Abstract
Chromosome 13 consomic and congenic rat strains were analyzed to investigate the pattern of genomic pathway utilization involved in protection against salt-sensitive hypertension and renal injury. Introgression of the entire Brown-Norway chromosome 13 (consomic SS-13BN) or nonoverlapping segments of this chromosome (congenic strains, 16 Mbp in D13Rat151–D13Rat197 or 14 Mbp in D13Rat111–D13Got22) into the genome of the Dahl salt-sensitive rat attenuated salt-induced hypertension and proteinuria. mRNA abundance profiles in the renal cortex and the renal medulla from rats receiving 0.4% or 8% NaCl diets revealed two important features of pathway recruitment in these rat strains. First, the two congenic strains shared alterations in several pathways compared with Dahl salt-sensitive rats, despite the fact that the genomic segments introgressed in the two congenic strains did not overlap. Second, even though the genomic segment introgressed in each congenic strain was a part of the chromosome introgressed in the consomic strain, pathways altered in each congenic strain were not simply a subset of those altered in the consomic. Supporting the relevance of the mRNA data, differential expression of oxidative stress-related genes among the four strains of rats was associated with differences in urinary excretion of lipid peroxidation products. The findings suggest that different genetic alterations might converge to influence shared pathways in protection from hypertension, and that, depending on the genomic context, the same genetic alteration might diverge to affect different pathways.
Keywords: gene expression, consomic rats, congenic rats, blood pressure, kidney
consomic and congenic strains of animals have been commonly used to identify genes containing sequence variations that contribute to a phenotype in animal models (3, 5, 7, 14). The general idea is to reduce the genetic difference between a disease strain and a protected strain to a small region of the genome to facilitate the identification of the gene(s) responsible for normal and diseased phenotypes.
As important is the identification of the biological pathways and the regulatory networks that underlie the disease development. It is now generally accepted that complex diseases such as hypertension cannot be explained by a single gene or even a single biological pathway. Instead, networks of interacting pathways are likely involved. This is illustrated by our previous work on molecular networks underlying Dahl salt-sensitive hypertension, which has led to several novel discoveries regarding the overall network (9) and specific components of the network (11, 20, 22, 23).
The effort to identify genomic pathways and regulatory networks, however, is hampered by poor understanding of the general nature of such complex interactions. In particular, several broadly relevant questions crucial to the use of consomic and congenic strains for pathway and network analysis remain largely unanswered. Would a genomic region or a gene always impact on a defined set of pathways, regardless of the composition of the rest of the genome? Should a narrow genomic region be expected to affect a subset of the pathways that are affected by a larger genomic region that encompasses the narrow region? If two or more distinct genomic regions are identified to be associated with a disease phenotype, does it necessarily mean that two or more distinct sets of pathways must be involved? Investigators have made various assumptions on these questions as they embark on the challenging task of searching for genomic pathways and regulatory networks, yet the validity of any of these critical assumptions remains largely untested.
The Dahl salt-sensitive (SS) rat is a genetic model of human salt-sensitive forms of hypertension and renal injury (1, 19, 24). The development and characterization of consomic and congenic strains derived from SS rats and salt-resistant Brown-Norway (BN) rats (http://pga.mcw.edu/) have yielded a series of new insights into the genetics and pathophysiology of Dahl salt-sensitive hypertension and renal injury. Introgression of eight different BN chromosomes, including chromosome 13, or several nonoverlapping regions of BN chromosome 13, into the SS genome significantly attenuates salt-induced hypertension and/or renal injury (2, 13, 16). Several biological pathways have been found that may contribute to the protection from salt-induced hypertension and/or renal injury observed in the consomic SS-13BN strain compared with SS rats. Examples include attenuated oxidative stress (20), reduced local levels of glucocorticoids (11), and improved fumarate metabolism (22, 23).
In the present study, we used SS rats, consomic SS-13BN rats, and two congenic rat strains to examine the dynamics of genomic and functional pathway recruitment and utilization resulting from chromosome segment substitution and to determine whether shared or independent pathways contribute to the antihypertensive effects of different regions of chromosome 13. Renal cortical and medullary gene expression profiles were analyzed in the four strains of rats at 6–7 wk of age fed a 0.4% NaCl diet since weaning or an 8% NaCl diet for 3 and 7 days. Effect of the high-salt diet on arterial blood pressure was examined at 6–9 wk of age and then at 9–12 wk of age to determine whether genomic expression differences in young rats in response to a high-salt diet would predict the blood pressure and renal injury responses observed at a later age. The result indicated several important features of pathway utilization that would be relevant to the understanding of Dahl salt-sensitive hypertension and renal injury as well as other complex disease phenotypes.
MATERIALS AND METHODS
Consomic and congenic rats.
