Abstract
Background
Gibbons or small apes inhabit tropical and subtropical rain forests in Southeast Asia and adjacent regions, and are, next to great apes, our closest living relatives. With up to 16 species, gibbons form the most diverse group of living hominoids, but the number of taxa, their phylogenetic relationships and their phylogeography is controversial. To further the discussion of these issues we analyzed the complete mitochondrial cytochrome b gene from 85 individuals representing all gibbon species, including most subspecies.
Results
Based on phylogenetic tree reconstructions, several monophyletic clades were detected, corresponding to genera, species and subspecies. A significantly supported branching pattern was obtained for members of the genus Nomascus but not for the genus Hylobates. The phylogenetic relationships among the four genera were also not well resolved. Nevertheless, the new data permitted the estimation of divergence ages for all taxa for the first time and showed that most lineages emerged during four short time periods. In the first, between ~6.7 and ~8.3 mya, the four gibbon genera diverged from each other. In the second (~3.0 - ~3.9 mya) and in the third period (~1.3 - ~1.8 mya), Hylobates and Hoolock differentiated. Finally, between ~0.5 and ~1.1 mya, Hylobates lar diverged into subspecies. In contrast, differentiation of Nomascus into species and subspecies was a continuous and prolonged process lasting from ~4.2 until ~0.4 mya.
Conclusions
Although relationships among gibbon taxa on various levels remain unresolved, the present study provides a more complete view of the evolutionary and biogeographic history of the hylobatid family, and a more solid genetic basis for the taxonomic classification of the surviving taxa. We also show that mtDNA constitutes a useful marker for the accurate identification of individual gibbons, a tool which is urgently required to locate hunting hotspots and select individuals for captive breeding programs. Further studies including nuclear sequence data are necessary to completely understand the phylogeny and phylogeography of gibbons.
Background
Gibbons, family Hylobatidae, are small arboreal apes, which inhabit tropical and subtropical rainforests of Southeast Asia and adjacent regions (Figure 1). Together with humans and great apes, they belong to the primate superfamily Hominoidea [1-4]. Among hominoids, gibbons were the first to branch off and they display a set of morphological and behavioural characteristics distinctly different from great apes and humans [1,5,6]. Most prominent in this respect is the predominantly monogamous life style, their territorial calls, and the typical brachiating locomotion [1,4-7]. Due to their extensive karyotypic diversity [8-11], gibbons provide an excellent model organism to study chromosomal rearrangements and, hence, to better understand human diseases caused by such alterations.
Figure 1.
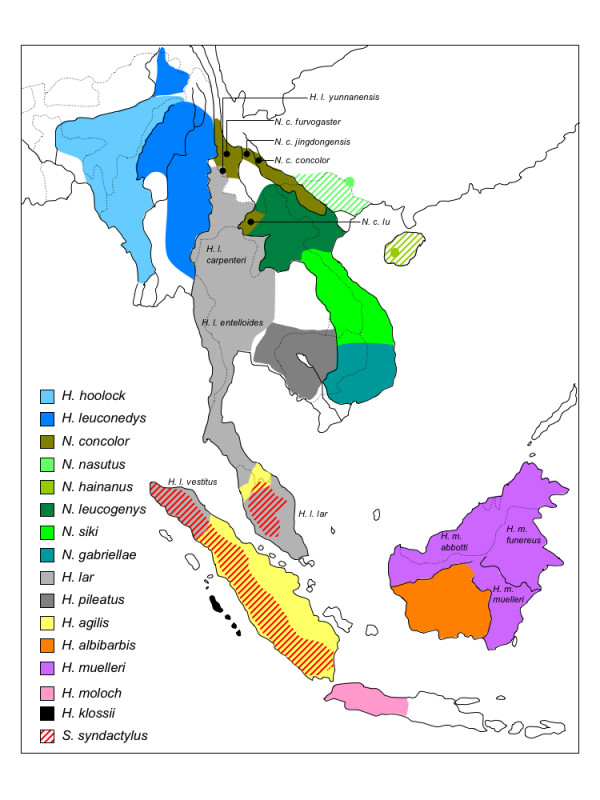
Geographical distribution of gibbons based on [2,5,23,41]. Dotted and solid lines indicate country borders and major rivers, respectively. Historical distribution of N. hainanus and N. nasutus is hatched.
Although in several aspects unique among primates and with up to 16 species the most diverse group of apes, gibbons are still in the shadow of great apes in respect of scientific studies, conservation efforts and public awareness. However, many gibbon species are on the brink of extinction and most of them are classified as "Endangered" or even "Critically Endangered" [12]. With approximately 20 individuals left in its native habitat, the Hainan gibbon (Nomascus hainanus) is the rarest primate in the world [6,13,14]. Responsible for this critical situation is habitat loss and hunting, which both have seriously reduced gibbon populations throughout their range [15,16]. Hence, much more attention has to be drawn on the gibbons' situation and extensive conservation actions are urgently required to save them from extinction [16].
While gibbons are widely considered to form a monophyletic clade, there is no consensus about the phylogeny and taxonomy within the family. Although various studies based on morphology, behaviour, vocalisation, protein electrophoresis, karyotyping and DNA sequencing were conducted [3-5,7,17-35], neither a congruent phylogeny nor a consistent taxonomic classification was obtained. Moreover, incomplete taxon sampling as well as misidentified specimens resulted in only fragmentary or even false conclusions. Accordingly, the classification of gibbon taxa at various taxonomic levels as well as their phylogenetic relationships remain disputed and a consensus is far from being available.
For example, in early studies, small apes were divided into two genera, with one (Symphalangus) including the siamang, and the other (Hylobates) all the remaining species [17,36]. Later on, the family was split into four major clades, which were recognized as subgenera [2,5,21] and eventually as genera [4,16,29,37,38]. This division is now widely accepted and takes into account the fact that species within each of the four major clades share a number of characteristics, most importantly a distinctive diploid chromosome number: Hoolock (2n = 38), Hylobates (2n = 44), Symphalangus (2n = 50) and Nomascus (2n = 52) [8]. Similarly, the number of species and subspecies is a matter of debate as well. While Symphalangus is consistently regarded as monotypic, the two Hoolock subspecies were recently elevated to species [38]. In Nomascus originally only one species was recognized [17,18,20,39], but in current classifications four to six species were suggested [2,4,12,16,34]. In contrast, the genus Hylobates already comprised at least four species in early classifications [17,39], but recent studies proposed six or seven species [2,4,16]. Due to this incongruence we follow the most recent classification of the IUCN Red List [12] with a total of 16 gibbon species (Table 1).
Table 1.
Common names, IUCN classification and proposed classification of gibbons.
| Common name | IUCN classification [12] | Proposed classification |
|---|---|---|
| Kloss' s gibbon | Hylobates klossii | Hylobates klossii |
| Eastern Müller's Bornean gibbon | Hylobates muelleri muelleri | Hylobates muelleri* |
| Northern Müller's Bornean gibbon | Hylobates muelleri funereus | Hylobates funereus* |
| Abbott's Müller's Bornean gibbon | Hylobates muelleri abbotti | Hylobates abbotti* |
| Agile gibbon | Hylobates agilis | Hylobates agilis* |
| Bornean white-bearded gibbon | Hylobates albibarbis | Hylobates albibarbis |
| Malayan lar gibbon | Hylobates lar lar | Hylobates lar lar* |
| Sumatran lar gibbon | Hylobates lar vestitus | Hylobates lar vestitus* |
| Mainland lar gibbon | Hylobates lar entelloides | Hylobates lar entelloides* |
| Carpenter's lar gibbon | Hylobates lar carpenteri | Hylobates lar carpenteri* |
| Yunnan lar gibbon | Hylobates lar yunnanensis | Hylobates lar yunnanensis* |
| Silvery Javan gibbon | Hylobates moloch | Hylobates moloch* |
| Pileated gibbon | Hylobates pileatus | Hylobates pileatus |
| Western hoolock gibbon | Hoolock hoolock | Hoolock hoolock |
| Eastern hoolock gibbon | Hoolock leuconedys | Hoolock leuconedys |
| Siamang | Symphalangus syndactylus | Symphalangus syndactylus* |
| Hainan gibbon | Nomascus hainanus | Nomascus hainanus |
| Cao-vit crested gibbon | Nomascus nasutus | Nomascus nasutus |
| Black crested gibbon | Nomascus concolor concolor | Nomascus concolor concolor* |
| West Yunnan black crested gibbon | Nomascus concolor furvogaster | Nomascus concolor concolor* |
| Central Yunnan black crested gibbon | Nomascus concolor jingdongensis | Nomascus concolor concolor* |
| Laotian black crested gibbon | Nomascus concolor lu | Nomascus concolor lu* |
| Northern white-cheeked gibbon | Nomascus leucogenys | Nomascus leucogenys* |
| Southern white-cheeked gibbon | Nomascus siki | Nomascus siki* |
| Red-cheeked gibbon | Nomascus gabriellae | Nomascus gabriellae |
| 16 species, 12 subspecies | 18 species, 7 subspecies | |
*further research required
In the present study, we analyse the complete mitochondrial cytochrome b (cytb) gene from 85 individuals, which represent all gibbon genera and species, and most subspecies. Based on our data, we are able to 1) provide the most complete phylogeny of gibbons on all taxonomic levels, 2) estimate divergence times between lineages, 3) establish a reliable classification, 4) elucidate gibbon phylogeography, and 5) provide a tool for the species identification of gibbon individuals.
