Abstract
The design of combinatorial libraries for molecular recognition requires extensive diversity to provide high affinity binding to myriad epitopes while maintaining a high degree of functionality to enable inclusion of binders in the limited screenable library size. In the current work, we directly compare minimal and maximal amino acid diversity libraries in the context of the 10th type III domain of human fibronectin. Libraries with either serine/tyrosine or full 20 amino acid diversity were created, pooled and screened for binding to rabbit and goat immunoglobulin G (IgG), and affinity matured by directed evolution. Multiple picomolar binders to rabbit IgG and nanomolar binders to goat IgG were engineered with peak affinities of 51 ± 4 pM and 1.2 ± 0.4 nM, respectively. Sequence analysis reveals that 93% of the selected BC and FG loops, including those from the highest affinity clones, originate from the full diversity library. Thus, with a modest initial library size (∼1 × 108) and an efficient affinity maturation scheme, more extensive diversity is superior to a binary serine/tyrosine code for the generation of picomolar to low nanomolar binders in the fibronectin domain. The highest affinity binders demonstrated utility in affinity purification of IgG from serum and as detection reagents in flow cytometry.
Keywords: fibronectin type III domain, Fn3, immunoglobulin G, protein engineering, synthetic library
Introduction
The 10th type III domain of human fibronectin (Fn3) is a validated scaffold for molecular recognition (Koide et al., 1998; Koide and Koide, 2007). The 10 kDa, cysteine-free beta-sandwich fold contains three solvent exposed loops, termed BC, DE and FG for the beta strands they connect. Binders have been engineered using a variety of combinatorial libraries including diversification of the amino acid sequence in one (Koide et al., 2002; Richards et al., 2003; Lipovsek et al., 2007), two (Koide et al., 1998; Karatan et al., 2004; Lipovsek et al., 2007; Olson et al., 2008) and three loops (Xu et al., 2002; Parker et al., 2005; Koide et al., 2007; Gilbreth et al., 2008; Hackel et al., 2008). Ultra high affinity binding is enabled with three-loop diversification, but the resultant expansion of theoretical sequence space necessitates efficient library design. Early molecular recognition library designs incorporated all 20 natural amino acids. However, it has been demonstrated that tyrosine can dominate molecular recognition in antibody domains (Fellouse et al., 2004, 2006) and that a serine/tyrosine binary code is sufficient to generate nanomolar affinity interactions in an antigen-binding fragment (Fellouse et al., 2005). This library approach has been successfully applied to generate low- to mid-nanomolar binders with the Fn3 scaffold (Koide et al., 2007). Yet while the binary amino acid code greatly reduces theoretical sequence space enabling isolation of functional clones, it also reduces the potential structural and chemical complementarity of the binder–target interaction. As such, it is not clear which library design is superior for the generation of high affinity binders. A hybrid design with all 20 amino acids yet biased toward tyrosine, serine and glycine was investigated in an antibody library (Fellouse et al., 2007); this library yielded more high affinity binders than a strictly serine/tyrosine library. This comparison was performed with an antibody domain binding to a single target. The current study directly compares full diversity with serine/tyrosine diversity in the context of the Fn3 domain in two binder engineering campaigns.
In parallel with the study of library design, we sought to develop useful reagents with advantageous biophysical properties. High-yield bacterial expression enables inexpensive production of Fn3 domains. The absence of lysines near the engineered binding surface and the cysteine-free sequence permits both amine- and thiol-based conjugation chemistries for immobilization or fluorophore coupling. The small, single-domain architecture facilitates multifunctional protein fusions. Engineered Fn3 domains have demonstrated utility as detection agents. Biotinylated αvβ3 integrin binders were used as a primary label in flow cytometry (Richards et al., 2003). Alkaline phosphatase fusions of Src-binding Fn3 domains were effective in western blotting (Karatan et al., 2004). The Src binders were also effective in pull-down experiments; estrogen receptor binders were effective in affinity chromatography (Huang et al., 2006). Thus, Fn3 domains that bind immunoglobulin G (IgG) may be broadly useful in these applications as well as in protein microarrays and as adaptors for nanoparticles or other supramolecular assemblies. In this study, high affinity binders to both goat and rabbit IgG were engineered. The resultant binders are characterized in terms of affinity, stability and specificity and are demonstrated as useful reagents for purification and detection.
Results
Library design and construction
Two libraries, NNB and YS, were constructed for comparison. In both libraries, the BC, DE and FG loop sequences of Fn3 were diversified. The NNB library has been previously described (Hackel et al., 2008). Eight, five and 10 residues in the BC, DE and FG loops were randomized using NNB codons; four loop lengths, selected based on phylogenetic occurrence, were included in each loop. The YS library diversified 9, 5 and 10 residues in the BC, DE and FG loops. The BC and FG loops were randomized between serine and tyrosine. BC loop diversity was extended to include Y31 because it is a large side chain with potential steric conflicts and inclusion of serine only increases the theoretical diversity 2-fold. The DE loop was diversified with a wild-type bias to improve structural integrity while enabling some diversity to either eliminate detrimental interactions or provide beneficial interactions. Biased nucleotides were synthesized to yield a library with ∼50% wild-type and 50% of the other 19 amino acids at G52, S53, S55 and T56. K54 was randomized using an NNB codon because the large, charged side chain is potentially sterically and electrostatically detrimental. Four loop lengths were allowed in each loop. Library design is summarized in Table I.
Table I.
Library designs
| Library | Design |
Diversity |
||||
|---|---|---|---|---|---|---|
| BC | DE | FG | BC | DE | FG | |
| WT | DAPAVTVRY | GSKST | GRGDSPASSK | 1 | 1 | 1 |
| NNB | X6–9Y | X4–7 | X5,6,8,10 | 5 × 1011 | 1 × 109 | 1 × 1013 |
| YS | (S|Y)7–10 | gsX0,1,3st | (S|Y)6,7,8,10 | 2 × 103 | 1 × 109 | 1 × 103 |
WT, wild-type sequence; X, any amino acid; S|Y, either serine or tyrosine; g, 50% glycine, 50% any other amino acid (analogous for s, t); subscripts refer to the number of amino acids.
