Summary
The cellular and viral determinants required for HIV-1 infection of nondividing cells have been a subject of intense scrutiny. Here we identify the 68 kDa subunit of cleavage factor Im, CPSF6, as an inhibitor of HIV-1 infection. When enriched in the cytoplasm by high level expression or mutation, CPSF6 prevents nuclear entry of the virus. Similar to TRIM5 and Fv1 type restrictions, CPSF6 targets the viral capsid (CA). N74D mutation of the HIV-1 CA leads to a loss of interaction with CPSF6 and evasion of the nuclear import restriction. Interestingly, N74D mutation of CA changes HIV-1 nucleoporin (NUP) requirements. Whereas wild-type HIV-1 requires NUP153, N74D HIV-1 mimics the NUP requirements of feline immunodeficiency virus (FIV) and is more sensitive to NUP155 depletion. These findings reveal a remarkable flexibility in HIV-1 nuclear transport and highlight a single residue in CA as essential in regulating interactions with NUPs.
Introduction
The synthesis of viral DNA (vDNA) from an RNA genome precursor and the insertion of the linear vDNA into the host cell chromatin are defining characteristics of retroviral replication. While the contributions of virion enzymatic proteins in reverse transcription and integration have been elegantly elaborated, the interaction of retroviruses with the host cell environment during early replication is less well understood. Progression from reverse transcription to integration requires the transport of a mega-Dalton complex of nascent vDNA and associated virion proteins, comprising the retroviral preintegration complex (PIC), across the nuclear membrane. Too large for passive diffusion through nuclear pore complexes, PICs are presumably dependent on host cell mechanisms to enter the nucleus (Fassati, 2006). Gammaretroviruses, such as murine leukemia virus (MLV), generally require progression through mitosis to integrate their genomes (Lewis and Emerman, 1994; Roe et al., 1993), but have also been observed to infect nonproliferating monocytes that are stimulated to differentiate (Jarrosson-Wuilleme et al., 2006). Lentiviruses, in contrast, integrate their genomes in both dividing cells as well as terminally differentiated cells, such as macrophages (Fassati, 2006).
The mechanisms exploited by lentiviruses, in particular HIV-1, to infect nondividing cells have been a subject of debate. Lentiviral PICs have been proposed to enter the nucleus via nuclear localization signal (NLS)-dependent and -independent pathways. A number of viral determinants, including matrix (MA), integrase (IN), Vpr, and discontinuous, triple-stranded vDNA present at the HIV-1 central polypurine tract, have been suggested to play key roles in nuclear entry (Bouyac-Bertoia et al., 2001; Bukrinsky et al., 1993; de Noronha et al., 2001; Gallay et al., 1997; Gallay et al., 1995a; Gallay et al., 1995b; Haffar et al., 2000; Heinzinger et al., 1994; Popov et al., 1998; Zennou et al., 2000). While these elements are either essential or can enhance the infection of dividing and nondividing cells, their specific contributions to nuclear entry have been questioned (Bukrinsky, 2004; Dvorin et al., 2002; Freed et al., 1995; Freed and Martin, 1994; Limon et al., 2002; Yamashita and Emerman, 2005, 2006).
One HIV-1 “nuclear-entry” determinant that has received scrutiny is the capsid (CA) protein, which comprises the core shell of mature retrovirus particles. CA dissociates from the HIV-1 reverse-transcription complex (RTC) prior to nuclear entry (Fassati and Goff, 2001; McDonald et al., 2002). The mechanism by which HIV-1 separates from its CA core before accessing the nuclear pore is unclear, but data suggest that substantial levels of CA may remain associated (Arhel et al., 2007; Dismuke and Aiken, 2006). Chimeric retroviruses in which HIV-1 CA is replaced with MLV CA are unable to infect nondividing cells (Yamashita and Emerman, 2004). Specific point mutations in CA can also impair the ability of HIV-1 to infect nondividing transformed cells and primary human macrophages (Yamashita et al., 2007).
HIV-1 CA mutants impaired in the infection of nondividing cells have a spectrum of phenotypes. For example, CA mutant Q63A/Q67A is impaired for nuclear entry and retains elevated levels of PIC-associated CA protein (Dismuke and Aiken, 2006). CA mutant T54A/N57A efficiently delivers its viral genome to the nucleus of nondividing cells but fails to integrate (Yamashita et al., 2007). Collectively such data suggest HIV-1 core dissociation, nuclear transport, and integration are tightly coupled processes.