Results
From all 85 gibbons, we successfully generated sequences of the complete mitochondrial cytb gene (1,140 bp). A contamination of our dataset with nuclear pseudogenes (numts) can be regarded as minimal, because no multiple amplifications of different copies were detected by direct sequencing. All sequences were correctly transcribed, and identical sequences were obtained for the same individual in cases where different material types were available. Moreover, no inconsistent positions were detected in alignments, which were assembled from overlapping sequences. Cross-contamination between individuals can be excluded as well, since all negative controls revealed no amplifications and randomly repeated PCRs for the same individual produced identical sequences.
Among the 85 individual gibbons studied, no identical haplotypes were detected. The cytb alignment comprising solely gibbons was characterized by 429 variable sites, of which 374 were parsimony-informative. In the complete alignment, which additionally contained great ape, human and hamadryas baboon representatives, we observed 565 variable sites, of which 462 were parsimony-informative.
Phylogenetic tree reconstructions based on maximum-parsimony (MP), neighbor-joining (NJ), maximum-likelihood (ML) and Bayesian algorithms revealed various strongly supported clades, which corresponded to genera, species and subspecies (Figure 2). All algorithms led to identical tree topologies, although several branching patterns gained only weak support. According to our reconstructions, hominoids diverged into a clade consisting of gibbons, and another with great apes and human. Among the latter, Pongo split off first, followed by Gorilla, before finally Pan and Homo diverged. Within gibbons, a basal position of Nomascus and a sister grouping of Hylobates and Hoolock was indicated, but support for this branching pattern was relatively low (Table 2). Similarly, with the exception of a strongly supported H. agilis + H. albibarbis clade, also the relationships among the species of Hylobates were not well resolved. However, at least species monophylies were clearly confirmed, though a common origin of H. agilis was only weakly supported. The relationships among the subspecies of H. muelleri and H. lar were less resolved. In Hoolock, the two species H. hoolock and H. leuconedys clearly segregated into two distinct clades. Within Nomascus, relationships among species were completely resolved, suggesting a N. hainanus + N. nasutus clade as sister lineage to the remaining species. Among them, N. concolor branched off first, followed by the divergence of N. gabriellae and N. leucogenys/N. siki. The monophyly of N. leucogenys was significantly supported, but evidence for a common origin of N. siki individuals was not obtained. Within N. concolor, specimens identified as N. concolor lu formed a distinct clade, while the remaining subspecies clustered together without further subdivision. However, support for a reciprocal monophyly of both clades was relatively low.
Figure 2.
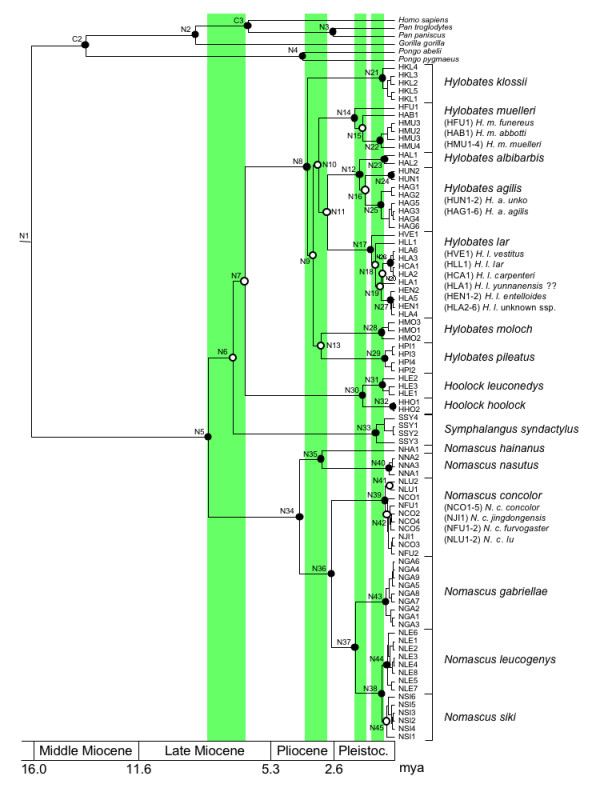
Ultrametric tree showing phylogenetic relationships and estimated divergence ages among studied gibbon individuals based on complete mitochondrial cytb sequence data. For individual codes see Additional File 1. Circles indicate bootstrap or posterior probability values (filled circles: >90%, >0.95, open circles: <70%, <0.80). Nodes of interest are arbitrarily numbered (N1-N45). C2 and C3 refer to two of the three nodes used for calibration (C1 not shown). Light green bars indicate the four radiations. A geological time scale is given below. Full details of age estimates and node supports are presented in Table 2.
Table 2.
Support values and Bayesian divergence date estimates (in mya)*.
| Node | Support values** | Divergence | Mean (95% CI) |
|---|---|---|---|
| C1 | Papio - Hominoidea | 24.04 (22.01-26.08) | |
| N1 | Hylobatidae - Hominidae | 16.26 (14.69-18.16) | |
| C2 | 96/92/92/0.99 | Pongo - Gorilla/Pan/Homo | 13.83 (13.28-14.41) |
| N2 | 91/93/98/1.0 | Gorilla - Pan/Homo | 8.90 (7.58-10.22) |
| C3 | 97/96/91/1.0 | Pan - Homo | 6.56 (6.01-7.08) |
| N3 | 100/91/97/0.99 | Pan troglodytes - P. paniscus | 2.74 (2.03-3.51) |
| N4 | 100/98/96/0.99 | Pongo pygmaeus - P. abelii | 4.12 (3.14-5.13) |
| N5 | 100/100/100/1.0 | Nomascus - Symphalangus/Hoolock/Hylobates | 8.34 (7.14-9.68) |
| N6 | 56/69/67/0.78 | Symphalangus - Hoolock/Hylobates | 7.22 (5.99-8.44) |
| N7 | 65/54/54/0.71 | Hoolock - Hylobates | 6.69 (5.56-7.88) |
| N8 | 100/93/94/0.99 | Hylobates klossii - H. pileatus/H. moloch/H. agilis/H. albibarbis/H. lar/H. muelleri | 3.91 (3.25-4.59) |
| N9 | <50/68/<50/<0.50 | H. pileatus/H. moloch - H. agilis/H. albibarbis/H. lar/H. muelleri | 3.65 (3.05-4.25) |
| N10 | <50/<50/<50/0.62 | H. muelleri - H. agilis/H. albibarbis/H. lar | 3.40 (2.81-3.99) |
| N11 | <50/53/<50/0.69 | H. agilis/H. albibarbis - H. lar | 3.02 (2.43-3.60) |
| N12 | 100/99/100/1.0 | H. agilis - H. albibarbis | 1.56 (1.19-1.98) |
| N13 | <50/52/<50/<0.50 | H. pileatus - H. moloch | 3.29 (2.64-3.97) |
| N14 | 96/96/98/1.0 | H. muelleri funereus - H. m. abbotti/H. m. muelleri | 1.78 (1.33-2.25) |
| N15 | 56/57/<50/<0.50 | H. muelleri abbotti - H. m. muelleri | 1.42 (1.02-1.81) |
| N16 | 63/<50/67/0.79 | H. agilis agilis - H. a. unko | 1.30 (0.95-1.68) |
| N17 | 100/100/99/1.0 | H. lar vestitus - H. l. lar/H. l. entelloides/H. l. carpenteri/H. l. yunnanensis | 1.05 (0.75-1.35) |
| N18 | <50/<50/50/0.76 | H. l. lar - H. entelloides/H. l. carpenteri/H. l. yunnanensis | 0.86 (0.60-1.13) |
| N19 | <50/63/65/0.79 | H. l. entelloides - H. l. carpenteri/H. l. yunnanensis | 0.62 (0.41-0.83) |
| N20 | <50/66/66/0.78 | H. l. carpenteri - H. l. yunnanensis | 0.52 (0.32-0.71) |
| N21 | 100/100/99/1.0 | MRCA H. klossii | 0.53 (0.29-0.81) |
| N22 | 99/96/97/1.0 | MRCA H. muelleri muelleri | 0.62 (0.38-0.88) |
| N23 | 100/100/100/1.0 | MRCA H. albibarbis | 0.44 (0.22-0.68) |
| N24 | 100/100/100/1.0 | MRCA H. agilis unko | 0.13 (0.02-0.25) |