The libraries of Fn3 genes were transformed into a yeast surface display library by homologous recombination with a vector including an N-terminal Aga2p protein for yeast surface tethering to Aga1p and a C-terminal c-myc epitope for full-length Fn3 detection. Electroporation yielded 21.5 × 107 and 25 × 107 transformants from the NNB and YS libraries, respectively. Sequencing and flow cytometry analysis (data not shown) indicate 34% and 60% of clones encode for full-length protein resulting in 7.3 × 107 and 15 × 107 Fn3 in each library.
Binder engineering
The efficacies of the NNB and YS libraries were compared for their ability to generate binders to goat IgG and rabbit IgG. The libraries were pooled to enable direct clone competition and eliminate any experimental bias. The libraries were sorted twice for binding to biotinylated IgG immobilized on streptavidin-coated magnetic beads followed by a fluorescence-activated cell sort (FACS) for c-myc+ clones that represent full-length Fn3. The selected population was diversified by both full gene mutagenesis and focused loop mutagenesis with shuffling as described (Hackel et al., 2008). The transformed mutants, mixed with the original clones, underwent two bead selections and an FACS followed by mutagenesis. Two bead selections were followed by an FACS for both c-myc and binding to biotinylated IgG, detected by streptavidin–fluorophore. Two additional rounds of mutagenesis and FACS with decreasing IgG concentrations were performed. At this point, the population sorted for binding to goat IgG exhibited binding to 500 pM goat IgG; the population sorted for binding to rabbit IgG exhibited binding to 50 pM rabbit IgG (data not shown).
Each intermediate population used for mutagenesis was also sorted three or four additional times without mutagenesis to identify the best clones during each round of affinity maturation. The naïve library was excluded from this analysis because the populations did not exhibit substantial binding until after mutagenesis. Several clones from each final population and two intermediate populations were sequenced (Table II).
Table II.
Binder sequences
| Goat IgG binders |
Rabbit IgG binders |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone | # | BC loop | DE loop | FG loop | Framework | Clone | # | BC loop | DE loop | FG loop | Framework |
| WT | — | DAPAVTVR | GSKST | GRGDSPASSK | — | WT | — | DAPAVTVR | GSKST | GRGDSPASSK | — |
| Round 1 | |||||||||||
| gI1.5.1 | 1 | ALPRSE | GIRS | AHKSVL | S2P, T58I | rI1.5.1 | 1 | VRPSYSRL | STATT | GYGGRRVQ | P51A, G61R, Y73C |
| gI1.5.2 | 1 | HSYYSYY | GFYST | MDGASPLQ | D7G | rI1.5.2 | 1 | ATTGKAPL | KGATA | SYYDYHS | Q46K, S60R |
| gI1.5.3 | 2 | KMRAAR | RFRS | GDGHGG | T58I | rI1.5.3 | 4 | VATSCL | ATWVK | HYDDTLS | — |
| gI1.5.4 | 1 | NLEIFPR | GIRS | RTRVI | P51S, T58I | rI1.5.4 | 2 | ATTGKTPL | RSAEM | HYDDTLS | — |
| gI1.5.5 | 1 | NLGIFPR | GIRS | RTRVI | T58I | rI1.5.8 | 1 | VATSCL | ATWVK | HYDDTLS | N42G |
| rI1.5.9 | 2 | ARASNPCL | ATWVK | GYGGKRVQ | Q46R, S60R, K98E | ||||||
| Round 2 | |||||||||||
| gI2.5.1 | 1 | TNLSSS | NWTS | SYGLVISN | T58I, V68A | rI2.5.1 | 1 | VRSPYRRL | RSARS | GYGGRRVQ | — |
| gI2.5.2 | 1 | ALPRSE | NWTS | SPGLVLGA | T58I | rI2.5.2 | 2 | VNGDSCL | ATWVK | GYGGKRVQ | S60R |
| gI2.5.3 | 2 | TRAYFAP | GSLSS | SYGLVITD | P51S, T58I, I88T | rI2.5.3 | 1 | VRPSYSRL | PTHFF | GYGGKRVQ | S60R |
| gI2.5.4 | 2 | YSSYSYY | GFRPT | YYSSSYY | T35A, E38G, S89P | rI2.5.4 | 1 | ARPSYSRL | ATWVK | GYGGKRVQ | — |
| gI2.5.5 | 1 | RMPVTD | Truncation | — | rI2.5.5 | 2 | VRPSYSRL | ATWVK | GYGGKRVQ | — | |
| gI2.5.8 | 1 | RLPRSA | NWTS | SPGLILGA | T58I, I90T | rI2.5.6 | 1 | VRPSYSRL | KGATV | GYGGKRVQ | P51S |
| gI2.5.9 | 1 | YCSYSYY | GFRSG | FDGVAF | — | rI2.5.9 | 1 | VRPSYSRL | ATWVK | GYGGERVQ | — |
| rI2.5.10 | 1 | VATSCL | RSATS | GYGGKRVQ | P51S | ||||||
| Round 3 | |||||||||||
| gI3.2.1 | 2 | TARMRSP | NWTS | SPGLILGA | T58I | rI3.6.2 | 1 | VRPSYSRL | RSWTS | GYGGKRVP | S60R |
| gI3.2.2 | 2 | ALPRSE | NWTS | SPGLVLGA | T58I | rI3.6.3 | 2 | SRARNACL | ATWVK | GYGGKRVQ | G61R |
| rI3.6.4 | 2 | VRPSYSRL | RSARS | GYGGERVQ | P51S, S60R | ||||||
| gI3.5.1 | 5 | TRAYFAP | GSLSS | SYGLVITD | P51S, T58I, I88T | rI3.6.5 | 1 | AHAPNPCL | ATWVK | GYGGKRVQ | S60R, I90T |
| rI3.6.6 | 2 | ATTGKAPL | KGATA | SYYDYHS | Q46K, S60R | ||||||
| rI3.6.10 | 1 | ARPSYSRL | GSAHV | GYGGKRVQ | P51S, S60R, G61R | ||||||
| Round 4 | |||||||||||
| gI4.5.1 | 5 | TRAYFAP | GSLSS | SYGLVITD | P51S, T58I, I88T | rI4.3.1 | 2 | ATTGKTPL | RSAEM | HYDDTLS | — |
| rI4.3.3 | 1 | ARASNPCL | ATWVK | GYGGKRVQ | S60R | ||||||
| rI4.3.4 | 1 | VNGDSCL | GSAHV | GYGGKRVQ | P51S, S60R, G61R, A74T | ||||||
| rI4.3.5 | 1 | AHAPNPCL | AAWVE | GYGGKRVQ | S60R | ||||||
| rI4.5.1 | 4 | ATTGKAPL | ATWVK | HYDDTLS | S60R, K98E | ||||||
| rI4.5.5 | 1 | ATTGKAPL | ATWVK | HYDDTLS | S60R | ||||||
Clones are named as (g/r)Ix.y.z where g or r indicates goat or rabbit species specificity, x is the number of mutagenesis steps, y the number of selections after the most recent mutagenesis and z the clone number in that population. # indicates the frequency of occurrence of the indicated clone.