The interaction of cellular factors with CA has been suggested to regulate HIV infection of nondividing cells (Yamashita et al., 2007). However, HIV-1 CA has not been directly associated with nuclear import factors. Defective tRNA species can facilitate the nuclear transport of HIV-1 RTCs in an in vitro system, possibly serving to tether the RTC to proteins that traffic to the nucleus (Zaitseva et al., 2006). How these tRNAs interact with the RTC has not yet been elucidated. Another study suggested that cyclophilin A (CypA) interactions, at least for certain CA mutant viruses, might regulate infection of nondividing cells after vDNA nuclear entry (Qi et al., 2008).
Genome-wide screens using RNA-interference (RNAi) technologies have revealed a wealth of host factors that, when depleted from cells, can limit HIV-1 infection (Brass et al., 2008; König et al., 2008; Zhou et al., 2008). The majority of these factors have not yet been carefully analyzed for direct interaction with HIV-1 or its infection pathway. One exception is TNPO3, a karyopherin known to transport S/R family proteins, which was identified in two functional screens to be required for HIV-1 infection at a step after reverse transcription (Brass et al., 2008; König et al., 2008). Subsequent work suggested that TNPO3 facilitates the nuclear transport of the PIC (Christ et al., 2008). Nuclear pore associated factors were also identified as possible HIV-1 cofactors in the same two screens. Although it is not known how these various NUP proteins interact with HIV-1, infection with HIV-1 mutants that evade restrictions can be used to identify co-factors that play a pivotal role in virus infection.
In this study, we sought to identify proteins that interfere with the early steps of HIV-1 infection through a functional screening approach using a cDNA expression library. We identified an S/R family protein, cleavage and polyadenylation factor 6 (CPSF6), that when C-terminally truncated, potently restricts HIV-1 and SIV infection, but not infection by other retroviruses. Cells ectopically expressing CPSF6 impaired the nuclear entry of HIV-1. A single amino acid substitution within CA bypasses this restriction. This mutant virus has provided a unique tool with which we have probed the nuclear pore requirements for HIV-1 infection and biologically linked CA to transport through the macromolecular structure.
Results
C-Terminally Truncated CPSF6 Identified in a Screen for HIV-1 Interfering Proteins
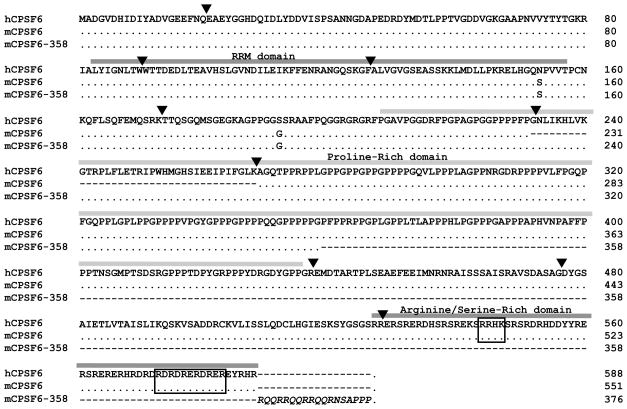
We identified cells resistant to infection with an HIV-1 vector using a cDNA expression screen (Supplemental Figure 1). Independent clones of these cells contained a cDNA encoding a splice variant of the 68 kDa component of cleavage factor (CF) Im (Ruegsegger et al., 1998), also known as CPSF6 (Figure 1). The cDNA present in the resistant cells contained an exon (exon 6 in humans) normally spliced from human and mouse mRNA encoding the 68 kDa form of CPSF6, and the cDNA was prematurely truncated within the subsequent exon as a result of random cDNA priming.
Figure 1.
Alignment of the predicted open reading frames of wild-type human CPSF6, wild-type mouse CPSF6 (68 kD splice variant), and mCPSF6-358. Human residues encoded by exon 6 are shown, though, as indicated for mCPSF6, this stretch is missing from the 68 kD form. Exon boundaries and protein domains are denoted by downward triangles and shaded boxes, respectively. Protein domains including the RNA-recognition motif (RRM) are labeled (Ruegsegger et al., 1998), and NLS determinants are boxed (Dettwiler et al., 2004). Amino acids in italics are acquired from the pMIGR1 vector and not needed for antiviral function (not shown).