| N25 | 99/96/94/1.0 | MRCA H. agilis agilis | 0.61 (0.36-0.89) |
| N26 | 95/98/92/1.0 | MRCA H. lar carpenteri | 0.17 (0.05-0.28) |
| N27 | 96/94/96/1.0 | MRCA H. lar entelloides | 0.18 (0.07-0.31) |
| N28 | 100/100/94/1.0 | MRCA H. pileatus | 0.41 (0.21-0.64) |
| N29 | 100/100/100/1.0 | MRCA H. moloch | 0.56 (0.30-0.84) |
| N30 | 100/100/100/1.0 | Hoolock hoolock - H. leuconedys | 1.42 (0.97-1.90) |
| N31 | 99/95/93/0.96 | MRCA H. leuconedys | 0.51 (0.28-0.80) |
| N32 | 100/100/100/1.0 | MRCA H. hoolock | 0.07 (0.00-0.17) |
| N33 | 100/99/99/1.0 | MRCA Symphalangus syndactylus | 0.83 (0.51-1.18) |
| N34 | 100/100/99/1.0 | Nomascus hainanus/N. nasutus - N. concolor/N. gabriellae/N. leucogenys/N. siki | 4.24 (3.46-5.06) |
| N35 | 91/92/92/0.99 | N. hainanus - N. nasutus | 3.25 (2.49-3.99) |
| N36 | 94/91/96/1.0 | N. concolor - N. gabriellae/N. leucogenys/N. siki | 2.83 (2.21-3.50) |
| N37 | 96/92/98/1.0 | N. gabriellae - N. leucogenys/N. siki | 1.74 (1.28-2.22) |
| N38 | 100/99/93/1.0 | N. leucogenys - N. siki | 0.55 (0.35-0.77) |
| N39 | 100/100/100/1.0 | N. concolor lu - N. c. concolor/N. c. furvogaster/N. c. jingdongensis | 0.43 (0.25-0.63) |
| N40 | 100/100/99/1.0 | MRCA N. nasutus | 0.23 (0.08-0.39) |
| N41 | <50/<50/67/0.75 | MRCA N. concolor lu | 0.19 (0.05-0.35) |
| N42 | 59/<50/<50/<0.50 | MRCA N. concolor concolor/N. c. furvogaster/N. jingdongensis | 0.32 (0.19-0.48) |
| N43 | 100/100/98/1.0 | MRCA N. gabriellae | 0.39 (0.21-0.57) |
| N44 | 92/91/98/1.0 | MRCA N. leucogenys | 0.33 (0.18-0.47) |
| N45 | <50/<50/<50/0.58 | MRCA N. siki | 0.38 (0.18-0.55) |
*Means and 95% credibility intervals (CI) are given for 48 nodes (see also Figure 2). Nodes used as calibrations are labelled with a "C", all others with an "N". MRCA denotes the most recent common ancestor. C1 not shown in Figure 2. **Support values as obtained from MP, NJ, ML and Bayesian reconstructions, respectively.
Based on divergence age estimates, gibbons separated from great apes and humans 16.26 million years ago (mya) (for 95% credibility intervals see Table 2). Within hominids, Pongo branched off first (13.83 mya), followed by Gorilla (8.90 mya), before finally Homo and Pan diverged from each other (6.56 mya). The differentiation of Pongo and Pan into species occurred 4.12 and 2.74 mya, respectively. In an initial radiation, gibbons diverged within a relative short time period of only 1.65 million years (6.69-8.34 mya) into four genera. Within Hylobates, most species diverged from each other between 3.02 and 3.90 mya. The only exception was the separation of H. albibarbis from H. agilis 1.56 mya, which was in the time frame of subspecies splits within H. muelleri (1.42-1.78 mya). Differentiation of H. lar into subspecies occurred even later (0.52-1.05 mya). The two Hoolock species diverged 1.42 mya from each other. In Nomascus, differentiation into species took place over a longer time period, lasting from 4.24 until 0.55 mya. The most recent species divergence within Nomascus occurred between N. siki and N. leucogenys (0.55 mya), which was in a similar range as the separation of N. concolor lu from the other N. concolor subspecies (0.43 mya).
Discussion
By analysing all species and most subspecies, the present study provides the most complete view into the evolutionary history of the gibbon family. However, as in earlier molecular studies on gibbons [26-35], relationships on various taxonomic levels are less resolved and partially contradict earlier findings. While the herein depicted branching pattern among genera is identical with that found in earlier studies using also cytb [32] or D-loop [29] sequences, it differs from another cytb-based study [28] in placing Nomascus and not Symphalangus as most basal genus. Studies based on mitochondrial ND3-ND4 sequences [31] or chromosomal rearrangements [8] suggest Hoolock as most ancestral lineage, and Nomascus together with either Hylobates [31] or Symphalangus [8] as the most recently diverged genera. For Hylobates, our data indicate a basal position of H. klossii, and a further division into a clade consisting of H. lar, H. muelleri, H. agilis and H. albibarbis, and another one with H. moloch and H. pileatus. Various branching patterns among Hylobates species are proposed [27,31,32,35], which all differ from our one, but respective support values are similarly low as in our study. In contrast, the relationships found among species of the genus Nomascus are well resolved and identical with that suggested by [30,31,33,34].
According to our and earlier data, relationships among gibbon genera and Hylobates species remain disputed, which most likely can be explained by the separation of respective lineages within relative short time periods. This becomes even more obvious when considering estimated divergence ages, which fall into four temporal windows. In the first, between ~6.7 and ~8.3 mya, the four gibbon genera originated. In a second radiation, between ~3.0 and ~3.9 mya, Hylobates split into various species, and in a third burst, between ~1.3 and ~1.8 mya, H. muelleri, the H. agilis + H. albibarbis clade and Hoolock further differentiated. Finally, in a fourth radiation, between ~0.5 and ~1.1 mya, H. lar diverged into subspecies. In contrast, speciation in Nomascus was a continuous process, lasting from 4.24 until 0.55 mya.
Taxonomic implications
Our data show that mitochondrial DNA (mtDNA) provides a powerful tool for the identification and taxonomic classification of gibbons, because taxa form strongly supported monophyletic clades, or at least appear to form distinct lineages in those cases where only one individual per taxon was tested. Moreover, most differentiation events fall into four temporal periods, which allow a hierarchical ranking as proposed by Goodman et al. [40], though the threshold for the recognition of a certain taxonomic unit whether genus, species, or subspecies remains disputed. Hence, to provide a more reliable classification, we compare divergence ages among gibbon lineages with those among other Asian primates and hominids.
