed this approach further by including sibship as a random intercept both in the HAPPY regression model (see Ref. 22) and in the threshold procedure (see Ref. 40).
The phenotype, area under the curve (AUC) of glucose after a glucose challenge, was first log-transformed from a skewed distribution to approximate residual normality. At each locus m we then modeled the effect of genetic variation on the transformed phenotype log(yi) of individual i as
| Eq. 1 |
where in the above expression xi(c) is the value of the covariate c for individual i, C is a set of covariates (see below), gi(m) is defined as above for additive and full genetic models (31), sk[i] is the effect of sibship k to which rat i belongs, sk ∼ N(0,σs2) and εi ∼ N(0,σ2) are random intercepts, and all other terms are estimated by the model. We defined C to include the following pretreatment covariates: location (2 buildings), injector (2 injectors), collector (person who collected the blood, 2 collectors), and the number of glucose injections the rat received (1 or 2; a small percentage of animals received a second glucose injection when the initial injection failed to lead to a glucose response). We fitted the model by restricted estimate maximum likelihood (REML) and calculated the nominal significance (as the log10 P value; logP) of association at each locus by a likelihood ratio test based on the full likelihoods at the REML estimates of the model in Eq. 1 against one that omits locus specific information but retains sibship and other effects (the null model). A region-wide significance threshold was then obtained by repeating the region scan on 200 sets of phenotypes generated by parametric bootstrap from the fitted null model and using the maximum logP's from those scans to fit a generalized extreme value distribution from which the upper 5% quantile was taken as the 5% region-wide significance threshold (18, 41). All models were fitted using the statistical language R (R-Development-Core-Team 2004) and the add-on packages lme4 and evd. For comparison with Johannesson et al. (18) we also performed a region-wide scan by fitting the model in Eq. 1 without the sibship effect sk. Thresholds for this secondary analysis were generated as above, i.e., by generating null phenotypes with a correlation structure based on sibship clustering [as in Johannesson et al. (18)], and, for further illustrative comparison, by permutation.
RESULTS
Phenotyping
Glucose during the IPGTT in HS and founders.
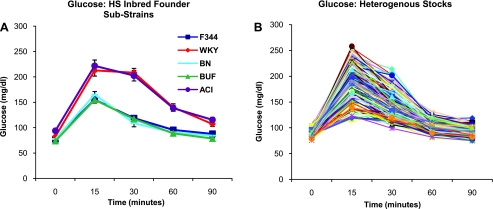
Based on a repeated-measures ANOVA in the inbred founder substrains, our results demonstrate a significant effect of strain (F4, 288 = 30.7, P < 1 × 10−16), time point (F4, 288 = 494.1, P < 1 × 10−16), and a strain by time-point interaction (F16, 288 = 17.7, P < 1 × 10−16). ACI and WKY rats exhibit significantly higher levels of glucose at 15, 30, and 60 min after a glucose challenge, while ACI rats continue to exhibit significantly higher levels of glucose 90 min after the challenge, relative to F344, BN, and BUF rats (P < 0.01, Tukey-Kramer; see Fig. 1A). WKY and ACI rats are not significantly different from each other at any time point. Glucose values for the 515 HS rats fell generally between the average values for the inbred founder sub-strains for all time points (see Fig. 1B).
Fig. 1.
Glucose response curves after a glucose challenge in inbred founders (A) and heterogeneous stock (HS) rats (B). Founder curves represent the mean of 6–14 animals within each inbred strain. Each line in the HS graphs represent an individual animal (200 representative HS rats are shown in graph).
Genetic Analysis
All but one (D1Rat77) of the 69 markers within the 67 Mb region on rat chromosome 1 were informative within the HS population, indicating the founder alleles segregate within the colony. The average heterozygosity in the HS population for these markers was 44%, indicating sufficient genetic diversity for fine-mapping within this region. These results also highlight the lack of inbreeding that has occurred within the HS rat colony, as least based on results for this region on chromosome 1.
Glucose tolerance.