The generation of consomic and congenic rat strains has been described previously (2, 16). The consomic strain SS-13BN was developed from selective breeding of the salt-resistant BN rat (BN/MCWi; BN) and the Dahl SS rat of the Medical College of Wisconsin (MCW) colony (SS/JrHsdMcwi; SS). The genome of SS-13BN contained homozygous SS alleles on all chromosomes, as confirmed by total genome scans, except chromosome 13, which was homozygous BN as confirmed by a scan at a higher density. Congenic strains SS.BN-(D13Rat111–D13Got22) and SS.BN-(D13rat151–D13rat197) were developed from SS and SS-13BN. The D13Rat111–D13Got22 segment (14 Mbp) and the D13rat151–D13rat197 segment (16 Mbp) of chromosome 13 were homozygous BN in the two congenic strains, respectively, while the remainder of chromosome 13 and all other chromosomes were homozygous SS. For simplicity, SS.BN-(D13Rat111–D13Got22) and SS.BN-(D13rat151–D13rat197) are referred to as Line 5 and Line 26, respectively, following the nomenclature used in the initial report of the congenic panel (16). The MCW Institutional Animal Care and Use Committee approved all experimental protocols.
Chronic measurement of arterial blood pressure.
Arterial blood pressure was measured in 8–17 male rats of each of the four strains, SS, consomic SS-13BN, congenic Line 5, and congenic Line 26, by radiotelemetry (25). All rats had been maintained on a 0.4% NaCl diet (Dyets, Bethlehem, PA) since weaning. Baseline blood pressure was recorded from 9:00 AM to noon for several days ∼1 wk after catheterization. The rats were then switched to an 8% NaCl diet (Dyets). Arterial blood pressure was monitored at several time points until day 14 on the high-salt diet. Rats were 5–6 or 8 wk old at the time of catheterization and 6–9 or 9–12 wk old when blood pressure was measured.
Urinary protein, albumin, and thiobarbituric acid-reactive substance.
Urine samples were collected from 6- to 7-wk-old male uninstrumented rats over several 24-h periods with metabolic cages. Urinary concentrations of protein and albumin were measured as described previously (9). Thiobarbituric acid-reactive substance (TBARS) was measured with an assay kit from Cayman Chemical.
Dietary protocol and sample preparation for microarray experiment.
A total of 72 rats, 18 for each of the 4 strains, were used for the gene expression study. Dietary treatment, tissue collection, and RNA extraction were performed as described previously (8, 9). Rats were male and 6–7 wk old. Tissues were harvested from rats on the 0.4% NaCl diet or at 3 or 7 days after the rats were switched to the 8% NaCl diet. A total of 72 individual RNA samples were generated for each of the two tissues, the renal cortex and the renal outer medulla (4 strains × 3 time points/strain × 6 rats/group = 72 samples). The quality of each total RNA sample was assessed with Agilent 2100 BioAnalyzer. The RNA Integrity Number was above 0.8 for all samples. Aliquots from every two samples were pooled. The pooled samples, three per group, were used for mRNA profiling experiments. The original individual samples, six per group, were used in real-time PCR experiments.
Affymetrix GeneChip analysis.
mRNA abundance profiles were analyzed with Affymetrix GeneChip rat genome 230 2.0 arrays, the 3′ IVT Expression kit, and the Hybridization, Wash, and Stain kit according to instructions from Affymetrix. A total of 72 arrays were hybridized (4 strains × 3 time points/strain × 3 pooled samples/group × 2 tissues = 72 arrays). Each array contained probes for 28,000 rat genes.
Briefly, 5 μg of total RNA was reverse transcribed with T7-oligo(dT) primers. Double-stranded cDNA was used as the template for 16 h of in vitro transcription to generate amplified RNA (aRNA). Biotinylated ribonucleotides were incorporated into aRNA during in vitro transcription. Purified and fragmented aRNA was hybridized to the array at 45°C for 16 h. Hybridized microarrays were washed and stained with the Affymetrix Fluidics Station-450 and scanned with the Affymetrix Scanner 30007G.
Signal intensities from each microarray were normalized to a median of 500 with the MAS algorithm. Normalized signal intensities were analyzed with Statistical Analysis of Microarray (SAM) and two-way ANOVA to identify genes differentially expressed between SS and each consomic or congenic rat strain over the time course of dietary salt treatment. Criteria for differential expression included false discovery rate (FDR) <10%, strain effect P < 0.01, absolute difference of signal intensity > 184, and “present” calls for at least 50% of the samples involved in the comparison. Pathways that the differentially expressed genes were involved in were identified and analyzed with version 7 of the Ingenuity Pathway Analysis (IPA) package, which contained 134 signaling pathways and 80 metabolic pathways.