Accordingly and concordant with recent classifications [4,12,16,29,34,37,38,41], the four major gibbon lineages are proposed as distinct genera (Table 1), since they split from each other in a similar time range as did colobine genera [[42,43], Roos C, Zinner D, Schwarz C, Nash SD, Xing J, Batzer MA, Leendertz FH, Ziegler T, Perwitasari-Farajallah D, Nadler T, Walter L, Osterholz M: Nuclear versus mitochondrial DNA: evidence for hybridization in colobine monkeys, submitted] or African great apes and human [40,42]. Most species of Hylobates and Nomascus emerged in or around the second radiation, which is on the same time scale as species splits within Pongo and Pan, and the separation of species groups within Macaca [44,45] and Trachypithecus [46]. Thus, taxa originating in this time period should be recognized as distinct species (H. moloch, H. pileatus, H. klossii, H. lar, H. muelleri, H. agilis/H. albibarbis, H. hoolock/H. leuconedys, N. nasutus, N. hainanus, N. concolor, N. gabriellae/N. leucogenys/N. siki), and might be even classified as species groups. Further differentiation events among gibbons occurred in the third time period, which is in a similar window as several speciation events within macaques [44,45]. Accordingly, H. leuconedys and H. albibarbis should be separated from H. hoolock and H. agilis on species level, respectively, and the three subspecies of H. muelleri could be considered for elevation to species level. Moreover, H. agilis is divided into two clades, which refer to individuals identified by pelage coloration as H. agilis agilis and H. agilis unko. However, in a recent work based on a larger number of individuals a reciprocal monophyly of both lineages is doubted [47], and, hence, we provisionally recognize H. agilis as monotypic. For H. lar, only a few unambiguously identified specimens were available for our study, but these represent at least four of the five recognized subspecies, while the identity of the putative H. lar yunnanensis individual remains uncertain. Based on our data, H. lar subspecies form distinct lineages, which diverged relative recently. We provisionally accept all five subspecies, though ongoing studies might reject some or all of them. For N. concolor, our data indicate a separation of N. concolor lu from the remaining subspecies, which form a clade without further subdivision into taxa. Hence and concordant with Monda et al. [33] and Roos et al. [34], we provisionally classify N. concolor furvogaster and N. concolor jingdongensis as synonyms of N. concolor concolor, while we feel N. concolor lu is a separate subspecies. We further separate N. gabriellae from N. siki/N. leucogenys on species level, while it is questionable whether the latter two should be recognized as species or subspecies. Our study reveals a split between both taxa just 0.55 mya, which is in a similar range as the subspecies differentiation within H. lar or N. concolor. Hence, a separation of both taxa only on subspecies level would be indicated. However, both taxa show slight differences in vocalisation and facial colouration [4,5,15], and Carbone et al. [48] found a chromosomal inversion unique to N. leucogenys. Accordingly, we follow here the current view and recognize N. leucogenys and N. siki as distinct species. In summary, we recognize four gibbon genera with 18 species and seven subspecies (Table 1).
Biogeographic implications
Multiple radiations in the evolutionary history of gibbons suggest a complicated biogeographic pattern leading to the current distribution of gibbon taxa. Since gibbons are arboreal [7,39], radiations most likely were correlated with expanding forest habitats. In fact, the complete range of gibbons experienced complex geographical and environmental changes during the last ten million years. Notably, in the late Miocene as well as in the Plio- and Pleistocene, a series of dramatic climatic changes influenced the geography and vegetation in the region, leading to shifts in the extension and distribution of different habitat types [49-54]. In particular, periods of maximum glaciation might have reduced rainforest cover, resulting in the appearance of more open and deciduous vegetation types in many parts of the region [[52-57], but see [58]]. Moreover, due to the alternately falling and rising sea water levels during the several glacial and interglacial periods [59-64], connections and separations of landmasses were common, and repeated migration between islands and today's mainland was possible [65-67].
By combining the available information, we develop the following dispersal scenario for gibbons, which is in general agreement with that proposed by Chatterjee [32,68], Harrison et al. [69], and Jablonski and Chaplin [70], but which differs substantially from them in some aspects. Accordingly, gibbons most likely originated on the Asian mainland, because all four gibbon genera occur there. Specifically, the Hengduan mountains in the border region of today's Burma, India and China might have been a possible diversification hotspot [71,72]. In the region, all the larger Southeast Asian rivers (Mekong, Salween, Yangtze) rise, which are all well known as barriers for arboreal primates [54]. Although these rivers changed their courses several times, their upper reaches in the Hengduan mountains exist at least since the early Miocene [73]. Recently, the Hengduan mountains were also proposed as a region of differentiation for colobine monkeys, and, most interestingly, respective splitting events occurred on a similar time scale as in gibbons [Roos C, Zinner D, Schwarz C, Nash SD, Xing J, Batzer MA, Leendertz FH, Ziegler T, Perwitasari-Farajallah D, Nadler T, Walter L, Osterholz M: Nuclear versus mitochondrial DNA: evidence for hybridization in colobine monkeys, submitted]. In fact, in the late Miocene, widely distributed rain forest habitats promoted range extension for arboreal primates [50,54]. Accordingly, in the late Miocene, Nomascus invaded the region east of the Mekong, Hoolock entered the region west of the Salween, and Hylobates and Symphalangus migrated into the area in-between and later on into Sundaland.
Hylobates successfully colonized large parts of Sundaland, but also survived on the Asian mainland. Shortly after its arrival in Sundaland in the Pliocene, populations on the Asian mainland, the Malay peninsula, Sumatra, Borneo, Java and the Mentawai archipelago became isolated. At the same time, various species groups of the genera Macaca and Trachypithecus diverged [44-46], indicating dramatic environmental changes. In fact, this time period was characterized by global warming and sea levels similar to today [54,61-63], which prevented migration between landmasses and, thus, promoted speciation due to vicariance. Whether Symphalangus experienced a similar range expansion in Sundaland like Hylobates, remains questionable. Today the genus appears only on Sumatra and the Malay peninsula, and fossil data provide only evidence for its historical occurrence on Java and Sumatra [69]. In the early Pleistocene, further differentiation in Hylobates occurred on Borneo and Sumatra, and in Hoolock on the mainland which is on a similar time scale when macaque species diverged [44,45], and which might has been triggered by the shrinking of forest habitats due to cold phases [[74], but see [58]]. Notably, H. albibarbis is mitochondrially closer related to Sumatran H. agilis than to the other Bornean gibbons, and acoustic, morphological and chromosomal data suggest an intermediate position [2,5,47,75]. Accordingly, H. albibarbis might be the product of an ancient hybridization event, in which proto-H. agilis invaded Borneo during sea level lowstands [61-64], and successfully reproduced with proto-H. muelleri. As we find mtDNA of proto-H. agilis in H. albibarbis, female introgression is the most likely hybridization scenario, which is in agreement with recent findings, that gibbon females disperse over longer distances than males [76]. Finally, in a last range expansion in the early to middle Pleistocene, H. lar colonized, starting from its Sumatran refuge, the Malaysian peninsular and mainland Southeast Asia [see also [70]].
In contrast to the biogeographic pattern found in Hylobates and to the scenario proposed by Chatterjee [32,68], for Nomascus not a radiation but a successive migration from North to South over a long time period becomes evident. Based on our data, Nomascus originated in the border region of Vietnam and China in the early Pliocene and it took to the early Pleistocene until the genus reached the southern extend of its current distribution in southern Vietnam and Cambodia.
Conservation implications
All gibbon species are on the brink of extinction and, with the exception of H. leuconedys (Vulnerable), are classified as "Endangered" or even "Critically Endangered" [12,16]. With approximately 20 individuals left in its native habitat, the Hainan gibbon (N. hainanus) is the rarest primate in the world [6,13,14], and the situation for its closest relative, the Cao-vit crested gibbon (N. nasutus) with approximately 100 individuals left [12,77], as well as for other gibbon species, the situation is alarming. Reasons for the decline of gibbons are manifold, but habitat loss due to forest clearance for agricultural use, oil palm or rubber plantations, gold mining, or charcoal and timber production, as well as illegal hunting for food and sport, and the trade for pets or medicine are major threats to wild gibbon populations [15,16].
To save gibbons from extinction, urgent actions are required to prevent ongoing habitat destruction and hunting, and to build up a viable gene pool in captivity for later release purposes. Specifically, to prevent or at least reduce hunting, hunting hotspots have to be identified. Therefore, it is crucial to confirm the taxon identity and if possible the geographical origin of confiscated gibbons or their remains. Similarly, to avoid artificial hybrids, only gibbons with clear taxon identity should be considered for reproduction in zoos or rescue centres. Finally, if captive gibbons are reintroduced into the wild, it has to be ascertained that these gibbons are pure individuals and of the same taxon as those, which naturally occur in the area they are to be released.