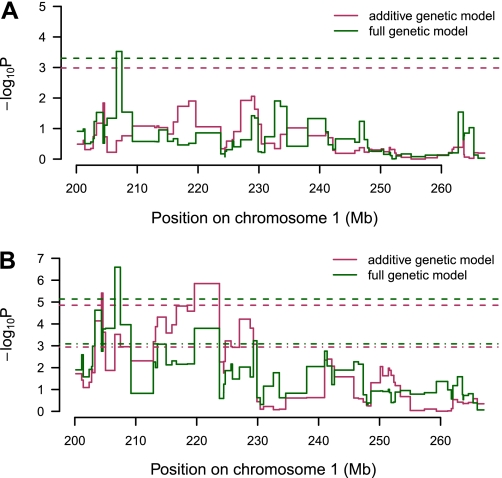
Using haplotype reconstruction and mixed model linear regression, which takes into account sibship relationships, we identified a single sharp peak (marker interval between D1Rat112 and D1Rat323, 206.57–207.48Mb; logP=3.52) for Glucose_AUC above the region-wide 5% significance threshold, which were logP = 2.98 and 3.30 for the full and additive models, respectively, where logP is defined as -log10(P value) (see Fig. 2A). This locus has been named Gluco56 and has Rat Genome Database identification number 2314011.
Fig. 2.
Mapping of Glucose_AUC within a 67 Mb region previously implicated for glucose tolerance on rat chromosome 1. The x-axis shows position in Mb, and the y-axis gives the -log10P value of association. Dashed lines represent region-wide 5% significance thresholds for additive (red) and full (additive + dominance; green) genetic models. A: results based on a mixed-model analysis incorporating family structure in both locus model and threshold procedure. B: results based on an analysis excluding family structure from the locus model with thresholds calculated so that family structure is accounted for (upper dashed lines) and ignored (lower dashed lines). AUC, area under the curve.
For comparison with the HS rat study of Johannesson et al. (18), we also analyzed the data using a model that accounted for sibship effects in the thresholds but not the genetic model. Under that analysis model (shown in Fig. 2B), the logPs are inflated because family structure is not accounted for in the single locus regression, but the thresholds are also made stricter to reflect that inflation (logP = 4.86 and 5.13 for additive and full models). We further calculated thresholds by unrestricted permutation in Fig. 2B to illustrate the inflated significance that would be attributed to peaks in a naïve analysis that ignores family structure (logP = 2.94 and 3.08) (see Ref. 4). When family structure is absent from the regression model but retained when determining significance thresholds, an additional peak at 220 Mb becomes significant. Given the marginal significance of this association, however, and following Valdar et al. (40), when possible we prefer the more conservative mixed model for our region-wide scan (Fig. 2A).
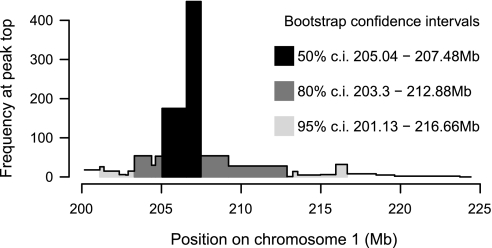
To estimate how much the location of the single peak varies with respect to finite sampling we performed positional bootstrapping after Visscher et al. (43). We ran 1,000 positional bootstraps of the region from 200.17–224.42 Mb. This region was chosen to avoid picking up associations from neighboring potential QTL peaks that would otherwise make the confidence interval meaningless. Of the 1,000 bootstrap samples, 623 were strongly associated to the locus as the peak marker interval in Fig. 2A or the interval upstream (darkest color, Fig. 3A; 205.04–207.48 Mb). Despite this relative concentration in the 2.44 Mb region, the 95% confidence interval for this region is considerably wider (lightest color in Fig. 3A; 201.13–216.66 Mb).
Fig. 3.
Positional bootstrapping of the significant peak from Fig. 2A. Histogram shows the number of times (y-axis) each marker interval (x-axis, locations in Mb) represented the strongest association in the subregion (205.04–207.48 Mb) when the analysis was applied to each of 1,000 bootstrapped sets of rats. Shading indicates the central quantile regions: e.g., for at least 50% of bootstraps the highest association was within 205.04–207.48 Mb.
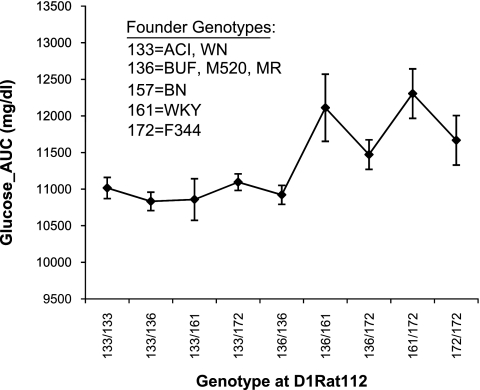
To characterize the association at this locus, we performed a one-way ANOVA of the phenotype, Glucose_AUC, against the genotype at the peak marker, D1Rat112. As expected, there is evidence for an effect of genotype on the glucose phenotype (F9,479 = 4.24, P = 2.7 × 10−5), as illustrated in Fig. 4.