Both the goat and rabbit IgG binder engineering yielded one dominant clone after four rounds of mutagenesis, although extensive sequence diversity appears in intermediate populations.
Sequence analysis reveals a strong preference for the NNB library. Since loop shuffling during mutagenesis can recombine loops from multiple sources, loop sequences were analyzed individually. Thirty-nine of 42 clones (93%) have BC loops of NNB origin. Ninety-three percent have FG loops of NNB origin. The DE loop, which was fully randomized in the NNB library and biased toward wild-type in the YS library, exhibited less preference; 65% of clones had DE loops more likely to originate from NNB whereas 23% were more likely to originate from the wild-type bias of YS and 13% were ambivalent. In fact, only a single clone, gI2.5.4, retained all three loops of apparent YS library origin.
Framework mutations
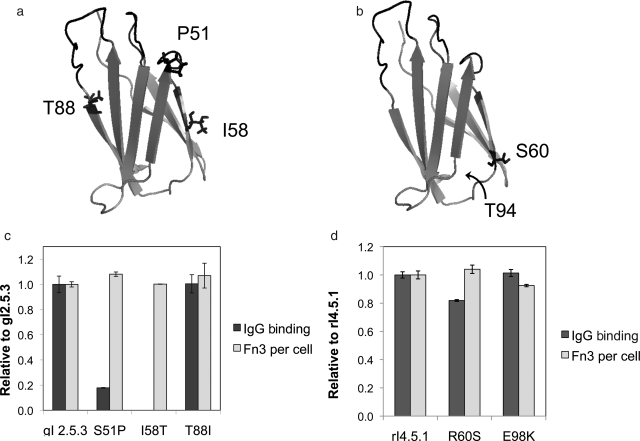
The dominant clones, gI2.5.3 (which is identical to gI3.5.1 and gI4.5.1) and rI4.5.1, each contain multiple framework mutations (Fig. 1a and b). The impact of these mutations was investigated in terms of binding and stability. Each framework mutation in the two clones was individually reverted to the wild-type side chain, and clonal cultures were assayed for stability and binding. The relative number of Fn3 molecules displayed on the yeast surface after induction at 37°, which correlates to protein stability (Shusta et al., 1999), was determined by flow cytometry. Binding to 10 nM goat IgG and 100 pM rabbit IgG was assayed by flow cytometry. None of the gI2.5.3 framework mutations significantly impact stability (Fig. 1) but two of three are important for binding. Reversion of isoleucine to threonine at amino acid 58 ablates binding at 10 nM IgG whereas reversion of serine to proline at position 51 decreases binding 5-fold. Conversely, threonine and isoleucine at position 88 yield indistinguishable results.
Fig. 1.
gI2.5.3 and rI4.5.1 framework mutations. (a) P51, T58 and I88 are presented in the wild-type Fn3 structure 1TTG (Main et al., 1992). (b) S60 is presented in the wild-type Fn3 structure. E98 was not included in this structure, but the C-terminal amino acid of the structure, T94, is indicated. (c) The indicated clone, with a c-myc epitope tag, was displayed on the surface of yeast with 37° induction. Cells were labeled with chicken anti-c-myc antibody followed by bovine anti-chicken phycoerythrin conjugate and analyzed by flow cytometry. The phycoerythrin signals above background were normalized to the gI2.5.3 sample and are presented as Fn3 per cell. Cells were labeled with 10 nM goat IgG-FITC conjugate and analyzed by flow cytometry. The FITC signal above background was divided by the relative number of Fn3 per cell, normalized to the gI2.5.3 sample, and is presented as IgG binding. Values are mean ± standard deviation of at least duplicate experiments. (d) As in (c) except samples were labeled with 100 pM rabbit IgG and were normalized to rI4.5.1.
The E98K reversion of rI4.5.1, which was also observed as clone rI4.5.5, does not significantly affect binding or stability. The R60S reversion maintains stability but has slightly decreased binding.
Affinity analysis
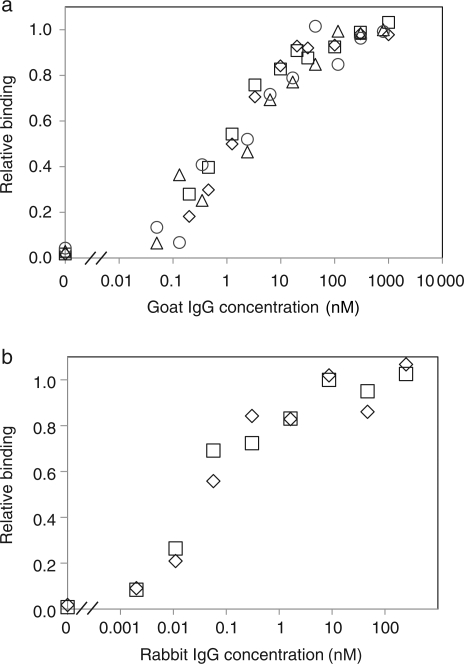
The affinities of several clones were determined by titration using yeast surface display and soluble IgG. Although avidity effects resulting from multivalent display of Fn3 binding to homodimeric IgG can enable improved binding at low concentrations relative to a monovalent interaction, it has been demonstrated that the concentration of half-maximal binding corresponds to the monovalent equilibrium dissociation constant, Kd. The highest affinity goat IgG binder, gI2.5.3T88I, has a Kd of 1.2 ± 0.4 nM (Fig. 2, Table III). Clones gI2.5.2 and gI2.5.4, the YS clone, have mid-nanomolar affinities. The highest affinity rabbit IgG binder, rI4.5.5, exhibits 51 ± 4 pM affinity (Fig. 2, Table III). Other clones demonstrate picomolar to low nanomolar affinities.