An accurate taxonomic identification of gibbons based on vocal data or pelage colouration is sometimes complicated [4,5]. In this respect, mtDNA analysis might be a promising tool. As shown in our study, gibbon taxa can be diagnosed through mtDNA, and, hence, a secure identification can easily be obtained. Yet since mtDNA is only maternally inherited, possible hybrids will not be detected in such analysis, so that additional markers should be studied as well.
Conclusions
Due to a nearly complete taxon sampling, the present study provides the most comprehensive insights into the evolutionary and biogeographic history of the hylobatid family. Based on estimated divergence ages and unresolved relationships among gibbon taxa on various levels, several radiation-like splitting events are indicated, which suggest a complex biogeographic history of gibbons. Presumably, most of these differentiation events occurred in wave-like range expansions in Sundaland and the Asian mainland followed by vicariance effects, most likely caused by alternately shrinking and expanding rain forest habitats and by repeated separations and connections of landmasses. In contrast, in the region east of the Mekong river gibbons underwent a successive North-to-South migration. Our study also shows that mtDNA provides a solid platform for the taxonomic classification of gibbons and that mtDNA can be successfully applied to accurately identify the species affiliation of gibbon individuals, which is urgently required for conservation purposes. However, to completely understand the phylogeny and phylogeography of gibbons, to identify hybrids in captivity, or to trace possible ancient hybridization events as it might be indicated for H. albibarbis, further studies including extended mitochondrial as well as autosomal, X and Y chromosomal sequence data, are necessary.
Methods
Sample Collection
A total of 85 specimens representing all species and most subspecies of hylobatids were included in our study. Blood, tissue, faecal or hair samples were collected during field surveys, in zoos or rescue centres, or from museum specimens between 1995 and 2008 (Additional file 1). Blood and hair samples were taken during routine health checks by veterinarians. Tissue samples were obtained only from deceased animals. Taxon identity of individuals was ascertained by pelage coloration, morphology and if possible by vocalization and geographic origin. With the exception of some H. lar individuals for which subspecies identity could not be traced, only clearly identified specimens were included in our study. Fresh tissue or faecal samples were preserved in 80-90% ethanol and dry samples (tissue, museum skins, hair samples) were placed in plastic bags without any additive. Samples were stored at ambient temperature for up to six months before further processing.
Laboratory Methods
Total genomic DNA was extracted with the DNeasy Blood & Tissue and QIAamp DNA Stool Mini kits from Qiagen. When hair follicle cells were used, up to three hairs were directly implemented into the PCR reaction. From high-quality DNA, the complete mitochondrial cytb gene was PCR-amplified in a single fragment with the primers 5'-AATGATATGAAAAACCATCGTTGTA-3' and 5'-TTCATTTCCGGCTTACAAGAC-3'. For low-quality DNA, extracted from faeces or museums material, two to seven overlapping PCR products were amplified with primers constructed on the basis of sequences from conspecifics (respective primers are available from the authors upon request). For all amplifications, wax-mediated hot-start PCRs were performed for 40 cycles, each with a denaturation step at 92°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 0.5-1.5 min, followed by a final extension step at 72°C for 5 min. The results of the PCR amplifications were checked on 1% agarose gels. Subsequently, PCR products were cleaned with the Qiagen Gel Extraction kit and sequenced on an ABI 3130xl sequencer using the BigDye Cycle Sequencing kit. Sequences were assembled with Geneious v4.6.1 [78] and checked for their potential to be correctly transcribed. Gibbon haplotypes were deposited at GenBank and are available under the accession numbers GU321245-GU321329 (see also Additional file 1).
To prevent cross-species contaminations, laboratory procedures followed described standards [46]. To exclude contaminations of the dataset with numts, we mainly used material in which nuclear DNA is highly degraded (faeces, museum tissue) [79,80]. Moreover, the applied primers are known to amplify solely the mitochondrial copy of the gene in hylobatids [34], and for cross-validation purposes, for some specimens, sequences were generated using different material types (blood, faeces).
Statistical Methods
For phylogenetic reconstructions, we expanded our dataset with orthologous sequences from various hominids (Homo, Pan, Gorilla, Pongo) and Papio hamadryas, which was used as outgroup. Phylogenetic trees were constructed with MP and NJ algorithms as implemented in PAUP v4.0b10 [81] as well as with ML and Bayesian algorithms, using the programs GARLI v0.951 [82] and MrBayes v3.1.2 [83,84]. For MP analysis, all characters were treated as unordered and equally weighted throughout. A heuristic search was performed with the maximum number of trees set to 100. For NJ and ML reconstructions, the optimal nucleotide substitution model (GTR + Γ) was chosen using Akaike information criterion (AIC) as implemented in MODELTEST v3.7 [85]. Relative support of internal nodes was performed by bootstrap analyses with 10,000 (MP, NJ) or 500 replications (ML). In GARLI, only the model specification settings were adjusted according to the dataset, while all other settings were left at their default value. ML majority-rule consensus trees were calculated in PAUP. For Bayesian reconstructions, the dataset was partitioned into codon positions, each with its own substitution model. We used four Markov Chain Monte Carlo (MCMC) chains with the default temperature of 0.1. Four repetitions were run for 10,000,000 generations with tree and parameter sampling occurring every 100 generations. The first 25% of samples were discarded as burnin, leaving 75,001 trees per run. Posterior probabilities for each split and a phylogram with mean branch lengths were calculated from the posterior density of trees.
To estimate divergence times, a Bayesian MCMC method, which employs a relaxed molecular clock approach [86], as implemented in BEAST v1.4.8 [87], was used. A relaxed lognormal model of lineage variation and a Yule prior for branching rates was assumed. The alignment was partitioned into codon positions, and the substitution model, rate heterogeneity and base frequencies were unlinked across codon positions. Optimal nucleotide substitution models were chosen using AIC in MODELTEST.
For calibrations we used the fossil-based divergence between Homo and Pan, which was dated at 6 - 7 mya [88-90], the separation of Pongo from the Homo/Pan lineage ~14 mya [91], and the divergence of hominoids and cercopithecoids ~23 mya [92,93]. Instead of hardbounded calibration points, we used the published dates as a normal distribution prior for the respective node. For the Homo - Pan divergence, this translates into a normal distribution with a mean of 6.5 mya and a standard deviation (SD) of 0.5 mya, for the separation of Pongo from the Homo/Pan clade into a mean of 14.0 mya and a SD of 1.0 mya, and for the hominoid - cercopithecoid divergence into a mean of 23 mya and a SD of 2 mya.
Since the estimation of phylogenetic relationships was not the main aim of this analysis, for the calculation an a-priori fixed tree topology as obtained from NJ reconstructions using the GTR + Γ model (Figure 2) was implemented. Four replicates were run for 10,000,000 generations with tree and parameter sampling occurring every 100 generations. The adequacy of a 10% burnin and convergence of all parameters were assessed by visual inspection of the trace of the parameters across generations using TRACER v1.4.1 [94]. Subsequently, the sampling distributions were combined (25% burnin) using the software LogCombiner v1.4.8, and a consensus chronogram with node height distribution was generated and visualized with TreeAnnotator v1.4.8 and FigTree v1.2.2 [95].
Authors' contributions
VNT collected samples, did laboratory work, analysed the data, and wrote the paper. ARM, TG, LM, TZ, MA, PM, and TN collected samples and wrote the paper. LW analysed data, and wrote the paper. CR designed the study, collected samples, did laboratory work, analysed data, and wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
Origin, material type, sample provider/collector and GenBank accession numbers of studied gibbon specimens.
Contributor Information
Van Ngoc Thinh, Email: vanthinhngoc@yahoo.com.
Alan R Mootnick, Email: alan@gibboncenter.org.
Thomas Geissmann, Email: thomas.geissmann@aim.uzh.ch.
Ming Li, Email: lim@ioz.ac.cn.
Thomas Ziegler, Email: tziegl@dpz.gwdg.de.
Muhammad Agil, Email: rhinogil@googlemail.com.
Pierre Moisson, Email: pierre.Moisson@agglo-mulhouse.fr.
Tilo Nadler, Email: t.nadler@mail.hut.edu.vn.
Lutz Walter, Email: lwalter@gwdg.de.
Christian Roos, Email: croos@dpz.eu.
