Abstract
The increasing use of high-frequency (kHz), long-duration (days) intracranial monitoring from multiple electrodes during pre-surgical evaluation for epilepsy produces large amounts of data that are challenging to store and maintain. Descriptive metadata and clinical annotations of these large data sets also pose challenges to simple, often manual, methods of data analysis. The problems of reliable communication of metadata and annotations between programs, the maintenance of the meanings within that information over long time periods, and the flexibility to re-sort data for analysis place differing demands on data structures and algorithms. Solutions to these individual problem domains (communication, storage and analysis) can be configured to provide easy translation and clarity across the domains. The Multi-scale Annotation Format (MAF) provides an integrated metadata and annotation environment that maximizes code reuse, minimizes error probability and encourages future changes by reducing the tendency to over-fit information technology solutions to current problems. An example of a graphical utility for generating and evaluating metadata and annotations for “big data” files is presented.
Introduction
Multi-scale electrophysiology requires collection and analysis of data over a wide range of spatial and temporal scales, satisfying the criteria of “big data” [1]. Multi-scale data creates a range of difficult technical issues including efficient data formats and storage solutions. Several of these needs specific to the acquisition, storage, and analysis of data for systems electrophysiology spurred the creation of a new file format, Multi-scale Electrophysiology Format (MEF) [2]. In addition to the raw data, another set of problems arises concerning the handling of the data that describe or are attached to the raw data; i.e., metadata and annotations. While some aspects of metadata and annotations from multi-scale recordings are similar to those for all types of data acquisition (e.g., subject identification, dates, filenames), several other aspects are unique to multi-scale acquisition from humans, such as patient privacy issues and scalability. The difficulties surrounding human multi-scale data acquisition (e.g., cost, ethics, supremacy of patient needs over scientific questions) emphasize another issue regarding the handling of scientific data: analyses commonly involve data that were collected over multiple years on an array of recording systems by different people asking a range of questions who are often located at different institutions. This creates a range of technical problems, including the problem of communicating the conditions under which data were acquired, variations in data acquisition parameters (e.g., sampling frequency) and the sheer numbers of observations inherent in such large data files. These challenges can be grouped into three problem domains: communication between users and applications, storage across time and institutions, and data analysis. Software solutions within each domain have been optimized to solve the problems that dominate that particular domain, but a new problem arises when research questions bridge multiple domains. Smaller, more focused data sets often avoid dealing with these problems by a range of simpler solutions that may not scale to multi-terabyte data files. (Figure 1)
Figure 1.
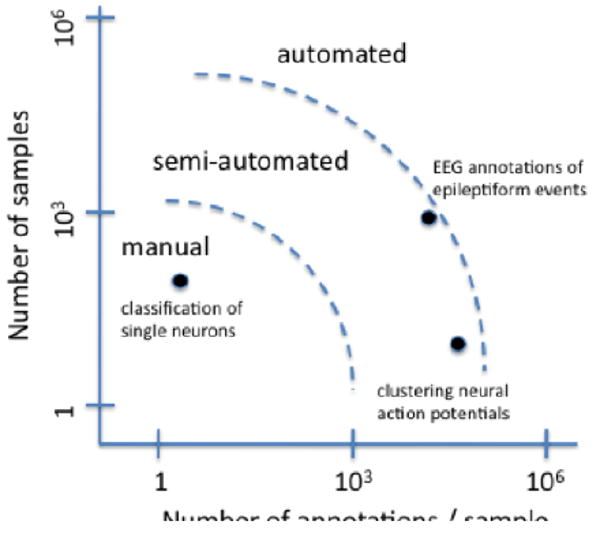
Increasing amounts of annotations and metadata require increasingly automated solutions to data management. In addition, the timeframe in which the annotations must be generated or retrieved also affects the degree of automation required.
For example, consider computing the spectral power within a fixed bandwidth during each second for data recorded from 50 patients, where data from 300 channels/patient were obtained for several days. Furthermore, imagine that some patients were recorded at one sampling frequency, others at a second frequency and others at a third. Finally, imagine that the results are to be sorted based on the anatomical location of each electrode. If the scope of the project involved fewer channels, patients or shorter recordings, many of the difficulties in this scenario could be handled manually (e.g., a lab notebook or a spreadsheet). Several solutions to different parts of this problem would seem reasonable: each file could be filtered for its specific sampling frequency, each file could be filtered and down-sampled to the lowest sampling frequency, the files or the results could be moved to folders based on recording location anatomy, or file-specific information could be stored in and extracted from the header of each file. This particular analysis, however, involves 15,000 files, many of which could have sizes exceeding one terabyte. In any given file, a user might want to note hundreds or even thousands of events, such as the presence of EEG spikes or artifacts, changes in behavioral state, where the terms used might not be standardized across patients or hospitals.