Fig. 2.
Affinity titrations of gI2.5.3T88I and rI4.5.5. Yeasts displaying gI2.5.3T88I (a) or rI4.5.5 (b) were incubated at 22° with the indicated concentration of FITC-conjugated goat IgG or AlexaFluor633-conjugated rabbit IgG. The cells were washed and analyzed by flow cytometry. The signals relative to non-displaying yeast were normalized to the maximum signal.
Table III.
Binding affinity
| Clone | Amino acid sequence |
Kd (nM) | |||
|---|---|---|---|---|---|
| BC | DE | FG | Framework | ||
| Goat IgG binders | |||||
| gI2.5.3T88I | TRAYFAP | GSLSS | SYGLVITD | P51S, T58I | 1.2 ± 0.4 |
| gI2.5.2 | ALPRSE | NWTS | SPGLVLGA | T58I | 32 ± 21 |
| gI2.5.4 | YSSYSYY | GFRPT | YYSSSYY | T35A, E38G, S89P | 35 ± 16 |
| Rabbit IgG binders | |||||
| rI4.5.5 | ATTGKAPL | ATWVK | HYDDTLS | S60R | 0.051 ± 0.004 |
| rI4.3.1 | ATTGKTPL | RSAEM | HYDDTLS | — | 0.117 ± 0.006 |
| rI3.6.6 | ATTGKAPL | KGATA | SYYDYHS | Q46K, S60R | 0.187 ± 0.034 |
| rI4.3.4 | VNGDSCL | GSAHV | GYGGKRVQ | P51S,S60R, G61R,A74T | 0.30 ± 0.12 |
| rI3.6.4 | VRPSYSRL | RSARS | GYGGERVQ | P51S, S60R | 0.63 ± 0.07 |
| rI4.3.3 | ARASNPCL | ATWVK | GYGGKRVQ | S60R | 1.08 ± 0.38 |
Kd, equilibrium dissociation constant at 22°.
Stability analysis
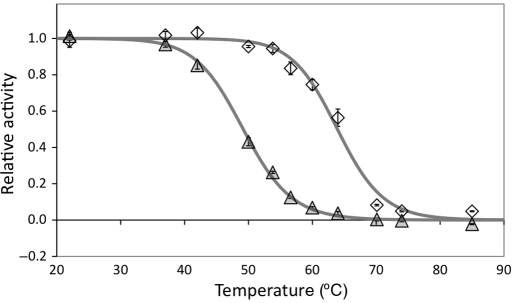
Thermal stabilities were analyzed using a previously validated yeast surface display assay (Orr et al., 2003; Hackel et al., 2008). The highest affinity binders, gI2.5.3T88I and rI4.5.5, have midpoints of thermal denaturation of 63.9 ± 0.3° and 49.1 ± 0.5°, respectively (Fig. 3). The serine/tyrosine clone gI2.5.4 has a T1/2 of 57.3 ± 0.9°. rI4.3.4, the second highest affinity rabbit IgG binder without sequence homology to rI4.5.5, has a T1/2 of 44.8 ± 1.9°. gI2.5.2 has a midpoint of denaturation of 59.3 ± 2.5°. rI3.6.4, which has FG loop homology to rI4.3.4 but novel BC and DE loops, has a T1/2 of 49.5 ± 0.5°.
Fig. 3.
Thermal stability. Yeasts displaying gI2.5.3T88I (diamonds) or rI4.5.5 (triangles) were incubated at the indicated temperature for 30 min, returned to room temperature, labeled with FITC-conjugated IgG and analyzed by flow cytometry. The FITC signal relative to non-displaying cells was normalized by the 22° sample. Data represent mean ± standard deviation of triplicate samples.
Specificity analysis
The specificity of binding was assayed by flow cytometry. None of the tested clones bind the non-cognate proteins lysozyme or streptavidin (Table IV). rI3.6.4, rI4.3.4 and rI4.5.5 exhibit unique specificity for rabbit IgG with no detectable binding to bovine, chicken, goat, human or mouse IgG. Conversely, all three goat IgG binders tested also bind bovine IgG. Clone gI2.5.3T88I also binds mouse IgG.
Table IV.
Binding specificity
| Target | gI2.5.2 | gI2.5.3T88I | gI2.5.4 | rI3.6.4 | rI4.3.4 | rI4.5.5 |
|---|---|---|---|---|---|---|
| Bovine IgG | + | + | + | − | − | − |
| Chicken IgG | − | − | − | − | − | − |
| Goat IgG | + | + | + | − | − | − |
| Human IgG | − | − | − | − | − | − |
| Mouse IgG | − | + | − | − | − | − |
| Rabbit IgG | − | − | − | + | + | + |
| Lysozyme | − | − | − | − | − | − |
| Streptavidin | − | − | − | − | − | − |
+, binding; −, no binding.
Yeasts displaying the indicated clone were labeled with 100 nM protein and analyzed by flow cytometry. The presence (+) or absence (−) of binding is indicated.