Real-time PCR.
Real-time PCR analysis was performed in the original individual samples (n = 6) with the use of SYBR Green chemistry (17) to verify some of the array data. Primers and reagents were from SABiosciences. 18S rRNA was used as the normalizer.
Statistics.
Data were analyzed with analysis of variance (ANOVA) and are reported as means ± SE. P < 0.05 was considered significant.
RESULTS
Introgression of two regions of BN chromosome 13 attenuated salt-induced hypertension and renal injury.
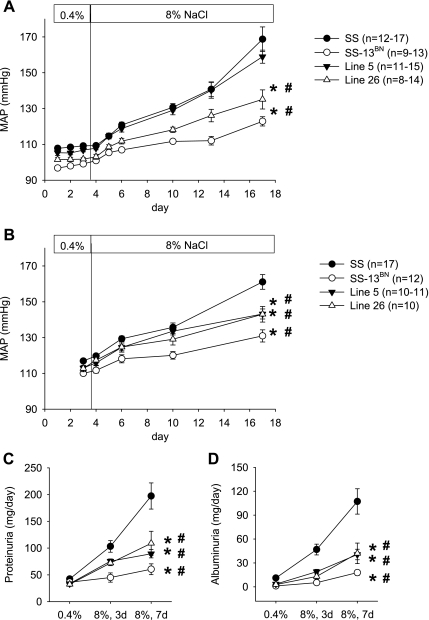
Salt-induced hypertension was significantly attenuated in consomic SS-13BN and congenic Line 26 in male rats 6–9 wk of age (Fig. 1A). After 14 days on the 8% NaCl diet, mean arterial blood pressure (MAP) increased by 60 mmHg in SS rats, 24 mmHg in SS-13BN rats, and 33 mmHg in Line 26 rats. A two-way repeated-measures ANOVA analysis indicated significant strain effects, diet effects, and strain-diet interactions when SS was compared with SS-13BN or Line 26. The difference between SS and SS-13BN or Line 26 was also observed in male rats 9–12 wk of age (Fig. 1B).
Fig. 1.
Substitution of chromosome 13 or 2 segments of chromosome 13 attenuated salt-induced hypertension, proteinuria, and albuminuria in Dahl salt-sensitive (SS) rats. A: mean arterial blood pressure (MAP) of rats 6–9 wk of age. B: MAP of rats 9–12 wk of age. C: proteinuria. D: albuminuria. Line 5 and Line 26, 2 congenic strains of rat as described in materials and methods. *P < 0.05 vs. SS over the time course, #P < 0.05 for strain-diet interaction vs. SS; based on 2-way repeated-measures ANOVA.
MAP did not differ significantly at the age of 6–9 wk between SS and Line 5 rats (Fig. 1A). The development of salt-induced hypertension, however, was significantly attenuated in 9- to 12-wk-old congenic Line 5 rats. MAP was increased by 44 mmHg in SS rats and only 30 mmHg in Line 5 rats after 14 days on the 8% NaCl diet (Fig. 1B).
Proteinuria and albuminuria were significantly ameliorated in SS-13BN, Line 5, and Line 26 rats compared with SS rats at 6–7 wk of age (Fig. 1, C and D).
Gene expression profiles were consistent with phenotypic characteristics.
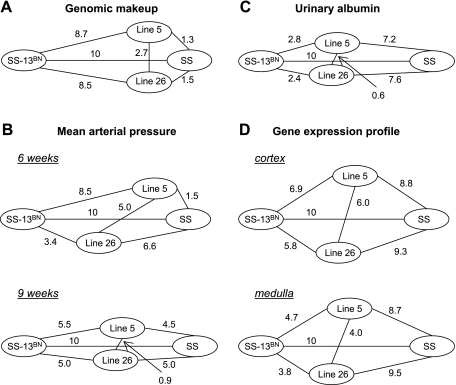
A new approach to examining the correlation between genotype, gene expression, and phenotype was developed, and the result is shown in Fig. 2. Congenic Line 5 and Line 26 rats resembled SS rats more closely than they did SS-13BN rats in terms of genomic makeup (Fig. 2A). The genomic segment that contained sequence differences between SS-13BN, Line 5, Line 26, and SS rats is ∼111, 14, and 16 Mbp, respectively. Line 5 and Line 26 contain sequence differences from SS-13BN in 97 and 95 Mbp of chromosome 13, respectively, while Line 5 and Line 26 differ from each other in segments of 30 Mbp.
Fig. 2.
Magnitude of differences in genomic makeup (A), MAP (B), urinary albumin excretion (C), and total gene expression profiles (D) among SS, SS-13BN, congenic Line 5, and congenic Line 26. See materials and methods and results for the calculation method. The strain differences were scaled so that the distance between SS and SS-13BN was a fixed value of 10.