Acknowledgements
We are grateful to the following colleagues, zoos, and institutions for providing permits or valuable gibbon materials: Claudia Barelli, Gareth Goldthorpe, Andrew Kitchener, Nicolas Lormée, Annette Schrod, Chris Smeenk, Berlin Zoo, Duisburg Zoo, Leipzig Zoo, Munich Zoo, Nuremberg Zoo, Rostock Zoo, Schwerin Zoo, Wuppertal Zoo, Zurich Zoo, Besancon Zoo, Plock Zoo, Banham Zoo, Bristol Zoo, Howletts Wild Animal Park, Paignton Zoo, Twycross Zoo, Beijing Zoo, Dhaka Zoo, Jakarta Zoo, Taman Safari, Singapore Zoo, Perth Zoo, Louisiana Purchase Gardens and Zoo, Bogor Agricultural University, the Indonesian Institute for Science, Museum für Naturkunde Berlin, National Museums Scotland Edinburgh, Natural History Museum Leiden, National Museum of Natural History Washington, Institute of Zoology of the Chinese Academy of Sciences Beijing, Bawangling National Nature Reserve, Bokeo Nature Reserve, Cat Tien National Park, Phong Nha-Ke Bang National Park and Khao Yai National Park. No international or national rules and regulations have been violated during sampling, and shipping. Many thanks also to Christiane Schwarz for her excellent laboratory work, and to Colin Groves and two anonymous reviewers for valuable comments on an earlier version of the manuscript. This study was financially supported by the German Primate Center, and the Biodiversitäts-Pakt of the Wissenschaftsgemeinschaft Gottfried-Wilhelm Leibniz.
References
- Fleagle JG. Primate Adaptations and Evolution. San Diego: Academic Press; 1999. [Google Scholar]
- Groves CP. Primate Taxonomy. Washington: Smithsonian Institution Press; 2001. [Google Scholar]
- Geissmann T. Taxonomy and evolution of gibbons. Evol Anthropol. 2002;11:28–31. doi: 10.1002/evan.10047. [DOI] [Google Scholar]
- Mootnick AR. Gibbon (Hylobatidae) species identification recommended for rescue or breeding centers. Primate Conserv. 2006;21:103–138. doi: 10.1896/0898-6207.21.1.103. [DOI] [Google Scholar]
- Geissmann T. Gibbon systematics and species identification. Int Zoo News. 1995;42:65–77. [Google Scholar]
- Cunningham C, Mootnick AR. Gibbons. Curr Biol. 2009;19:R543–R544. doi: 10.1016/j.cub.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Geissmann T. Duet-splitting and the evolution of gibbon songs. Biol Rev. 2002;77:57–76. doi: 10.1017/s1464793101005826. [DOI] [PubMed] [Google Scholar]
- Müller S, Hollatz M, Wienberg J. Chromosomal phylogeny and evolution of gibbons (Hylobatidae) Hum Genet. 2003;113:493–501. doi: 10.1007/s00439-003-0997-2. [DOI] [PubMed] [Google Scholar]
- Roberto R, Capozzi O, Wilson RK, Mardis ER, Lomiento M, Tuzun E, Cheng Z, Mootnick AR, Archidiacono N, Rocchi M, Eichler EE. Molecular refinement of gibbon genome rearrangements. Genome Res. 2007;17:249–257. doi: 10.1101/gr.6052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misceo D, Capozzi O, Roberto R, Dell'Oglio MP, Rocchi M, Stanyon R, Archidiancono N. Tracking the complex flow of chromosome rearrangements from the Hominoidea ancestor to extant Hylobates and Nomascus gibbons by high-resolution synteny mapping. Genome Res. 2008;18:1530–1537. doi: 10.1101/gr.078295.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giriajan S, Chen L, Graves T, Marques-Bonet T, Ventura M, Fronick C, Fulton L, Rocchi M, Fulton RS, Wilson RK, Mardis ER, Eichler EE. Sequencing human-gibbon breakpoints of synteny reveals mosaic new insertions at rearrangement sites. Genome Res. 2009;19:178–190. doi: 10.1101/gr.086041.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN Red List of Threatened Species 2009.2. 2009. http://www.iucnredlist.org
- Chan BPL, Fellowes JR, Geissmann T, Zhang J. Hainan Gibbon Status Survey and Conservation Action Plan. Version 1. Hong Kong: Kadoorie Farm & Botanic Garden; 2005. [Google Scholar]
- Mootnick AR, Xiaoming W, Moisson P, Chan BPL, Fellowes JR, Nadler T. In: Primates in Peril: The World's 25 Most Endangered Primates, 2006-2008. Mittermeier RA, Ratsimbazafy J, Rylands AB, Williamson L, Oates JF, Mbora D, Ganzhorn JU, Rodriguez-Luna E, Palacios E, Heymann EW, Cecilia M, Kierulff M, Yongcheng L, Supriatna J, Roos C, Walker S, Aguiar JM, editor. Arlington: IUCN/SSC Primate Specialist Group (PSG), International Primatological Society (IPS), and Conservation International (CI); 2007. Hainan Gibbon, Nomascus hainanus (Thomas, 1892) pp. 16–17. [Google Scholar]
- Geissmann T, Dang Nguyen Xuan, Lormée N, Momberg F. Vietnam Primate Conservation Status Review. Part 1: Gibbon. Hanoi: Fauna & Flora International, Indochina Programme; 2000. [Google Scholar]
- Geissmann T. Status reassessment of the gibbons: results of the Asian primate red list workshop 2006. Gibbon J. 2007;3:5–15. [Google Scholar]
- Napier JR, Napier PH. A Handbook of Living Primates. London: Academic Press; 1967. [Google Scholar]
- Groves CP. In: Gibbon and Siamang. Rumbaugh DM, editor. Basel: Karger; 1972. Systematics and phylogeny of gibbons; pp. 1–89. [Google Scholar]
- Bruce EJ, Ayala FJ. Phylogenetic relationships between man and the apes: eletrophoretic evidence. Evolution. 1979;33:1040–1056. doi: 10.2307/2407465. [DOI] [PubMed] [Google Scholar]
- Haimoff EH, Chivers DJ, Gittins SP, Whitten AJ. A phylogeny of gibbons (Hylobates spp.) based on morphological and behavioural characters. Folia Primatol. 1982;39:213–237. doi: 10.1159/000156079. [DOI] [PubMed] [Google Scholar]
- Prouty LA, Buchanan PD, Pollitzer WS, Mootnick AR. Bunopithecus: A genus-level taxon for the hoolock gibbon (Hylobates hoolock) Am J Primatol. 1983;5:83–87. doi: 10.1002/ajp.1350050110. [DOI] [PubMed] [Google Scholar]
- Creel N, Preuschoft H. In: The Lesser Apes. Evolutionary and Behavioural Biology. Preuschoft H, Chivers DJ, Brockelman WY, Creel N, editor. Edinburgh: Edinburgh University Press; 1984. Systematics of the lesser apes: A quantitative taxonomic analysis of craniometric and other variables; pp. 562–613. [Google Scholar]
- Marshall JT, Sugardjito J. In: Comparative Primate Biology, Systematics, Evolution and Anatomy. Swindler DR, Erwin J, editor. Vol. 1. New York: Liss; 1986. Gibbon systematics; pp. 137–185. [Google Scholar]
- Shafer DA. In: Current Perspectives in Primate Biology. Taub DM, King FA, editor. New York: Van Nostrand Reinhold Co; 1986. Evolutionary cytogenetics of the siabon (gibbon-siamang) hybrid apes; pp. 226–239. [Google Scholar]
- Liu R, Shi L, Chen Y. A study on the chromosomes of white-browed gibbon (Hylobates hoolock leuconedys) Acta Theriol Sinica. 1987;7:1–7. [Google Scholar]
- Garza JC, Woodruff DS. A phylogenetic study of the gibbons (Hylobates) using DNA obtained non-invasively from hair. Mol Phylogenet Evol. 1992;1:202–210. doi: 10.1016/1055-7903(92)90016-A. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Hayasaka K, Takenaka O, Horai S. Molecular phylogeny of gibbons inferred from mitochondrial DNA sequences: preliminary report. J Mol Evol. 1995;41:359–365. doi: 10.1007/BF01215183. [DOI] [PubMed] [Google Scholar]
- Hall LM, Jones DS, Wood BA. Evolution of the gibbon subgenera inferred from cytochrome b DNA sequence data. Mol Phylogenet Evol. 1998;10:281–286. doi: 10.1006/mpev.1998.0539. [DOI] [PubMed] [Google Scholar]
- Roos C, Geissmann T. Molecular phylogeny of the major hylobatid divisions. Mol Phylogenet Evol. 2001;19:486–494. doi: 10.1006/mpev.2001.0939. [DOI] [PubMed] [Google Scholar]
- Roos C. In: Conservation of Primates in Vietnam. Nadler T, Streicher U, Ha Thang Long, editor. Hanoi: Endangered Primate Rescue Center, Frankfurt Zoological Society; 2004. Molecular evolution and systematics of Vietnamese primates; pp. 23–28. [Google Scholar]