Utility in affinity purification
The utility of the engineered Fn3 domains in affinity purification from complex mixtures was investigated. Fn3 domains were produced in Escherichia coli and biotinylated on exposed amines. To increase biotinylation potential, a two-lysine tail was included. Also, the two engineered lysines in rI4.5.5 were reverted to serine (rI4.5.5K27S/K56S) to avoid biotinylation of the paratope. Affinity analysis reveals that these mutations only increase the Kd 3-fold thereby maintaining picomolar affinity.
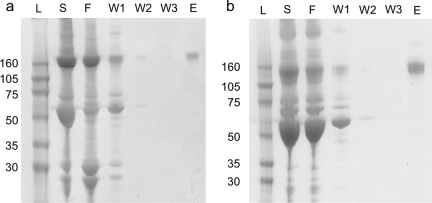
Goat or rabbit serum was applied to a column containing biotinylated Fn3 (gI2.5.3T88I or rI4.5.5K27S/K56S) immobilized on streptavidin agarose. The column was washed in PBS and bound protein was eluted with 0.1 M glycine pH 2.5. Elution fractions contain pure IgG of the expected 150 kDa molecular weight (Fig. 4).
Fig. 4.
Affinity purification. Goat (a) or rabbit (b) serum was purified on an affinity column composed of streptavidin–agarose and biotinylated Fn3 (gI2.5.3T88I or rI4.5.5K27S/K56S). The serum (S), flowthrough (F), washes (W1–W3) and elution (E) were separated by polyacrylamide gel electrophoresis and stained with SimplyBlue SafeStain. L indicates the protein ladder.
Utility as detection reagents
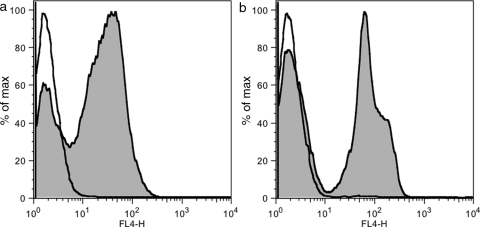
The fluorophore DyLight633 was conjugated to amines on gI2.5.3T88I and rI4.5.5K27S/K56S. These fluorophore–Fn3 conjugates were used as secondary reagents in flow cytometry. Yeast displaying HA–Fn3–c-myc (irrelevant Fn3 clone) were labeled with anti-HA goat IgG followed by gI2.5.3T88I-DyLight633 and analyzed by flow cytometry. The cells displaying HA–Fn3–c-myc are clearly labeled whereas cells that lost plasmid (see Materials and methods) or those without primary antibody have only background signal (Fig. 5a). Likewise, rI4.5.5K27S/K56S-DyLight633 effectively labels yeast initially labeled by anti-c-myc rabbit IgG (Fig. 5b).
Fig. 5.
Flow cytometry detection. (a) Yeasts displaying HA–Fn3–c-myc (irrelevant Fn3 clone) were labeled with PBS (empty) or anti-HA goat IgG (shaded) followed by DyLight633-conjugated gI2.5.3T88I and analyzed by flow cytometry. (b) As in (a) except anti-c-myc rabbit IgG and rI4.5.5K27S/K56S were used. Note that the two peaks in the IgG-labeled samples correspond to cells with and without plasmid, which serves as an effective internal control.
Discussion
Library creation and screenable throughput limit the number of clones that can be analyzed in a combinatorial library. Thus, the aim of library design is to enable sufficient shape and chemical diversity, within this limited sequence space, to provide high affinity binding to any desired epitope. Full amino acid diversity enables better shape and chemical complementarity, but the vastness of sequence space may include sufficient non-functional sequences such as to reduce the overall frequency of binders in the screened library. Conversely, well-designed reduced diversity can improve the frequency of binders, but may not be able to mediate an interaction of equally high affinity. The current work provides direct competition of minimal and maximal diversity libraries in the fibronectin scaffold for molecular recognition of immunoglobulin G. Multiple binders of picomolar affinity for rabbit IgG and multiple binders with nanomolar affinity for goat IgG were isolated with a dominant preference for clones from the full diversity library. It is important to note that this comparison includes an effective dual mutagenesis approach for the evolution of lead clones enabling a more efficient broad search of sequence space. Although a serine/tyrosine binary code is effective in isolating binders of mid-nanomolar affinity (Fellouse et al., 2005; Koide et al., 2007), extensive diversity is more effective at generating low nanomolar to picomolar binding. Nevertheless, it remains true that amino acids have varied functionality and some, in particular tyrosine, are better suited for molecular recognition. Thus, the current results support the growing opinion that the ideal synthetic library design is a hybrid of full diversity and a binary serine/tyrosine code in which tyrosine is present at an increased level but more extensive alternatives than serine are included (Koide and Sidhu, 2009).
The relative lack of library dominance in the DE loop (65% full diversity, 23% wild-type bias, 13% ambivalent) supports the hypothesis that the serine/tyrosine versus full diversity comparison was not largely affected by differences in the DE loop. In fact, given the prevalence of full diversity BC and FG loops in the selected binders, the increased presence of wild-type biased DE loops from the other library suggests that a wild-type bias is superior to random full diversity in this loop. This result is expected given its position on the edge of the diversified region as well as the ability of previously engineered Fn3 domains to bind with fully wild-type DE loops as previously discussed (Hackel et al., 2008).
Conformational diversity via loop length variation can enhance full chemical diversity (Lipovsek et al., 2007; Hackel et al., 2008) and partially compensate for constrained chemical diversity (Fellouse et al., 2007; Koide et al., 2007; Gilbreth et al., 2008). The current selected clones further demonstrate the importance of loop length diversity. BC loops of wild-type length and with one or two deletions are observed multiple times. DE loops of wild-type length and one amino acid shorter occur. FG loops of all possible (shorter) lengths except wild-type are observed. Thus, the available loop lengths appear to provide an efficient coverage of conformational space. Although additional lengths could expand the binding capacity of the libraries, particularly the chemically constrained serine/tyrosine library, the efficacy of other lengths is suspect. The range of loop lengths observed in fibronectin type III domains across phylogeny (Hackel et al., 2008) is included in the libraries and it is noteworthy that the longest allowed lengths in each loop were not observed in the sequenced binders. Longer loops are observed in nature as well as previously published binders and, thus, are tolerable in the scaffold. However, one could speculate that longer loops could be less structured resulting in entropic penalty upon binding. This phenomenon will require further monitoring as the collection of engineered fibronectin domains increases.