The euclidean distance between every two strains of rats was calculated for MAP and urinary albumin excretion based on the data shown in Fig. 1. The method of calculation was similar to that described previously (8). Line 26 appeared to resemble SS-13BN more closely than SS in regard to salt-induced changes in MAP (Fig. 2B). Salt-induced hypertension in Line 5 was closer to SS at 6–9 wk of age but moved closer to SS-13BN at 9–12 wk of age (Fig. 2B). In terms of renal injury as reflected by urinary albumin excretion, Line 5 and Line 26 resembled SS-13BN much more closely than they did SS, as shown in Fig. 2C.
Approximately 20,000 genes were detectable in the renal cortex or medulla in the GeneChip analysis of the four rat strains on the 0.4% NaCl diet or after 3 or 7 days on the 8% NaCl diet. The euclidean distance between every two strains of rats was calculated based on the signal intensities of the 20,000 genes. The result showed that mRNA expression profiles in congenic Line 5 and Line 26 resembled SS-13BN more closely than they did SS in both the renal cortex and medulla (Fig. 2D). Expression profiles in Line 26 were more similar to SS-13BN than Line 5 was (Fig. 2D).
Differentially expressed genes and pathway analysis.
According to the criteria described in materials and methods, 1,259, 977, and 1,165 genes in the renal cortex were differentially expressed in SS-13BN, Line 5, and Line 26 rats, respectively, compared with SS rats over the entire time course of dietary salt exposure. In the renal medulla, 3,647, 2,903, and 3,513 genes were identified for SS-13BN, Line 5, and Line 26, respectively. Lists of genes differentially expressed between SS and each consomic or congenic strain are shown in Supplemental Tables S1–S6.1 Differentially expressed genes that are located in the substituted genomic region in each strain are highlighted in yellow.
Real-time PCR analysis of 15 genes and 18S rRNA was performed in 108 samples, involving 3,456 PCR reactions, and largely confirmed the differential expression indicated by the array analysis. The genes to be validated were selected based on their pathway affiliation or genomic location, not on the degree of differential expression. The overall level of concordance between the array analysis and the real-time PCR analysis was 80% (Supplemental Table S7). The P values and differential expression calls shown in Supplemental Table S7 represent the strain effect in a two-way ANOVA analysis of strain and diet effects.
Genes differentially expressed between SS and SS-13BN, Line 5, and Line 26 rats fell, respectively, into 85, 91, and 67 metabolic or signaling pathways in the renal cortex and 93, 72, and 83 in the renal medulla according to IPA. The number of statistically overrepresented pathways ranged from 4 to 16. All pathways are shown in Supplemental Tables S8–S13.
Statistical overrepresentation of a pathway based on IPA suggested a greater likelihood that the function of the pathway was affected. Lack of overrepresentation, however, did not rule out the involvement of a pathway. Changes in rate-limiting steps would change the function of a pathway even if many other genes in the pathway were not differentially expressed. Accordingly, the subsequent analysis described below was performed for overrepresented pathways as well as all pathways.
Several pathways shared by congenic Lines 5 and 26.
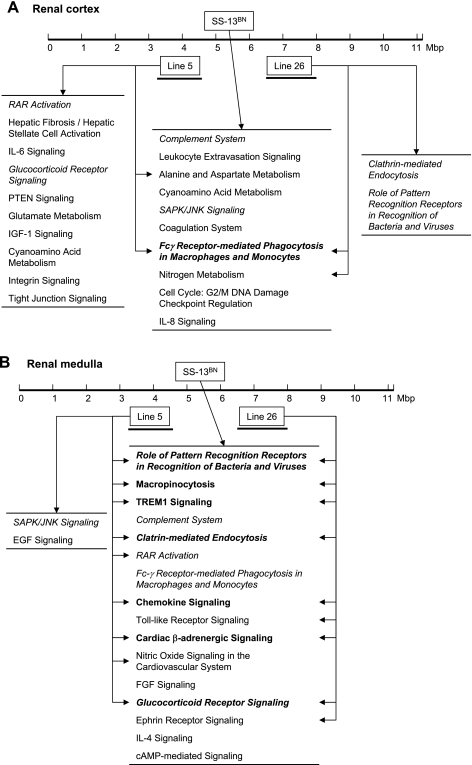
The 14-Mbp and 16-Mbp genomic segments introgressed in congenic Line 5 and Line 26, respectively, did not overlap (16). In other words, the two strains had distinct genomic differences compared with SS. However, the development of hypertension was attenuated in both strains, and, as shown in Fig. 3, gene expression profiles indicated that Line 5 and Line 26 shared alterations in several overrepresented pathways compared with SS. Shared alterations in pathways were particularly abundant in the renal outer medulla (Fig. 3B), where 7 of the 18 overrepresented pathways were found in common. In the renal cortex (Fig. 3A), a single shared pathway related to Fcγ receptor-mediated phagocytosis in macrophages and monocytes was identified.