Fig. 4.
Effect plot for marker D1Rat112 on phenotype Glucose_AUC in HS rats. SSLP genotypes are shown on the x-axis (note that 4 alleles were found in the HS for this marker). Glucose_AUC is shown on the y-axis, with means ± SE for animals within each genotype group plotted on the graph. Founder allele sizes are listed in the graph. Note that the 133 allele (ACI and WN) appears to protect against high glucose, while HS animals with either the 161 (WKY) the 172 (F344) allele exhibit significantly higher glucose levels during the glucose tolerance test than those with the other alleles, except when heterozygous with the protective 133 allele. The BN allele at this locus, 157, was not represented within the HS colony.
DISCUSSION
We have shown that HS rats are a useful tool for rapidly fine-mapping a diabetic trait. We demonstrate here that at least two of the HS founders (ACI and WKY) exhibit glucose intolerance and that HS rats exhibit large variation in response to a glucose challenge. We have used a conservative statistical method to fine-map a 67 Mb region on rat chromosome 1, previously implicated in glucose tolerance and type 2 diabetes. Using this mixed model analysis, which accounts for the complex family structure of the HS, we have identified a novel 2.44 Mb locus within this region. These results indicate that HS rats will be a useful tool for uncovering novel genes involved in diabetes and emphasize the importance of taking into account family structure when analyzing highly recombinant populations.
The statistical method we used accounts for the complex family structure of highly recombinant populations such as the HS. Importantly, HS animals are related to each other by different degrees (sibs, half-sibs, cousins, etc.), and this imposes a cluster structure in the data that, if not accounted for analytically, results in spurious associations in addition to genuine QTLs (13, 40). A previous study using the HS rat population accounted for family structure by employing a significance threshold that takes into account these family relationships (18). In the current study, we have developed this approach further by not only taking family into account when determining the significance threshold, but also by including family as a random intercept in the regression model. By employing this conservative technique, we identify a single locus within this chromosome 1 region. When family is not used in the regression model, but instead is used only to determine the significance threshold, one other peak (at 220 Mb) passes the significance threshold. However, as this peak is only marginally significant, we prefer the more conservative mixed-model approach. Potential confirmation of the second peak will require an increase in the number of animals used.
To assign a confidence interval to this single peak, we used positional bootstrapping. Markers within a 2.44 Mb region, between 205.04 and 207.48 Mb, were identified as the peak marker in >50% of 1,000 bootstrap samples. Neighboring markers spanning the region from 201.13 to 216.66 Mb were identified as the peak marker in the remaining bootstrap samples. However, these neighboring markers were identified at very low frequencies (see Fig. 3), thereby strongly suggesting that this QTL most likely lies within the 2.44 Mb region between 205.04 and 207.48 Mb. We have confirmed the strength of the association at this locus using an ANOVA at the peak marker, D1Rat112.
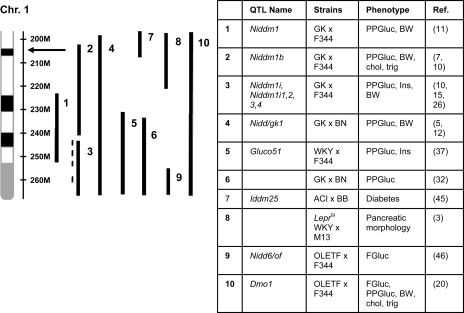
This 2.44 Mb region lies at the distal end of a region that has previously been identified for glucose regulation in multiple F2 linkage studies in the rat, including two HS founders (see Fig. 5). Recently, congenic and expression studies have shown that multiple loci within this region play a role in glycemic control (5, 7, 10, 15, 26, 44). Furthermore, the homologous region in humans has been identified in several linkage studies (6, 8, 14, 19, 27, 28, 42, 47), and three genes in this region have recently been identified in human genome wide association studies (GWAS) (Tcf7l2, HHEX and IDE) (34–36, 38, 49). Using the HS rat, we have identified a single 2.44 Mb locus within this region for Glucose_AUC. This novel locus does not overlap fine-mapped loci previously identified using congenic animals of the type 2 diabetic GK rat (5, 10, 15). Interestingly, however, this locus overlaps a large QTL identified in the ACI founder for type 1 diabetes (45), as well as a locus identified for pancreatic morphology in the type 2 diabetic Leprfa/Leprfa WKY rat (3) (see Fig. 5). In the current study, we show that the ACI allele (133) appears to confer protection against high glucose at this locus, while both the WKY (161) and F344 (172) alleles associate with glucose intolerance. These results suggest that the F344 allele is acting through transgressive segregation, a relatively common phenomenon (33). Considering the complexity of this rich region, it is not surprising that using a resource with multiple alleles would uncover a novel locus within this region. We expect that uncovering the causative gene or genes within this region will elucidate a novel gene involved in glucose regulation.