The efficacy of the engineered Fn3 domains in purification and detection further exemplifies the utility of this alternative scaffold for biotechnology applications and provides two useful high affinity reagents. The small size, single-domain architecture and lack of disulfide bonds in Fn3 domains provide potential benefits over antibody-based reagents. Moreover, their facile production in bacteria and ease of site-specific labeling through amines or thiols simplify their use.
Materials and methods
Fn3 library construction
Oligonucleotides were purchased from MWG Biotech (High Point, NC, USA) and IDT DNA Technologies (Coralville, IA, USA). The NNB library was previously constructed as described (Hackel et al., 2008). In the NNB library, the oligonucleotides encoding the BC, DE and FG loops were replaced by NNB codons; specifically, the DNA encoding amino acids 23–30 (DAPAVTVR), 52–56 (GSKST) and 77–86 (GRGDSPASSK) were replaced with (NNB)x where x = 6, 7, 8 or 9 for the BC loop, x = 4, 5, 6 or 7 for the DE loop and x = 5, 6, 8 or 10 for the FG loop. The YS library replaced the DNA encoding amino acids 23–31 (DAPAVTVRY) and 77–86 with TMY codons that encode for serine and tyrosine in equal frequency. Loop lengths of 7, 8, 9 or 10 in the BC loop and 6, 7, 8 or 10 in the FG loop were included. Earlier selections from the NNB library rarely yielded FG loops of five amino acids, thus the YS library was constructed with a seven amino acid option rather than five. The DNA encoding amino acids 52–56 was replaced by a set of biased codons designed to yield 50% wild-type amino acid at G52, S53, S55 and T56. K54, because of its potential for steric and electrostatic hindrance of binding, was replaced by NNBx where x = 0, 1 or 3. The degenerate portion of the oligonucleotide was ggBtcBNNBtcBacB where a, c, g and t represent mixtures of 70% of the indicated nucleotide and 10% of each of the other three. Full Fn3 genes were constructed by sequential annealing and extension of eight overlapping oligonucleotides. A 50 μl reaction was prepared with 0.2 μM oligonucleotide A (a2, b3, c6 or d7), 0.4 μM oligonucleotide B (a1, b4, c5 or d8), 1× polymerase buffer, 0.2 mM deoxynucleotide triphosphates, 1 mM MgSO4, 1 U KOD HotStart DNA Polymerase (Novagen, Madison, WI, USA), 1 M betaine and 3% dimethyl sulfoxide. The mixture was denatured at 95° for 2 min, cycled 10 times through 94° for 30 s, 58° for 30 s and 68° for 1 min, and finally extended at 68° for 10 min. Forty microliters of products (a1 + a2, b3 + b4, c5 + c6, d7 + d8) were combined and thermally cycled at identical conditions. The appropriate strand (sense for a1 + a2 + b3 + b4 and anti-sense for c5 + c6 + d7 + d8) was amplified with 0.4 μM primer (p1 or p8) in a 100 µl reaction. The products were combined and thermally cycled under identical conditions. Full Fn3 genes were purified on an agarose gel, amplified by p1 and p8 in 100 μl reactions and concentrated with PelletPaint (Novagen). The plasmid acceptor vector pCTf1f4 (Lipovsek et al., 2007) was digested with NcoI, NdeI and SmaI (New England Biolabs, Ipswich, MA, USA). Multiple aliquots of ∼10 μg of Fn3 gene and ∼3 μg of plasmid vector were combined with 50–100 μl of electrocompetent EBY100 and electroporated at 0.54 kV and 25 μF. Homologous recombination of the linearized vector and degenerate insert yielded intact plasmid. Cells were grown in YPD (10 g/l yeast extract, 20 g/l peptone, 20 g/l glucose) for 1 h at 30°, 250 rpm. The total number of transformants was determined by serial dilution plating on SD-CAA plates (0.1 M sodium phosphate, pH 6.0, 182 g/l sorbitol, 6.7 g/l yeast nitrogen base, 5 g/l casamino acids, 20 g/l glucose). The library was propagated in SD-CAA, pH 5.3 (0.07 M sodium citrate pH 5.3, 6.7 g/l yeast nitrogen base, 5 g/l casamino acids, 20 g/l glucose, 0.1 g/l kanamycin, 100 kU/l penicillin and 0.1 g/l streptomycin) at 30°, 250 rpm.
Binder selection and affinity maturation
Yeasts were grown in SD-CAA at 30°, 250 rpm to logarithmic phase, pelleted and resuspended to 1 × 107 cells/ml in SG-CAA (0.1 M sodium phosphate, pH 6.0, 6.7 g/l yeast nitrogen base, 5 g/l casamino acids, 19 g/l galactose, 1 g/l glucose, 0.1 g/l kanamycin, 100 kU/l penicillin and 0.1 g/l streptomycin) to induce protein expression. Induced cells were grown at 30°, 250 rpm for 8–24 h.
Magnetic bead sorts consisted of a negative selection for clones that do not bind streptavidin-coated beads followed by a positive selection for clones that bind biotinylated IgG complexed to streptavidin-coated beads as described (Ackerman et al., 2009). Biotinylated goat or rabbit IgG 0.75 μg (Rockland Immunochemicals, Gilbertsville, PA, USA) was added to 4 × 106 streptavidin-coated magnetic Dynabeads (Invitrogen, Carlsbad, CA, USA) in 1 ml PBSA (0.01 M sodium phosphate, pH 7.4, 0.137 M NaCl, 1 g/l bovine serum albumin) and incubated at 4° for 12–24 h. Beads were washed using a Dynal magnet with PBSA. Yeasts displaying Fn3 were washed, resuspended in PBSA and incubated with 4 × 106 IgG-free streptavidin beads for 2–12 h at 4°. A magnet was applied to the cell/bead mixture and unbound cells were collected. The washed IgG-labeled beads were added to these cells and incubated at 4° for 2–12 h. The beads were applied to the magnet and washed with PBSA. The beads and attached cells were transferred to SD-CAA for growth.