- Takacs Z, Morales JC, Geissmann T, Melnick DJ. A complete species-level phylogeny of the Hylobatidae based on mitochodrial ND3-ND4 gene sequence. Mol Phylogenet Evol. 2005;36:456–467. doi: 10.1016/j.ympev.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Chatterjee HJ. Phylogeny and biogeography of gibbons: a dispersal-vicariance analysis. Int J Primatol. 2006;27:699–712. doi: 10.1007/s10764-006-9044-1. [DOI] [Google Scholar]
- Monda K, Simmons RE, Kressirer P, Su B, Woodruff DS. Mitochondrial DNA hypervariable region-1 sequence variation and phylogeny of the concolor gibbons, Nomascus. Am J Primatol. 2007;69:1–22. doi: 10.1002/ajp.20439. [DOI] [PubMed] [Google Scholar]
- Roos C, Thanh Vu Ngoc, Walter L, Nadler T. Molecular systematics of Indochinese primates. Vietn J Primatol. 2007;1(1):41–53. [Google Scholar]
- Whittaker DJ, Morales JC, Melnick DJ. Resolution of the Hylobates phylogeny: congruence of mitochondrial D-loop sequences with molecular, behavioral, and morphological datasets. Mol Phylogenet Evol. 2007;45:620–628. doi: 10.1016/j.ympev.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Schultz AH. Observations on the growth, classification and evolutionary specialization of gibbons and siamangs. Human Biol. 1933;5:212–255. 385-428. [Google Scholar]
- Brandon-Jones D, Eudey AA, Geissmann T, Groves CP, Melnick DJ, Morales JC, Shekelle M, Stewart CB. Asian primate classification. Int J Primatol. 2004;25:97–164. doi: 10.1023/B:IJOP.0000014647.18720.32. [DOI] [Google Scholar]
- Mootnick AR, Groves CP. A new generic name for the hoolock gibbon (Hylobatidae) Int J Primatol. 2005;26:971–976. doi: 10.1007/s10764-005-5332-4. [DOI] [Google Scholar]
- Chivers DJ. In: Primate Conservation. Prince Rainier III of Monaco, Bourne GH, editor. London & New York: Academic Press; 1977. The lesser apes; pp. 539–598. [Google Scholar]
- Goodman M, Porter CA, Czelusniak J, Page SL, Schneider H, Shoshani J, Gunnell G, Groves CP. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol Phylogenet Evol. 1998;9:585–598. doi: 10.1006/mpev.1998.0495. [DOI] [PubMed] [Google Scholar]
- The Gibbon Research Lab. http://www.gibbons.de
- Raaum RL, Sterner KN, Noviello CM, Stewart CB, Disotell TR. Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence. J Hum Evol. 2005;48:237–257. doi: 10.1016/j.jhevol.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Sterner KN, Raaum RL, Zhang YP, Stewart CB, Disotell TR. Mitochondrial data support an odd-nosed colobine clade. Mol Phylogenet Evol. 2006;40:1–7. doi: 10.1016/j.ympev.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ. Paternal, maternal, and biparental molecular markers provide unique windows onto the evolutionary history of macaque monkeys. Evolution. 2003;57:1419–1435. doi: 10.1111/j.0014-3820.2003.tb00349.x. [DOI] [PubMed] [Google Scholar]
- Ziegler T, Abegg C, Meijaard E, Perwitasari-Farajallah D, Walter L, Hodges JK, Roos C. Molecular phylogeny and evolutionary history of Southeast Asian macaques forming the M. silenus group. Mol Phylogenet Evol. 2007;42:807–816. doi: 10.1016/j.ympev.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Roos C, Nadler T, Walter L. Mitochondrial phylogeny, taxonomy and biogeography of the silvered langur species group (Trachypithecus cristatus) Mol Phylogenet Evol. 2008;47:629–636. doi: 10.1016/j.ympev.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Hirai H, Hayano A, Tanaka H, Mootnick AR, Wijayanto H, Perwitasari-Farajallah D. In: The Gibbons. New Perspectives on Small Ape Socioecology and Population Biology. Lappan S, Whittaker DJ, editor. New York: Springer; 2009. Genetic differentiation of agile gibbons between Sumatra and Kalimantan in Indonesia; pp. 37–49. [Google Scholar]
- Carbone L, Mootnick A, Nadler T, Moisson P, Ryder O, Roos C, de Jong PJ. A chromosomal inversion unique to the northern white-cheeked gibbon. PLoS ONE. 2009;4:e4999. doi: 10.1371/journal.pone.0004999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudey AA. In: The Macaques: Studies in Ecology, Behavior and Evolution. Lindburg DG, editor. New York: Van Nostrand Rheinhold; 1980. Pleistocene glacial phenomena and the evolution of Asian macaques; pp. 52–83. [Google Scholar]
- Morley RJ, Flenley JR. In: Biogeographical Evolution of the Malay Archipelago. Whitmore TC, editor. Oxford: Oxford Scientific Publications; 1987. Late Cainozoic vegetational and environmental changes in the Malay Archipelago; pp. 50–59. [Google Scholar]
- Morley RJ. Origin and Evolution of Tropical Rain Forests. West Sussex: John Wiley & Sons; 2000. [Google Scholar]
- Bird MI, Taylor D, Hunt C. Palaeoenvironments of insular Southeast Asia during the last glacial period: a savanna corridor in Sundaland? Quaternary Sci Rev. 2005;24:2228–2242. doi: 10.1016/j.quascirev.2005.04.004. [DOI] [Google Scholar]
- Meijaard E. PhD thesis. Australian National University, Anthropology and Archaeology Department; 2004. Solving mammalian riddles. A reconstruction of the Tertiary and Quaternary distribution of mammals and their palaeoenvironments in island South-East Asia. [Google Scholar]
- Meijaard E, Groves CP. In: Primate Biogeography. Lehman SM, Fleagle JG, editor. New York: Springer; 2006. The geography of mammals and rivers in mainland Southeast Asia; pp. 305–329. full_text. [Google Scholar]
- Heaney LR. A synopsis of climatic and vegetational change in Southeast Asia. Climatic Change. 1991;19:53–61. doi: 10.1007/BF00142213. [DOI] [Google Scholar]
- Urushibara-Yoshino K, Yoshino M. Palaeoenvironmental change in Java island and its surrounding areas. J Quaternary Sci. 1997;12:435–442. doi: 10.1002/(SICI)1099-1417(199709/10)12:5<435::AID-JQS325>3.0.CO;2-E. [DOI] [Google Scholar]
- Kaars S van der. Pollen distribution in marine sediments from the south-eastern Indonesian waters. Palaeogeography, Palaeoclimatology, Palaeoecology. 2001;171:341–361. doi: 10.1016/S0031-0182(01)00253-X. [DOI] [Google Scholar]
- Cannon CH, Morley RJ, Bush ABG. The current refugial rainforests of Sundaland are unrepresentative of their biogeographic past and highly vulnerable to disturbance. Proc Natl Acad Sci USA. 2009;106:11188–11193. doi: 10.1073/pnas.0809865106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski NG, Whitfort MJ. Environmental changes during the Quaternary in East Asia and its consequences for mammals. Records of the Western Australian Museum. 1999;57:307–315. [Google Scholar]
- Meijaard E. Mammals of South-East Asian islands and their late Pleistocene environments. J Biogeography. 2003;30:1245–1257. doi: 10.1046/j.1365-2699.2003.00890.x. [DOI] [Google Scholar]
- Miller KG, Kominz MA, Browning JV, Wright JD, Mountain GS, Katz ME, Sugarman PJ, Cramer BS, Christie-Blick N, Pekar SF. The phanerozoic record of global sea-level change. Science. 2005;310:1293–1298. doi: 10.1126/science.1116412. [DOI] [PubMed] [Google Scholar]
- Lisiecki LE, Raymo ME. A Pliocene-Pleistocene stack of 57 globally distributed benthic δ18 O records. Paleoceanography. 2005;20:PA1003. doi: 10.1029/2004PA001071. [DOI] [Google Scholar]
- Naish TR, Wilson GS. Constraints on the amplitude of Mid-Pliocene (3.6-2.4?Ma) eustatic sea-level fluctuations from the New Zealand shallow-marine sediment record. Phil Trans R Soc A. 2009;367:169–187. doi: 10.1098/rsta.2008.0223. [DOI] [PubMed] [Google Scholar]
- Woodruff DS. Biogeography and conservation in Southeast Asia: how 2.7 million years of repeated environmental fluctuations affect today's patterns and the future of the remaining refugial-based biodiversity. Biodivers Conserv. in press .