Fig. 3.
Pathways overrepresented in the genes differentially expressed between SS and SS-13BN, congenic Line 5, and congenic Line 26. Bar at top represents length of chromosome 13 that was substituted in SS-13BN. Segments substituted in congenic Line 5 and Line 26 are indicated below the bar. Overrepresented pathways potentially affected in each consomic or congenic strain are shown. Pathways shared by all 3 strains are shown in bold. Pathways shared by the renal cortex and the renal medulla are shown in italics. Pathways were identified by Ingenuity Pathway Analysis.
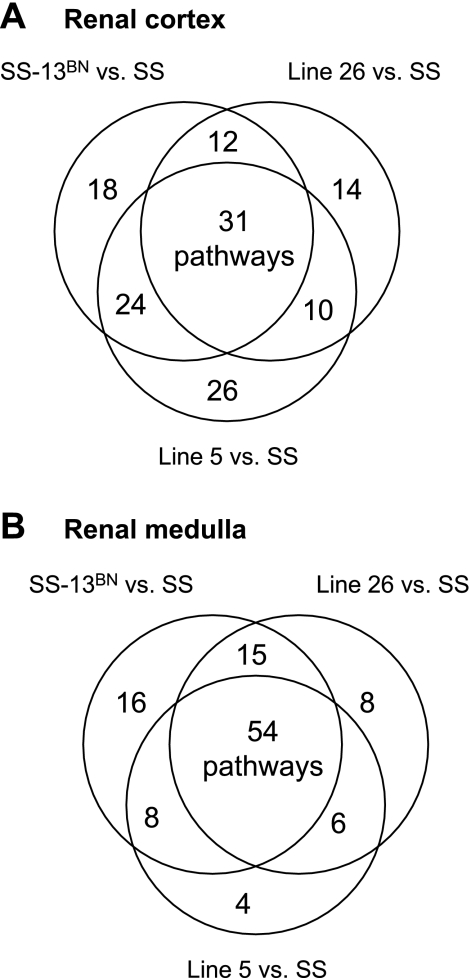
Substantial overlaps also existed when all pathways were examined, including those not overrepresented. Compared with SS, Line 5 and Line 26 shared 41 affected pathways in the renal cortex and 60 pathways in the renal medulla (Fig. 4). Not more than 29 pathways in the cortex and 28 in the medulla would be expected to be shared by Line 5 and Line 26 if the pathways were randomly selected.
Fig. 4.
Counts of all pathways in which 1 or more genes were differentially expressed between SS and SS-13BN, congenic Line 5, and congenic Line 26. Venn diagrams are used to show the number of pathways unique to each strain or shared by 2 or all 3 strains.
Pathways altered in a congenic strain were not just a subset of those altered in the consomic strain.
The genomic segment substituted in Line 5 or Line 26 was a part of chromosome 13 that was introgressed in SS-13BN. However, several pathways that were affected in Line 5 or Line 26 were not affected in SS-13BN. Ten of the 12 overrepresented pathways in the renal cortex in Line 5, and 2 of the 4 in Line 26, were not among the overrepresented pathways in SS-13BN (Fig. 3A). In the renal medulla, 2 of the 11 overrepresented pathways in Line 5 were not among the overrepresented pathways in SS-13BN (Fig. 3B).
When all affected pathways were examined, 40% and 36% of the pathways in the renal cortex of Line 5 and Line 26, respectively, were not among the pathways affected in SS-13BN (Fig. 4A). In the renal medulla, 14% and 17% of the pathways in Line 5 and Line 26, respectively, were outside of the pathways affected in SS-13BN (Fig. 4B).
Congenic Lines 5 and 26 recapitulated a substantial fraction of the pathways altered in the consomic SS-13BN.
Genes in several pathways were consistently affected in SS-13BN, Line 5, and Line 26 compared with SS. They included one overrepresented pathway in the renal cortex and seven in the renal medulla (Fig. 3). When all affected pathways were examined, including those not overrepresented, 31 of a total of 135 affected pathways in the renal cortex and 54 of 111 in the renal medulla were shared by all three strains (Fig. 4). Not more than 11 pathways in the cortex and 12 in the medulla would be expected to be shared by all three strains if the pathways were randomly selected.
Validation of functional changes suggested by gene expression profiles.