The naïve library was sorted twice (zero washes at 4°, one wash at 4°) with growth and induction after each sort. The resultant population was labeled with 150 nM mouse anti-c-myc antibody (clone 9E10, Covance, Denver, PA, USA) followed by 25 nM goat anti-mouse phycoerythrin conjugate (Invitrogen). Full-length Fn3 clones, represented by cells with a positive phycoerythrin signal, were selected via FACS on an FACS Aria (Becton Dickinson, Franklin Lakes, NJ, USA) or MoFlo (Dako Cytomation, Carpinteria, CA, USA). Plasmid DNA was extracted and mutagenized as described (Hackel et al., 2008). Error-prone PCR was performed on the full gene and each of the three loops; the mutated gene or shuffled combinations of the mutated loops were co-transformed with linearized plasmid vector to produce intact plasmid via homologous recombination. Transformed yeasts were grown in SD-CAA for further selection. The mutagenized population was sorted twice on magnetic beads (one wash at 4°, one wash at 22°) followed by c-myc+ FACS and further mutagenesis. After two magnetic bead sorts (one wash at 22°, two washes at 22°), binding to soluble IgG was assayed by flow cytometry. Yeast were labeled with 3.3 nM biotinylated IgG followed by 33 nM streptavidin–phycoerythrin conjugate. Cells with the highest phycoerythrin signal were collected by FACS and mutated. Remaining selections were performed with 20–500 pM biotinylated IgG and 67 nM chicken anti-c-myc followed by 150 nM streptavidin–fluorophore and 25 nM bovine anti-chicken phycoerythrin conjugate. Cells with the highest fluorophore:phycoerythrin ratio were selected by FACS.
DNA sequencing and point mutations
Multiple clones from several populations were sequenced. Plasmid DNA was isolated using the Zymoprep kit II (Zymo Research, Orange, CA, USA), cleaned using the Qiagen PCR Purification kit (Qiagen, Valencia, CA, USA) and transformed into DH5α (Invitrogen) or XL1-Blue E.coli (Stratagene, La Jolla, CA, USA). Individual clones were grown, miniprepped and sequenced using BigDye chemistry on an Applied Biosystems 3730.
Single amino acid mutations were introduced by standard site-directed mutagenesis using the QuikChange Mutagenesis Kit (Stratagene) according to the manufacturer's instructions. Clone construction was verified by DNA sequencing.
Affinity measurement
The plasmid for the clone of interest was transformed into yeast using the Frozen EZ Transformation Kit II (Zymo Research), and cells were grown and induced as for selection. Cells were washed in PBSA and resuspended in PBSA containing fluorophore-conjugated IgG over a range of concentrations. Sample volumes and cell densities were selected to ensure 10-fold excess of IgG relative to displayed Fn3. Samples were incubated at 22° for a sufficient time to ensure the approach to equilibrium was at least 98% complete. After incubation, cells were washed and analyzed on an FACS Calibur cytometer (Becton Dickinson). The relative binding was calculated by subtracting background signal, which was determined in an unlabeled control, and normalizing to the saturated signal at high concentrations. The equilibrium dissociation constant, Kd, was identified as the concentration corresponding to half-maximal binding.
Stability
The yeast surface display thermal denaturation assay (Orr et al., 2003) was performed as described (Hackel et al., 2008). Yeasts displaying the clone of interest were washed and resuspended in PBSA, incubated at 22–85° for 30 min and incubated on ice for 5 min. The cells were incubated in 20 nM fluorescein-conjugated IgG for 30 min, washed and analyzed on an Epics XL flow cytometer. The minimum and maximum fluorescence, the midpoint of thermal denaturation (T1/2) and the enthalpy of unfolding at T1/2 were determined by minimizing the sum of squared errors between experimental data and theoretical values according to a two-state unfolding equation.
Specificity
Yeasts displaying the clone of interest were incubated with 100 nM bovine IgG-phycoerythrin (Santa Cruz Biotechnology, Santa Cruz, CA, USA), AlexaFluor488 conjugates of streptavidin, chicken IgG or mouse IgG (Invitrogen) or fluorescein conjugates of goat IgG, human IgG or rabbit IgG (Sigma). Cells were incubated for 30 min, washed with PBSA and analyzed by flow cytometry. To test lysozyme binding, cells were incubated with 100 nM biotinylated lysozyme for 30 min, washed and resuspended in 25 nM AlexaFluor488-conjugated streptavidin. Cells were washed and analyzed by flow cytometry. Fluorophore signal was compared with both unlabeled and non-displaying cells.
Fn3 production
The Fn3 gene was digested with NheI and BamHI and transformed to a pET vector containing a HHHHHHKGSGK-encoding C-terminus. The six histidines enable metal affinity purification, and the pentapeptide provides two additional amines for chemical conjugation. The plasmid was transformed into Rosetta (DE3) E.coli (Novagen), which was grown in LB medium with 100 mg/l kanamycin and 34 mg/l chloramphenicol at 37°. Two hundred microliters of overnight culture were added to 100 ml of LB medium, grown to an optical density of 0.5 units, and induced with 0.5 mM IPTG overnight. Cells were pelleted, resuspended in lysis buffer [50 mM sodium phosphate, pH 8.0, 0.5 M NaCl, 5% glycerol, 5 mM CHAPS, 25 mM imidazole and 1× complete EDTA-free protease inhibitor cocktail (Roche, Indianapolis, IN, USA)] and exposed to four freeze-thaw cycles. The soluble fraction was clarified by centrifugation at 15 000g for 10 min and purified by metal affinity chromatography on TALON resin (Clontech, Mountain View, CA, USA).