Solutions to such problems could be based on somewhat divergent ways of looking at data in general (Figure 2). Storing information in an efficiently searchable format that also reduces the incidence of errors also has a long history that has centered on the concept of databases (e.g., GenBank) [3], virtually all of which use a handful of tested tools with a long track record. The SQL programming language lies at the heart of virtually all database software whether the vendor is Oracle, Microsoft or an open-source provider. Open Database Connectivity (ODBC) provides a widely accepted set of programming standards that provide easy access to database data across a range of programming languages. While storing metadata and annotations in a database provides users with increased access to their data in terms of searching and sorting and reduce probability of error or data corruption, these methods are of less use for data analysis or communication.
Figure 2.
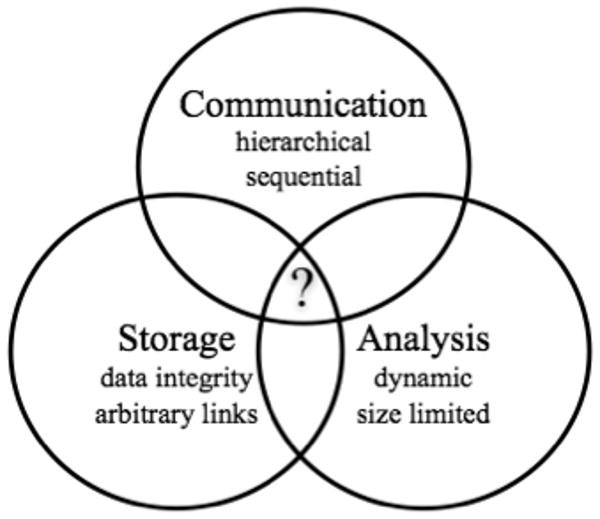
Problem domains involved in handling multi-scale electrophysiological data. An integrated solution would take advantage of the strengths of existing solutions that would allow easy translations between the domains.
The situation regarding the preferred programming language for data analysis is even less clear. Several programming languages have been used in systems electrophysiology (e.g., C, C++, R, Java, Fortran, Matlab, Python) in conjunction with range of operating systems (e.g., Windows, Mac OS X, Linux). The advent of web-based programming languages has added even more options. Several programming concepts, however, have attracted a great deal of attention in the software community, including object-oriented programming, software “design patterns”, and agile programming [4-6], but few have translated directly to improvements in either the storage or communication of scientific data.
Arranging information in a context-free manner for communication between computers or between human readers has often focused on methods to serialize data; that is, methods by which the meaning within data is maintained even when the data must be broken into a series of values for transmission along a communication channel (e.g., TCP/IP, XML, PDF). While optimal for maintaining the meaning within communications, these formats are rarely used in either databases or data analysis software. Providing XML storage of information allows users to store intermediate data analysis results when a connection to the database does not exist or when users do not want to persist intermediate results into the database.
In each problem domain, general solutions to a vast range of problems have been optimized through decades of research and products. Many of these solutions are freely available in the form of open-source software, which could provide the added benefit of freeing data from being tied to the particular software vendor that supplied the recording equipment; i.e., “vendor lock-in”. In this paper, we describe ongoing work to develop MAF, the Multi-scale Annotation Format, which attempts to find a set of software solutions within each problem domain that can interact with one another to provide an integrated efficient, fast, error-minimizing, reusable, open-source and scalable suite of software tools tailored to handling “big data” metadata and annotations.
Methods and Results
The difficulties involved in bridging technological domains can be seen in the diversity of names that each applies to a standardized description of contents (e.g., “schema/metadata” for databases, “DTD/schema” for XML, “API” for programs). MAF is described as a “format” to reduce the confusion with similar descriptions applying to any one of the three domains. MAF consists of three sub-formats that must be consistent with one another. This encourages use of a proven paradigm of “programming to an interface”, shifting the focus of software development from defining what elements a solution must have to describing what a solution must do. An interface can be thought of as a contract between programming language creators and users regarding the names of functions within a software package and the number and types of variables that those functions expect. One analogy is the three-pronged electrical outlet: the power company (analogous to software writers) need only worry about providing electrical power to electrical outlets, without considering how consumer electronics (that is, “users”) will use the electricity, and the users don't have to worry about how the power is generated. The producer and the user need only agree on the physical dimensions of the plug and the properties of the electrical current. This agreement becomes the “interface” between the producer and the consumer, freeing each to handle their respective tasks independently of the other. The goal of MAF can be stated more concisely as an attempt to find interfaces that provide solutions within problem domains that can interact flexibly and easily with the interfaces chosen in the other problem domains. “Programming to an interface” decouples the problem domains, allowing developers to improve a solution in one domain without affecting the existing solutions in the others. The language chosen for the development of MAF was Java written in the Eclipse IDE (Integrated Development Environment), because Java objects can be loaded easily into several different interactive environments, including Matlab [7], Jython [8] and Groovy [9].