Fig. 5.
Quantitative trait loci (QTLs) identified in multiple rat F2 crosses within a 67 Mb region on rat chromosome 1. Vertical bars to the right of chromosome 1 (shown from 200 Mb to the end of the chromosome) represent 95% confidence interval of the QTL. The black arrow indicates the location of the 2.44 Mb locus we have mapped using HS rats. Information for each QTL can be found by number in the accompanying table. Phenotypes in the table are abbreviated: PPGluc, postprandial glucose; FGluc, fasting glucose; BW, body weight; Chol, cholesterol; Trig, triglycerides; Ins, postprandial insulin.
The 2.44 Mb region for glucose tolerance contains 62 genes (see Supplementary Table S2), none of which have previously been identified for type 2 diabetes. Interestingly, several genes in this region play a role in oxidative phosphorylation, a key metabolic pathway in which ATP is produced from glucose and other nutrients. Increasingly, mitochondrial dysfunction has been linked with type 2 diabetes, and the underlying genetic components are only beginning to be understood (2, 24, 29). Our findings in the HS rat suggest this will be a useful model for uncovering at least some of these genes.
The next step in these studies will be to identify the potentially causative gene or genes within this fine-mapped region. While there is still a relatively large number of genes within this locus, the identification of causative genes is facilitated by having eight individual founder strains as opposed to only two, as found in F2 intercrosses or congenic strains. As shown previously, sequence information from the founders can be used to rapidly narrow potential candidate variants (48), which can then be verified in functional studies (9).
We have demonstrated that founders of the HS rat colony exhibit a high degree of variation for glucose tolerance, and that this variation is represented within the HS colony. Using a mixed model analysis, which accounts for family structure in the HS colony, we have been able to rapidly fine-map a 2.44 Mb locus involved in glucose tolerance. These results demonstrate the utility of the HS rat for rapidly fine-mapping metabolic traits, as well as emphasize the importance of taking into account family relationships in the analysis.
GRANTS
This work was funded by National Institute of Diabetes and Digestive and Kidney Diseases Grant K01 DK-076977 and by the Individualized Medicine Institute of the Medical College of Wisconsin.
DISCLOSURES
No conflicts of interest are declared by the authors.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Drs. Howard Jacob and Alan Attie for invaluable mentorship, Andrew Patzer for assistance with programming, Steve Stanhope for help with the software, and Nancy Schlick and Jamie Wendt Andrae for assistance with animal experiments.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Caballero A, Toro MA. Interrelations between effective population size and other pedigree tools for the management of conserved populations. Genet Res 75: 331–343, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Cho YM, Park KS, Lee HK. Genetic factors related to mitochondrial function and risk of diabetes mellitus. Diabetes Res Clin Pract 77, Suppl 1: S172–S177, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Chung WK, Zheng M, Chua M, Kershaw E, Power-Kehoe L, Tsuji M, Wu-Peng XS, Williams J, Chua SC, Jr, Leibel RL. Genetic modifiers of Leprfa associated with variability in insulin production and susceptibility to NIDDM. Genomics 41: 332–344, 1997 [DOI] [PubMed] [Google Scholar]
- 4.Churchill GA, Doerge RW. Naive application of permutation testing leads to inflated type I error rates. Genetics 178: 609–610, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins SC, Wallis RH, Wilder SP, Wallace KJ, Argoud K, Kaisaki PJ, Bihoreau MT, Gauguier D. Mapping diabetes QTL in an intercross derived from a congenic strain of the Brown Norway and Goto-Kakizaki rats. Mamm Genome 17: 538–547, 2006 [DOI] [PubMed] [Google Scholar]