Affinity purification
Purified Fn3 (gI2.5.3T88I and rI4.5.5K27S/K56S) was biotinylated using EZ-Link Sulfo-N-hydroxysuccinimide-LC-biotin (Pierce, Rockford, IL, USA) according to the manufacturer's instructions. Excess biotin was removed using a Zeba desalting spin column (Pierce). Biotinylated Fn3 was added to 1 ml of strepavidin–agarose (Pierce) in a column and washed. Goat or rabbit serum was added to the column and flowthrough was reapplied once. The column was washed with three 5 ml aliquots of PBS. Protein was eluted with 0.1 M glycine, pH 2.5. The original serum, flowthrough, washes and elution were separated by SDS–PAGE on 12% BisTris gel (Invitrogen) in the absence of dithiothreitol. The gel was stained with SimplyBlue SafeStain (Invitrogen) and imaged.
Detection
Purified Fn3 (gI2.5.3T88I and rI4.5.5K27S/K56S) was labeled with DyLight633 N-hydroxysuccinimide-ester (Pierce) according to the manufacturer's instructions. Unreacted dye was removed using a Zeba desalting spin column. Yeasts were induced to display an irrelevant Fn3 clone with the HA and c-myc epitopes. As in all yeast surface display, a fraction of this population does not display any Fn3 as a result of plasmid loss. These cells serve as an internal negative control. One million yeasts were incubated with 50 nM anti-HA goat IgG or anti-c-myc rabbit IgG (Genscript, Piscataway, NJ, USA), washed and incubated with 50 nM DyLight633-conjugated Fn3. Cells were washed and analyzed on an FACS Calibur cytometer.
Funding
The work was supported by the National Institutes of Health (CA96504 to K.D.W.); the Department of Defense (National Defense Science and Engineered Graduate Fellowship to B.J.H.) and the National Science Foundation (Graduate Fellowship to B.J.H.).
Acknowledgement
Assistance from the MIT Flow Cytometry Core Facility is appreciated.
Footnotes
Edited by Arne Skerra
References
- Ackerman M., Levary D., Tobon G., Hackel B., Orcutt K.D., Wittrup K.D. Biotechnol. Prog. 2009;25:774–783. doi: 10.1002/btpr.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellouse F., Wiesmann C., Sidhu S. Proc. Natl Acad. Sci. USA. 2004;101:12467–12472. doi: 10.1073/pnas.0401786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellouse F., Li B., Compaan D.M., Peden A.A., Hymowitz S.G., Sidhu S. J. Mol. Biol. 2005;348:1153–1162. doi: 10.1016/j.jmb.2005.03.041. [DOI] [PubMed] [Google Scholar]
- Fellouse F., Barthelemy P.A., Kelley R.F., Sidhu S. J. Mol. Biol. 2006;357:100–114. doi: 10.1016/j.jmb.2005.11.092. [DOI] [PubMed] [Google Scholar]
- Fellouse F., et al. J. Mol. Biol. 2007;373:924–940. doi: 10.1016/j.jmb.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Gilbreth R.N., Esaki K., Koide A., Sidhu S., Koide S. J. Mol. Biol. 2008;381:407–418. doi: 10.1016/j.jmb.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackel B., Kapila A., Wittrup K. J. Mol. Biol. 2008;381:1238–1252. doi: 10.1016/j.jmb.2008.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Koide A., Nettle K., Greene G., Koide S. Protein Expr. Purif. 2006;47:348–354. doi: 10.1016/j.pep.2005.10.021. [DOI] [PubMed] [Google Scholar]
- Karatan E., Merguerian M., Han Z., Scholle M.D., Koide S., Kay B.K. Chem. Biol. 2004;11:835–844. doi: 10.1016/j.chembiol.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Koide A., Koide S. Methods Mol. Biol. 2007;352:95–109. doi: 10.1385/1-59745-187-8:95. [DOI] [PubMed] [Google Scholar]
- Koide S., Sidhu S.S. ACS Chem. Biol. 2009;4:325–334. doi: 10.1021/cb800314v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A., Bailey C.W., Huang X., Koide S. J. Mol. Biol. 1998;284:1141–1151. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- Koide A., Abbatiello S., Rothgery L., Koide S. Proc. Natl Acad. Sci. USA. 2002;99:1253–1258. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A., Gilbreth R.N., Esaki K., Tereshko V., Koide S. Proc. Natl Acad. Sci. USA. 2007;104:6632–6637. doi: 10.1073/pnas.0700149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovsek D., Lippow S., Hackel B., Gregson M.W., Cheng P., Kapila A., Wittrup K. J. Mol. Biol. 2007;368:1024–1041. doi: 10.1016/j.jmb.2007.02.029. [DOI] [PubMed] [Google Scholar]
- Main A.L., Harvey T.S., Baron M., Boyd J., Campbell I.D. Cell. 1992;71:671–678. doi: 10.1016/0092-8674(92)90600-h. [DOI] [PubMed] [Google Scholar]
- Olson C.A., Liao H.I., Sun R., Roberts R.W. ACS Chem. Biol. 2008;3:480–485. doi: 10.1021/cb800069c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr B.A., Carr L.M., Wittrup K., Roy E.J., Kranz D.M. Biotechnol. Prog. 2003;19:631–638. doi: 10.1021/bp0200797. [DOI] [PubMed] [Google Scholar]
- Parker M., Chen Y., Danehy F., Dufu K., Ekstrom J., Getmanova E., Gokemeijer J., Xu L., Lipovsek D. Protein Eng. Des. Sel. 2005;18:435–444. doi: 10.1093/protein/gzi050. [DOI] [PubMed] [Google Scholar]
- Richards J., Miller M., Abend J., Koide A., Koide S., Dewhurst S. J. Mol. Biol. 2003;326:1475–1488. doi: 10.1016/s0022-2836(03)00082-2. [DOI] [PubMed] [Google Scholar]
- Shusta E.V., Kieke M.C., Parke E., Kranz D.M., Wittrup K.D. J. Mol. Biol. 1999;292:949–956. doi: 10.1006/jmbi.1999.3130. [DOI] [PubMed] [Google Scholar]
- Xu L., et al. Chem. Biol. 2002;9:933–942. doi: 10.1016/s1074-5521(02)00187-4. [DOI] [PubMed] [Google Scholar]