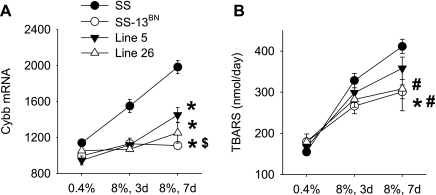
An important assumption in gene expression profiling studies is that mRNA differential expression could often predict changes in metabolic and/or functional pathways. A proof-of-principle study was performed. NAD(P)H oxidase contributes importantly to oxidative stress in SS rats compared with SS-13BN rats (20). Cybb encodes gp91phox, which is a key subunit of NAD(P)H oxidase. The GeneChip analysis indicated that Cybb was expressed at higher levels in the renal cortex of SS rats than in SS-13BN, Line 5, and Line 26 renal cortex. Expression in Line 5 was higher than in SS-13BN (Fig. 5A). A similar pattern of Cybb expression was observed in the renal medulla in the GeneChip analysis. Real-time PCR analysis confirmed the differential expression of Cybb in the renal cortex, but the differential expression in the renal medulla did not reach statistical significance (Supplemental Table S8). We determined urinary excretion of TBARS, an index of lipid peroxidation, in the four strains of rats to examine whether it would be consistent with renal oxidative stress suggested by the gene expression data. As shown in Fig. 5B, urinary excretion of TBARS was significantly reduced in SS-13BN compared with SS rats. A significant strain-diet interaction was also observed when Line 26 was compared with SS. TBARS in Line 5 tended to be reduced compared with SS, but the difference did not reach statistical significance.
Fig. 5.
Urinary thiobarbituric acid-reactive substance (TBARS) in SS, SS-13BN, Line 5, and Line 26. A: Cybb, encoding gp91phox, was expressed at a higher level in the renal cortex of SS rats. Data from the GeneChip analysis are shown; n = 3. *P < 0.05 vs. SS; $P < 0.05 vs. Line 5. B: TBARS excretion in 24-h urine; n = 12 for SS, 10 for SS-13BN and Line 5, and 6 for Line 26. *P < 0.05 vs. SS for strain effect; #P < 0.05 vs. SS for strain-diet interaction.
DISCUSSION
The present study suggests several important principles that might be generally applicable to genetic and mechanistic studies of complex physiological processes and diseases.
It appears that multiple, different genomic regions could affect the development of a complex disease by working through shared pathways. This is in agreement with a treelike paradigm that we previously proposed (11). A complex disease may involve many genes with sequence variations. The genes could impact on various pathways that gradually converge to form shared pathways and impact on the disease. In the present study, Line 5 and Line 26 had two distinct sets of genes introgressed from BN that contained sequence variations compared with SS. Yet, one-third of all pathways altered in the cortex and two-thirds in the medulla were common in Line 5 and Line 26. The shared pathways may represent important, common mechanisms leading to phenotypic changes shared by the two strains including, but probably not limited to, protection from salt-sensitive hypertension and renal injury. The pathways that were unique to each congenic strain are also interesting and may represent a more direct effect of the genes within the substituted region. The effects may include those that will eventually converge to affect shared phenotypes as well as those that affect phenotypes unique to each strain.
Another finding is that the effect of a genomic region on the biology of an organism appears to depend in part on the composition of the rest of the genome. The genomic segments substituted in Line 5 and Line 26 were part of the chromosome substituted in SS-13BN. In other words, SS-13BN contains all the genetic variations that exist in Line 5 and Line 26 compared with SS. Yet pathways altered in Line 5 and Line 26 were not just a subset of the pathways altered in SS-13BN. Each congenic line recapitulated a number of pathways altered in SS-13BN but also exhibited alterations in pathways that were apparently not affected in SS-13BN. This suggests that the effect of the genes located in the Line 5 or Line 26 region depends in part on the interaction between those genes and the genes located on the rest of chromosome 13. The latter was the only genomic region that could be different between SS-13BN and each congenic line.
The interregional interaction that we observed is consistent with the genome background effect that has been demonstrated previously. The effect of specific genes in mice has been shown to vary depending on the strain in which the gene is knocked out or deficient. For example, leptin deficiencies in C57BL/6J ob/ob mice and BALB/cJ ob/ob mice are associated with different disease profiles (18). The importance of genome background is also clearly demonstrated in the consomic rat panels showing that substitution of BN chromosomes into SS versus Fawn-Hooded Hypertensive rats have very different protective effects against hypertension (12, 13). The importance of biological context goes even beyond genome background, as a recent study showed that the effect of sequence variations on gene expression was largely cell type specific (6).