- Verstappen HT. In: Modern Quaternary Research in Southeast Asia. Barstra GJ, Casparie WA, editor. Rotterdam: Balkema; 1975. On paleo-climates and landform development in Malesia; pp. 3–35. [Google Scholar]
- Tougard C. Biogeography and migration routes of large mammal faunas in South-East Asia during the late middle Pleistocene: focus on the fossil and extant faunas from Thailand. Palaeogeography, Palaeoclimatology, Palaeoecology. 2001;168:337–358. doi: 10.1016/S0031-0182(00)00243-1. [DOI] [Google Scholar]
- Woodruff DS, Turner LM. The Indochinese-Sundaic zoogeographic transition: a description and analysis of terrestrial mammal species distributions. J Biogeography. 2009;36:803–821. doi: 10.1111/j.1365-2699.2008.02071.x. [DOI] [Google Scholar]
- Chatterjee HJ. In: Gibbons: New Perspectives on Small Ape Socioecology and Population Biology. Lappan S, Whittaker DJ, editor. New York: Springer; 2009. Evolutionary relationships among the gibbons: a biogeographic perspective; pp. 13–36. [Google Scholar]
- Harrison T, Krigbaum J, Manser J. In: Primate Biogeography. Lehman SM, Fleagle JG, editor. New York: Springer; 2006. Primate biogeography and ecology on the Sunda Shelf islands: a paleontological and zooarchaeological perspective; pp. 331–374. full_text. [Google Scholar]
- Jablonski NG, Chaplin G. In: Gibbons: New Perspectives on Small Ape Socioecology and Population Biology. Lappan S, Whittaker DJ, editor. New York: Springer; 2009. The fossil record of gibbons; pp. 111–130. [Google Scholar]
- Peng YZ, Pan RL, Jablonski NG. Classification and evolution of Asian colobines. Folia Primatol. 1993;60:106–117. doi: 10.1159/000156680. [DOI] [PubMed] [Google Scholar]
- Jablonski NG. Natural History of the Doucs and Snub-nosed Monkeys. New Jersey: World Scientific Publishing Company; 1998. [Google Scholar]
- Hallet B, Molnar P. Distorted drainage basins as markers of crustal strain east of the Himalayas. J Geophysical Res. 2001;106:13697–13709. doi: 10.1029/2000JB900335. [DOI] [Google Scholar]
- Singh AD, Srinivasan MS. Quaternary climate changes indicated by planktonic forminifera of northern Indian ocean. Curr Sci. 1993;64:908–915. [Google Scholar]
- Hirai H, Wijayanto H, Tanaka H, Mootnick AR, Hayano A, Perwitasari-Farajallah D, Iskandriati D, Sajuthi D. A whole-arm translocation (WAT8/9) separating Sumatran and Bornean agile gibbons, and its evolutionary features. Chromosome Res. 2005;13:123–133. doi: 10.1007/s10577-005-7475-8. [DOI] [PubMed] [Google Scholar]
- Lappan S. Patterns of dispersal in Sumatran siamangs (Symphalangus syndactylus): preliminary mtDNA evidence suggests more frequent male than female dispersal to adjacent groups. Am J Primatol. 2007;69:692–698. doi: 10.1002/ajp.20382. [DOI] [PubMed] [Google Scholar]
- Long Y, Nadler T. In: Primates in Peril: The World's 25 Most Endangered Primates 2008-2010. Mittermeier RA, Wallis J, Rylands AB, Ganzhorn JU, Oates JF, Williamson EA, Palacios E, Heymann EW, Kierulff MCM, Long Y, Supriatna J, Roos C, Walker S, Cortés-Ortiz L, Schwitzer C, editor. Arlington: IUCN/SSC Primate Specialist Group (PSG), International Primatological Society (IPS), and Conservation International (CI); 2009. Eastern black crested gibbon Nomascus nasutus (Kunkel d'Herculais, 1884) pp. 60–61. [Google Scholar]
- Drummond AJ, Kearse M, Heled J, Moir R, Thierer T, Ashton B, Wilson A, Stones-Havas S. Geneious, version 4.6.1. 2008. http://www.geneious.com
- Hofreiter M, Siedel H, Van Neer W, Vigilant L. Mitochondrial DNA sequence from an enigmatic gorilla population (Gorilla gorilla uellensis) Am J Phys Anthropol. 2003;121:361–368. doi: 10.1002/ajpa.10186. [DOI] [PubMed] [Google Scholar]
- Thalmann O, Hebler J, Poinar HN, Pääbo S, Vigilant L. Unreliable mtDNA data due to nuclear insertions: a cautionary tale from analysis of humans and other great apes. Mol Ecol. 2004;13:321–335. doi: 10.1046/j.1365-294X.2003.02070.x. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), Version 4.0b10. Sunderland: Sinauer Associates; 2003. [Google Scholar]
- Zwickl DJ. PhD thesis. Texas University, Austin; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence data sets under the maximum likelihood criterion. [Google Scholar]
- Huelsenbeck JP, Ronquist F, Nielsen R, Bollback JP. Bayesian inference of phylogeny and its impact on evolutionary biology. Science. 2001;294:2310–2314. doi: 10.1126/science.1065889. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:e214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignaud P, Duringer P, Mackaye HT, Likius A, Blondel C, Boisserie JR, De Bonis L, Eisenmann V, Etienne ME, Geraads D, Guy F, Lehmann T, Lihoreau F, Lopez-Martinez N, Mourer-Chauvire C, Otero O, Rage JC, Schuster M, Viriot L, Zazzo A, Brunet M. Geology and palaeontology of the upper Miocene Toros-Menalla hominid locality, Chad. Nature. 2002;418:152–155. doi: 10.1038/nature00880. [DOI] [PubMed] [Google Scholar]
- Brunet M, Guy F, Pilbeam D, Lieberman DE, Likius A, Mackaye HT, Ponce de León MS, Zollikofer CP, Vignaud P. New material of the earliest hominid from the upper Miocene of Chad. Nature. 2005;434:752–755. doi: 10.1038/nature03392. [DOI] [PubMed] [Google Scholar]
- Lebatard AE, Bourles DL, Duringer P, Jolivet M, Braucher R, Carcaillet J, Schuster M, Arnaud N, Monie P, Lihoreau F, Likius A, Mackaye HT, Vignaud P, Brunet M. Cosmogenic nuclide dating of Sahelanthropus tchadensis and Australopithecus bahrelghazali: Mio-Pliocene hominids from Chad. Proc Natl Acad Sci USA. 2008;105:3226–3231. doi: 10.1073/pnas.0708015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley J. In: The Primate Fossil Record. Hartwig WC, editor. Cambridge: Cambridge University Press; 2002. The hominoid radiation in Asia; pp. 369–384. [Google Scholar]
- Benefit BR, McCrossin ML. In: The Primate Fossil Record. Hartwig WC, editor. Cambridge: Cambridge University Press; 2002. The Victoriapithecidae, Cercopithecoidea; pp. 241–253. [Google Scholar]
- Young NM, MacLatchy L. The phylogenetic position of Mortopithecus. J Hum Evol. 2004;46:163–184. doi: 10.1016/j.jhevol.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v1.4.1: MCMC trace analysis tool. Institute of Evolutionary Biology, University of Edinburgh; 2007. [Google Scholar]
- Rambaut A. FigTree: Tree figure drawing tool, version 1.2.2. Institute of Evolutionary Biology, University of Edinburgh; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Origin, material type, sample provider/collector and GenBank accession numbers of studied gibbon specimens.