- 6.Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, O'Connell P, Stern MP. Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet 64: 1127–1140, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fakhrai-Rad H, Nikoshkov A, Kamel A, Fernstrom M, Zierath JR, Norgren S, Luthman H, Galli J. Insulin-degrading enzyme identified as a candidate diabetes susceptibility gene in GK rats. Hum Mol Genet 9: 2149–2158, 2000 [DOI] [PubMed] [Google Scholar]
- 8.Florez JC, Burtt N, de Bakker PI, Almgren P, Tuomi T, Holmkvist J, Gaudet D, Hudson TJ, Schaffner SF, Daly MJ, Hirschhorn JN, Groop L, Altshuler D. Haplotype structure and genotype-phenotype correlations of the sulfonylurea receptor and the islet ATP-sensitive potassium channel gene region. Diabetes 53: 1360–1368, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Fullerton JM, Willis-Owen SA, Yalcin B, Shifman S, Copley RR, Miller SR, Bhomra A, Davidson S, Oliver PL, Mott R, Flint J. Human-mouse quantitative trait locus concordance and the dissection of a human neuroticism locus. Biol Psychiatry 63: 874–883, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Galli J, Fakhrai-Rad H, Kamel A, Marcus C, Norgren S, Luthman H. Pathophysiological and genetic characterization of the major diabetes locus in GK rats. Diabetes 48: 2463–2470, 1999 [DOI] [PubMed] [Google Scholar]
- 11.Galli J, Li LS, Glaser A, Ostenson CG, Jiao H, Fakhrai-Rad H, Jacob HJ, Lander ES, Luthman H. Genetic analysis of non-insulin dependent diabetes mellitus in the GK rat. Nat Genet 12: 31–37, 1996 [DOI] [PubMed] [Google Scholar]
- 12.Gauguier D, Froguel P, Parent V, Bernard C, Bihoreau MT, Portha B, James MR, Penicaud L, Lathrop M, Ktorza A. Chromosomal mapping of genetic loci associated with non-insulin dependent diabetes in the GK rat. Nat Genet 12: 38–43, 1996. [DOI] [PubMed] [Google Scholar]
- 13.Ghazalpour A, Doss S, Kang H, Farber C, Wen PZ, Brozell A, Castellanos R, Eskin E, Smith DJ, Drake TA, Lusis AJ. High-resolution mapping of gene expression using association in an outbred mouse stock. PLoS Genet 4: e1000149, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh S, Watanabe RM, Valle TT, Hauser ER, Magnuson VL, Langefeld CD, Ally DS, Mohlke KL, Silander K, Kohtamaki K, Chines P, Balow J, Jr, Birznieks G, Chang J, Eldridge W, Erdos MR, Karanjawala ZE, Knapp JI, Kudelko K, Martin C, Morales-Mena A, Musick A, Musick T, Pfahl C, Porter R, Rayman JB. The Finland-United States investigation of non-insulin-dependent diabetes mellitus genetics (FUSION) study. I. An autosomal genome scan for genes that predispose to type 2 diabetes. Am J Hum Genet 67: 1174–1185, 2000 [PMC free article] [PubMed] [Google Scholar]
- 15.Granhall C, Park HB, Fakhrai-Rad H, Luthman H. High-resolution quantitative trait locus analysis reveals multiple diabetes susceptibility loci mapped to intervals<800 kb in the species-conserved Niddm1i of the GK rat. Genetics 174: 1565–1572, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen C, Spuhler K. Development of the National Institutes of Health genetically heterogeneous rat stock. Alcohol Clin Exp Res 8: 477–479, 1984 [DOI] [PubMed] [Google Scholar]
- 17.Ikeda H, Shino A, Matsuo T, Iwatsuka H, Suzuoki Z. A new genetically obese-hyperglycemic rat (Wistar fatty). Diabetes 30: 1045–1050, 1981 [DOI] [PubMed] [Google Scholar]