The findings of pathway convergence and interregional interaction suggest that a unidimensional approach is not appropriate for understanding the genetics and mechanisms underlying complex diseases. Multiple genes might affect a disease through common pathways, while each gene could still have its effects through other unique pathways. Moreover, a gene might affect a disease when it is situated within an “appropriate” genome, but might not have an effect when it is located within other genomes. As such, a “specific setting” of hypertension should no longer be defined as broad types of hypertension such as salt-induced versus spontaneous. Instead, the only meaningful way to define a specific setting of hypertension is to specify a detailed genomic makeup and a complete set of environmental factors, in keeping with the principles of individualized medicine. This is critical because, as demonstrated by the present study, the specific biological pathways that are involved in the development of hypertension and perhaps the therapeutic approach that is most appropriate may vary between even slightly different genomic contexts.
The present study also suggested several other points that are more specific to the Dahl model. Line 5 and Line 26 rats exhibited protection from salt-induced hypertension, proteinuria, and oxidative stress that was generally intermediate between SS-13BN and SS. Line 26 appeared to more closely resemble SS-13BN than Line 5 did since the protection from hypertension was not significant in Line 5 rats until 9–12 wk of age. The gene expression profile study was performed in young rats during the early phase of high-salt exposure. The design was intended for identification of gene expression changes that likely contribute to the development of phenotypic differences rather than consequences of large changes in phenotypes. The result indicated that the overall gene expression profiles among the four rat strains were generally consistent with phenotypic differences. The expression profiles in younger Line 5 rats appeared to be predictive of protection from hypertension at an older age. Such “delayed” predictivity could represent an interesting mechanism, but it is entirely speculative at the current stage of our investigation. In addition, modest differences in blood pressure and urinary protein did exist between the rat strains even at the young age, suggesting that some of the gene expression changes we observed could still be secondary to phenotypic differences.
Increased oxidative stress has been reported to be involved in the development of Dahl salt-sensitive hypertension and renal injury in previous studies (4, 15, 20, 23). The present study suggests that correction of these abnormalities might contribute to the protection seen in SS-13BN, Line 5, and Line 26 rats. It remains to be determined what genes within the substituted genomic regions initiate changes in oxidative stress pathways and how these pathways eventually contribute to salt-sensitive hypertension and renal injury. It would also be interesting to examine any interaction between these pathways and other pathways, such as local glucocorticoid metabolism (11) and fumarate metabolism (22, 23), that have been shown to contribute to the protection from salt-induced hypertension in SS-13BN rats.
mRNA profiling approaches provide near-genomewide coverage of genes. However, it is important to recognize that mRNA abundance does not always predict protein abundance or activity. Pathway activations suggested by mRNA abundance profiles should be further examined at protein, metabolite, and/or functional levels, an example of which was provided in the present study. High-throughput analysis of proteomes (21, 22), microRNAs (10, 21), and small metabolites would be valuable next steps, even though current technologies only allow the analysis of limited submetabolomes and subproteomes.
GRANTS
This study was supported by National Heart, Lung, and Blood Institute Grants HL-082798 (A. W. Cowley, Jr.), HL-077263, and HL-085267 (M. Liang).
DISCLOSURES
The authors are not aware of financial conflict(s) with the subject matter or materials discussed in this manuscript with any of the authors, or any of the authors’ academic institutions or employers.
Supplementary Material
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Cowley AW., Jr The genetic dissection of essential hypertension. Nat Rev Genet 7: 829–840, 2006 [DOI] [PubMed] [Google Scholar]
- 2.Cowley AW, Jr, Roman RJ, Kaldunski ML, Dumas P, Dickhout JG, Greene AS, Jacob HJ. Brown Norway chromosome 13 confers protection from high salt to consomic Dahl S rat. Hypertension 37: 456–461, 2001 [DOI] [PubMed] [Google Scholar]
- 3.Cowley AW, Jr, Liang M, Roman RJ, Greene AS, Jacob HJ. Consomic rat model systems for physiological genomics. Acta Physiol Scand 181: 585–592, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Cowley AW., Jr Renal medullary oxidative stress, pressure-natriuresis, and hypertension. Hypertension 52: 777–786, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng AY. Positional cloning of quantitative trait loci for blood pressure: how close are we?: a critical perspective. Hypertension 49: 740–747, 2007 [DOI] [PubMed] [Google Scholar]