Several factors influenced the choice of the communication format. The eXtensible Markup Language (XML) emphasizes clarity of information transmission and flexibility of content [10]. In the context of neuroimaging studies, the XML-based Clinical Data Exchange (XCEDE) schema was developed by the Biomedical Informatics Research Network (BIRN [11]) to document research and clinical studies. The schema easily represents “many-to-many” relationships (e.g., between data files and analyses, where one analysis can use multiple files, and one data file can be used in multiple analyses), which can pose a problem for hierarchical schema. XCEDE proposes assigning reference IDs to each element that can subsequently be used flexibly. This use of reference IDs to relate data elements of different types is a central organizing principal underlying MAF. The XML format used in MAF extends this theme into objects more directly related to experimental data collection (e.g., subjects, timestamps), leading to the XREDE (XML-based Research Data Exchange) schema (Figure 3). This approach integrated easily with the other problem domains, particularly with that of data storage, because it allows for representations of arbitrary relations between data elements. XML tags extend naturally to object-oriented classes representing subjects, recording episodes, analysis tasks, EEG events, etc. (Figure 4). In particular, we chose the JDOM package of XML parsers, an integrated set of objects that can be loaded easily into any Java-capable environment [12].
Figure 3.
A portion of the XREDE XML schema written in RELAX NG Compact format. The use of XML validation provides another check on data integrity, while documenting the relationships that must hold in both the data analysis classes and in the database.
Figure 4.
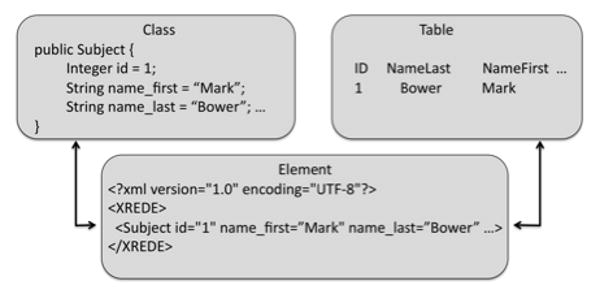
The Class-Element-Table integration for annotations. While it is possible to translate directly between objects and database storage, the use of XML validation in an intermediate state provides a check on data integrity, while documenting the relationships that must hold in both the data analysis classes and in the database.
Data storage technologies include a broad range of options: files within hierarchical folders, XML, spreadsheets and databases each offer advantages. Storing data within a system of hierarchical folders distributes information across the folder tree; the source of data in a file is a function of the chain of parent folders. This tends, however, to enforce, fixed relationships (e.g., patient/session/channel), does not prevent errors (e.g., if a file is misplaced or dropped) and is difficult to search. Spreadsheets, too, bias storage towards a fixed relationship, holding all information in a single table, and attach meaning to location within a table. As with hierarchical files, it is difficult to catch errors due to misplaced or dropped data. Databases were designed specifically to address these issues. Relational databases, in particular, minimize errors by enforcing data integrity rules, requiring all related information to be entered in a state that is consistent with the MAF database schema, preventing the deletion of data that are required by an existing element. Relational databases link related information via unique identification numbers, allowing arbitrary relations to be extracted after data have been entered and preventing accidental deletion of necessary data. Another advantage of database storage is that information related to each entry is stored in only one location; if an error is identified in an entry it only needs to be corrected in one place. MySQL, a popular relational database, was chosen as the initial database underlying MAF, because it is freely available, is easy to install and use, and publishes drivers for most ODBC standards [13]. Combined, MAF integrates an object-oriented class, an XML element and an MySQL table for each data component, along with translation methods for converting between the three (Figure 4).
The first project to be implemented using these guidelines was an interactive, Matlab-based data viewer for adding and visualizing neurological annotations to patient EEG, which is called “eeg_view” (Figure 5). Annotations are label-time stamp pairs associated with neurological events identified visually by an expert or automatically by a program that are generally associated with a specific analysis task. Annotations may be inspected visually, allowing users to accept or reject existing annotations and add events that were missed. The XREDE format can note the annotations that were rejected by the user, as well as which annotations were added manually, so that false positive detections can be counted. The user can load or generate events for multiple EEG files. Annotations are saved to an XML file corresponding to the MAF format. The XML file can be transferred directly to another program or stored to the database. Associating events with tasks in the database allows users to select different sets of events based on the method by which they were generated, combine events generated by different sources, or keep only those events that have been designated as the “gold standard”. The resulting database subsets are returned in XREDE format, which can then be opened by the MAF-capable EEG viewer.
Figure 5.