- 18.Johannesson M, Lopez-Aumatell R, Stridh P, Diez M, Tuncel J, Blazquez G, Martinez-Membrives E, Canete T, Vicens-Costa E, Graham D, Copley RR, Hernandez-Pliego P, Beyeen AD, Ockinger J, Fernandez-Santamaria C, Gulko PS, Brenner M, Tobena A, Guitart-Masip M, Gimenez-Llort L, Dominiczak A, Holmdahl R, Gauguier D, Olsson T, Mott R, Valdar W, Redei EE, Fernandez-Teruel A, Flint J. A resource for the simultaneous high-resolution mapping of multiple quantitative trait loci in rats: the NIH heterogeneous stock. Genome Res 19: 150–158, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun G, Song Y, Stein CM, Iyengar SK. An autosome-wide search using longitudinal data for loci linked to type 2 diabetes progression. BMC Genet 4, Suppl 1: S8, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanemoto N, Hishigaki H, Miyakita A, Oga K, Okuno S, Tsuji A, Takagi T, Takahashi E, Nakamura Y, Watanabe TK. Genetic dissection of “OLETF”, a rat model for non-insulin-dependent diabetes mellitus. Mamm Genome 9: 419–425, 1998 [DOI] [PubMed] [Google Scholar]
- 21.Katayama S, Inaba M, Maruno Y, Morita T, Awata T, Oka Y. Glucose intolerance in spontaneously hypertensive and Wistar-Kyoto rats: enhanced gene expression and synthesis of skeletal muscle glucose transporter 4. Hypertens Res 20: 279–286, 1997 [DOI] [PubMed] [Google Scholar]
- 22.Kennedy BW, Quinton M, van Arendonk JA. Estimation of effects of single genes on quantitative traits. J Anim Sci 70: 2000–2012, 1992 [DOI] [PubMed] [Google Scholar]
- 23.Kimura M, Crow JF. On maximum avoidance of inbreeding. Genet Res 4: 399, 1963 [Google Scholar]
- 24.Lacraz G, Giroix MH, Kassis N, Coulaud J, Galinier A, Noll C, Cornut M, Schmidlin F, Paul JL, Janel N, Irminger JC, Kergoat M, Portha B, Donath MY, Ehses JA, Homo-Delarche F. Islet endothelial activation and oxidative stress gene expression is reduced by IL-1Ra treatment in the type 2 diabetic GK rat. PLoS One 4: e6963, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lango H, Palmer CN, Morris AD, Zeggini E, Hattersley AT, McCarthy MI, Frayling TM, Weedon MN. Assessing the combined impact of 18 common genetic variants of modest effect sizes on type 2 diabetes risk. Diabetes 57: 3129–3135, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JM, Ortsater H, Fakhrai-Rad H, Galli J, Luthman H, Bergsten P. Phenotyping of individual pancreatic islets locates genetic defects in stimulus secretion coupling to Niddm1i within the major diabetes locus in GK rats. Diabetes 50: 2737–2743, 2001 [DOI] [PubMed] [Google Scholar]
- 27.Lindgren CM, Mahtani MM, Widen E, McCarthy MI, Daly MJ, Kirby A, Reeve MP, Kruglyak L, Parker A, Meyer J, Almgren P, Lehto M, Kanninen T, Tuomi T, Groop LC, Lander ES. Genomewide search for type 2 diabetes mellitus susceptibility loci in Finnish families: the Botnia study. Am J Hum Genet 70: 509–516, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meigs JB, Panhuysen CI, Myers RH, Wilson PW, Cupples LA. A genome-wide scan for loci linked to plasma levels of glucose and HbA(1c) in a community-based sample of Caucasian pedigrees: The Framingham Offspring Study. Diabetes 51: 833–840, 2002 [DOI] [PubMed] [Google Scholar]
- 29.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34: 267–273, 2003 [DOI] [PubMed] [Google Scholar]
- 30.Moreno C, Kennedy K, Andrae JW, Jacob HJ. Genome-wide scanning with SSLPs in the rat. Methods Mol Med 108: 131–138, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Mott R, Talbot CJ, Turri MG, Collins AC, Flint J. A method for fine mapping quantitative trait loci in outbred animal stocks. Proc Natl Acad Sci USA 97: 12649–12654, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobrega MA, Woods LC, Fleming S, Jacob HJ. Distinct genetic regulation of progression of diabetes and renal disease in the Goto-Kakizaki rat. Physiol Genomics 39: 38–46, 2009 [DOI] [PubMed] [Google Scholar]
- 33.Rieseberg LH, Widmer A, Arntz AM, Burke JM. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos Trans R Soc Lond B Biol Sci 358: 1141–1147, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336, 2007 [DOI] [PubMed] [Google Scholar]
- 35.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445: 881–885, 2007 [DOI] [PubMed] [Google Scholar]