- 6.Dimas AS, Deutsch S, Stranger BE, Montgomery SB, Borel C, Attar-Cohen H, Ingle C, Beazley C, Arcelus MG, Sekowska M, Gagnebin M, Nisbett J, Deloukas P, Dermitzakis ET, Antonarakis SE. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 325: 1246–1250, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joe B, Letwin NE, Garrett MR, Dhindaw S, Frank B, Sultana R, Verratti K, Rapp JP, Lee NH. Transcriptional profiling with a blood pressure QTL interval-specific oligonucleotide array. Physiol Genomics 23: 318–326, 2005 [DOI] [PubMed] [Google Scholar]
- 8.Liang M, Yuan B, Rute E, Greene AS, Olivier M, Cowley AW., Jr Insights into Dahl salt-sensitive hypertension revealed by temporal patterns of renal medullary gene expression. Physiol Genomics 12: 229–237, 2003 [DOI] [PubMed] [Google Scholar]
- 9.Liang M, Lee NH, Wang H, Greene AS, Kwitek AE, Kaldunski ML, Luu TV, Frank BC, Bugenhagen S, Jacob HJ, Cowley AW., Jr Molecular networks in Dahl salt-sensitive hypertension based on transcriptome analysis of a panel of consomic rats. Physiol Genomics 34: 54–64, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Liang M, Liu Y, Mladinov D, Cowley AW, Jr, Trivedi H, Fang Y, Xu X, Ding X, Tian Z. MicroRNA: a new frontier in kidney and blood pressure research. Am J Physiol Renal Physiol 297: F553–F558, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, Singh RJ, Usa K, Netzel BC, Liang M. Renal medullary 11β-hydroxysteroid dehydrogenase type 1 in Dahl salt-sensitive hypertension. Physiol Genomics 36: 52–58, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, Cowley AW, Jr, Jacob HJ. Chromosomal mapping of the genetic basis of hypertension and renal disease in FHH rats. Am J Physiol Renal Physiol 293: F1905–F1914, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Mattson DL, Dwinell MR, Greene AS, Kwitek AE, Roman RJ, Jacob HJ, Cowley AW., Jr Chromosome substitution reveals the genetic basis of Dahl salt-sensitive hypertension and renal disease. Am J Physiol Renal Physiol 295: F837–F842, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride MW, Charchar FJ, Graham D, Miller WH, Strahorn P, Carr FJ, Dominiczak AF. Functional genomics in rodent models of hypertension. J Physiol 554: 56–63, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng S, Cason GW, Gannon AW, Racusen LC, Manning RD., Jr Oxidative stress in Dahl salt-sensitive hypertension. Hypertension 41: 1346–1352, 2003 [DOI] [PubMed] [Google Scholar]
- 16.Moreno C, Kaldunski ML, Wang T, Roman RJ, Greene AS, Lazar J, Jacob HJ, Cowley AW., Jr Multiple blood pressure loci on rat chromosome 13 attenuate development of hypertension in the Dahl S hypertensive rat. Physiol Genomics 31: 228–235, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Morrison J, Knoll K, Hessner MJ, Liang M. Effect of high glucose on gene expression in mesangial cells: upregulation of the thiol pathway is an adaptational response. Physiol Genomics 17: 271–282, 2004 [DOI] [PubMed] [Google Scholar]
- 18.Qiu J, Ogus S, Mounzih K, Ewart-Toland A, Chehab FF. Leptin-deficient mice backcrossed to the BALB/cJ genetic background have reduced adiposity, enhanced fertility, normal body temperature, and severe diabetes. Endocrinology 142: 3421–3425, 2001 [DOI] [PubMed] [Google Scholar]
- 19.Rapp JP. Dahl salt-susceptible and salt-resistant rats. Hypertension 4: 753–763, 1982 [DOI] [PubMed] [Google Scholar]
- 20.Taylor NE, Glocka P, Liang M, Cowley AW., Jr NADPH oxidase in the renal medulla causes oxidative stress and contributes to salt-sensitive hypertension in Dahl S rats. Hypertension 47: 692–698, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Tian Z, Greene AS, Pietrusz JL, Matus IR, Liang M. MicroRNA-target pairs in the rat kidney identified by microRNA microarray, proteomic, and bioinformatic analysis. Genome Res 18: 404–411, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian Z, Greene AS, Usa K, Matus IR, Bauwens J, Pietrusz JL, Cowley AW, Jr, Liang M. Renal regional proteomes in young Dahl salt-sensitive rats. Hypertension 51: 899–904, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Tian Z, Liu Y, Usa K, Mladinov D, Fang Y, Ding X, Greene AS, Cowley AW, Jr, Liang M. Novel role of fumarate metabolism in Dahl salt-sensitive hypertension. Hypertension 54: 255–260, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobian L. Salt and hypertension. Lessons from animal models that relate to human hypertension. Hypertension 17: I52–I58, 1991 [DOI] [PubMed] [Google Scholar]
- 25.Williams JM, Sarkis A, Hoagland KM, Fredrich K, Ryan RP, Moreno C, Lopez B, Lazar J, Fenoy FJ, Sharma M, Garrett MR, Jacob HJ, Roman RJ. Transfer of the CYP4A region of chromosome 5 from Lewis to Dahl S rats attenuates renal injury. Am J Physiol Renal Physiol 295: F1764–F1777, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.