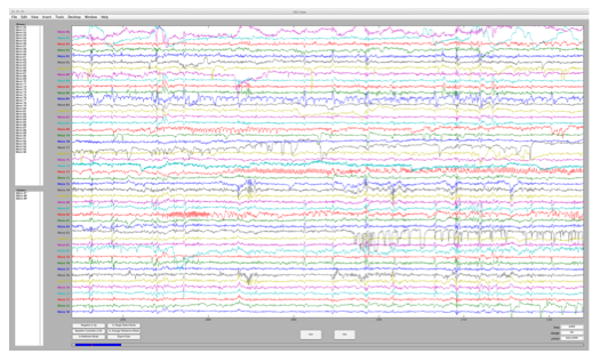
Screen capture from the multi-scale EEG viewer. The viewer provides an interactive display of continuously-recorded macro- and microelectrode files and user-defined annotations. Annotations are stored in a database via an XML file.
Discussion
It should be stressed that the technologies selected and combined in this paper represent only one possible solution to the problem of managing metadata and annotations in big data. Many more solutions are possible with current technology, and future developments promise to expand these opportunities. For example, the current choice of technologies described in this paper underutilizes the power of browser-based technologies, e.g., Google Web Toolkit, which allows existing Java code to run within a browser without modifications [14]. Recent developments in database technology offer several promising technologies: object-relational map databases utilize an intermediate description of classes to automate the storage and retrieval of data within objects into a relational database (e.g., Hibernate [15] and DataNucleus [16]); object-oriented databases store the objects themselves into a modified relational database (e.g., db4o [17]); distributed databases allow storage of very large data objects (e.g., Project Voldemort [18]), which could be particularly useful for multi-scale electrophysiology. The suite of technologies chosen should offer real-world assistance to challenges in ways that bridge the problem domains, while providing for their own future replacement; technology lock-in can be just as insidious a problem as vendor lock-in. Solutions that rely on a feature specific to one particular technology should be avoided.
Conclusion
Multi-scale electrophysiology poses challenges both in terms of the amount of data and the complexity of analyses. Attempting to solve these challenges by focusing either on a single problem or one particular technology often creates new problems in transferring solutions from one task to the next. General-purpose solutions exist to the problems of communication, storage and analysis of metadata and annotations that can alleviate the problems of complexity and lack of portability, but their use requires integration of software tools tailored to both clinical and research needs. The Multi-scale Annotation Format (MAF) integrates proven solutions within each of the problem domains of communication, storage and analysis in the forms of XML, relational databases and object-oriented programming by promoting a class-element-table approach to data representation. These proven technologies not only offer solutions to current problems, but also modularize the problem domains, providing the opportunity for future improvements without impacting existing experimental information. The programming API, XREDE schema and MySQL database description can be found at the website for the Mayo Systems Electrophysiology Lab (MSEL) and are freely available under GNU open-source licensing, along with documentation describing the integrated use of these tools [19].
Acknowledgments
This work was supported by the National Institutes of Health (Grant K23 NS47495), State of Minnesota Partnership Grant and Epilepsy Therapy Development Project grant from the Epilepsy Foundation of America.
References
- 1.Howe D, Costanzo M, Fey P, Gojobori T, Hannick L, Hide W, Hill DP, Kania R, Schaeffer M, St Pierre S, Twigger S, White O, Yon Rhee S. Big data: The future of biocuration. Nature. 2008 Sep 4;455(7209):47–50. doi: 10.1038/455047a. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkmann BH, Bower MR, Stengel KA, Worrell GA, Stead M. Large-scale Electrophysiology: Acquisition, Compression, Encryption, and Storage of Big Data. J Neurosci Methods. 2009 May 30;180(1):185–92. doi: 10.1016/j.jneumeth.2009.03.022. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson DA, Boguski MS, Lipman DJ, Ostell J, Ouellette BF, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Research. 1999;27(1):12–17. doi: 10.1093/nar/27.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckel B. Thinking in Java. Prentice Hall; 2006. [Google Scholar]
- 5.Gamma E, Helm R, Johnson R, Vlissides JM. Design Patterns: Elements of Reusable Object-Oriented Software. Addison-Wesley; 1994. [Google Scholar]
- 6.Kuchana P. Software Architecture Design Patterns in Java. Auerbach Publications; 2004. [Google Scholar]
- 7.Matlab is a registered trademark of Mathworks, Inc., Natick, MA. http://www.mathworks.com.
- 8.http://www.jython.org
- 9.http://groovy.codehaus.org/
- 10.http://www.w3.org/XML
- 11.http://www.nbirn.net
- 12.http://www.jdom.org/
- 13.http://www.mysql.com/
- 14.http://code.google.com/webtoolkit/
- 15.http://www.hibernate.org
- 16.http://www.datanucleus.org
- 17.http://www.db4o.org
- 18.http://www.project-voldemort.org
- 19.http://mayoresearch.mayo.edu/mayo/research/msel/