- 37.Solberg Woods LC, Ahmadiyeh N, Baum A, Shimomura K, Li Q, Steiner DF, Turek FW, Takahashi JS, Churchill GA, Redei EE. Identification of genetic loci involved in diabetes using a rat model of depression. Mamm Genome 20: 486–497, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinthorsdottir V, Thorleifsson G, Reynisdottir I, Benediktsson R, Jonsdottir T, Walters GB, Styrkarsdottir U, Gretarsdottir S, Emilsson V, Ghosh S, Baker A, Snorradottir S, Bjarnason H, Ng MC, Hansen T, Bagger Y, Wilensky RL, Reilly MP, Adeyemo A, Chen Y, Zhou J, Gudnason V, Chen G, Huang H, Lashley K, Doumatey A, So WY, Ma RC, Andersen G, Borch-Johnsen K, Jorgensen T, van Vliet-Ostaptchouk JV, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Rotimi C, Gurney M, Chan JC, Pedersen O, Sigurdsson G, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet 39: 770–775, 2007 [DOI] [PubMed] [Google Scholar]
- 39.Talbot CJ, Radcliffe RA, Fullerton J, Hitzemann R, Wehner JM, Flint J. Fine scale mapping of a genetic locus for conditioned fear. Mamm Genome 14: 223–230, 2003 [DOI] [PubMed] [Google Scholar]
- 40.Valdar W, Holmes CC, Mott R, Flint J. Mapping in structured populations by resample model averaging. Genetics 182: 1263–1277, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet 38: 879–887, 2006 [DOI] [PubMed] [Google Scholar]
- 42.Vionnet N, Hani EH, Lesage S, Philippi A, Hager J, Varret M, Stoffel M, Tanizawa Y, Chiu KC, Glaser B, Permutt MA, Passa P, Demenais F, Froguel P. Genetics of NIDDM in France: studies with 19 candidate genes in affected sib pairs. Diabetes 46: 1062–1068, 1997 [DOI] [PubMed] [Google Scholar]
- 43.Visscher PM, Thompson R, Haley CS. Confidence intervals in QTL mapping by bootstrapping. Genetics 143: 1013–1020, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wallis RH, Collins SC, Kaisaki PJ, Argoud K, Wilder SP, Wallace KJ, Ria M, Ktorza A, Rorsman P, Bihoreau MT, Gauguier D. Pathophysiological, genetic and gene expression features of a novel rodent model of the cardio-metabolic syndrome. PLoS ONE 3: e2962, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallis RH, Wang K, Marandi L, Hsieh E, Ning T, Chao GY, Sarmiento J, Paterson AD, Poussier P. Type 1 diabetes in the BB rat: a polygenic disease. Diabetes 58: 1007–1017, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei S, Wei K, Moralejo DH, Ogino T, Koike G, Jacob HJ, Sugiura K, Sasaki Y, Yamada T, Matsumoto K. Mapping and characterization of quantitative trait loci for non-insulin-dependent diabetes mellitus with an improved genetic map in the Otsuka Long-Evans Tokushima fatty rat. Mamm Genome 10: 249–258, 1999 [DOI] [PubMed] [Google Scholar]
- 47.Wiltshire S, Hattersley AT, Hitman GA, Walker M, Levy JC, Sampson M, O'Rahilly S, Frayling TM, Bell JI, Lathrop GM, Bennett A, Dhillon R, Fletcher C, Groves CJ, Jones E, Prestwich P, Simecek N, Rao PV, Wishart M, Bottazzo GF, Foxon R, Howell S, Smedley D, Cardon LR, Menzel S, McCarthy MI. A genomewide scan for loci predisposing to type 2 diabetes in a UK population (the Diabetes UK Warren 2 Repository): analysis of 573 pedigrees provides independent replication of a susceptibility locus on chromosome 1q. Am J Hum Genet 69: 553–569, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yalcin B, Flint J, Mott R. Using progenitor strain information to identify quantitative trait nucleotides in outbred mice. Genetics 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeggini E, Weedon MN, Lindgren CM, Frayling TM, Elliott KS, Lango H, Timpson NJ, Perry JR, Rayner NW, Freathy RM, Barrett JC, Shields B, Morris AP, Ellard S, Groves CJ, Harries LW, Marchini JL, Owen KR, Knight B, Cardon LR, Walker M, Hitman GA, Morris AD, Doney AS, McCarthy MI, Hattersley AT. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 316: 1336–1341, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.