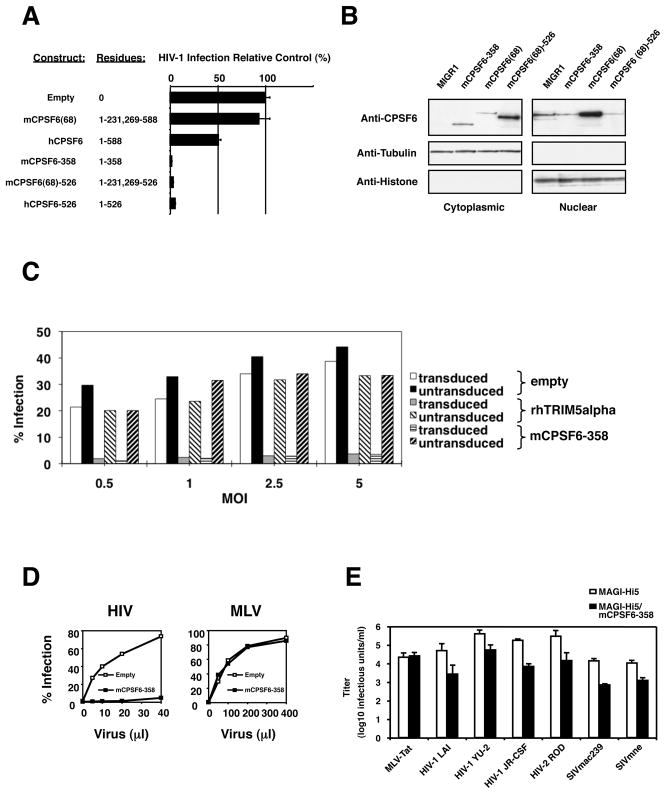
NIH3T3.hCycT1 cells stably expressing the truncated 358 amino acid protein obtained in the screen, mCPSF6-358, strongly restricted infection by the single-round vector HIV-RFP/VSV-G (Figure 2A). Elevated expression of the 68 kDa mouse CPSF6 did not impair susceptibility to the same virus; while stable expression of full-length human CPSF6 including exon 6 impaired infection. Truncation of mouse or human CPSF6 after residue 526, which removes the C-terminal Arginine/Serine-rich domain and elements required for nuclear localization (Figure 1), was sufficient to enhance antiviral function (Figure 2A). The truncated forms of CPSF6 that restrict HIV-1 are enriched in the cytoplasm (Figure 2B and Supplemental Figure 2) while endogenous CPSF6 is predominantly nuclear. Truncated CPSF6 also restricted HIV-1 in human cells. Expression of mCPSF6-358 in primary human CD4+ T cells was as effective as rhesus (rh) TRIM5alpha in rendering these cells resistant to HIV-1 infection (Figure 2C).
Figure 2. C-terminally truncated CPSF6 restricts HIV-1 infection.
(A) Infection of NIH3T3.hCycT1 cells stably expressing different CPSF6 molecules by HIV-RFP/VSV-G.
(B) Upper panels: Subcellular localization of endogenous, ectopically-expressed wild-type, and mutant CPSF6 molecules in NIH3T3.hCycT1 cell lines that express the mutant proteins. Western blot analysis was done with polyclonal antisera to determine the distribution of CPSF6 proteins in cells that contain only the empty MIGR1 vector versus those that express mCPSF6-358, wild-type mCPSF6(68), or the minimally truncated mCPSF6(68)-526. Lower panels: The blots were reprobed with tubulin and histone antibodies to validate cell fractionation.
(C) mCPSF6-358 blocks HIV-1 infection in primary T cells. CD4+ T cells activated through the TCR by using anti-CD3 and anti-CD28 as previously described (Unutmaz et al., 1999) were transduced with lentiviral vectors expressing IRES-HSA, rhTRIM5alpha-IRES-HSA, or mCPSF6-358-IRES-HSA. The infected cells were expanded in IL-2 containing media for 4 days and superinfected at different multiplicities of infection (m.o.i.) by HIV-eGFP/VSV-G. Numbers represent % of GFP+ cells over HSA+ cells day 4 post superinfection.
(D) Infection of HeLa cells expressing mCPSF6-358 with HIV-RFP/VSV-G and MX-RFP/VSV-G vectors. The percent of the cells that were infected was measured by FACS for RFP expression; the amounts of the virus containing supernatants is shown on the x-axis.
(E) Replication competent primate lentiviruses are inhibited by mCPSF6-358. Infections were performed under single-cycle conditions using MAGI cells expressing CCR5. Cells that expressed mCPSF6-358 are shown by the black bars. MLV expressing HIV-1 Tat and pseudotyped with VSV-G was used as a control.
Despite the antiviral properties of mCPSF6-358, endogenous CPSF6 did not appear to function as a cofactor for HIV-1 infection. Depletion of cellular CPSF6 via RNA interference did not impair HIV-RFP/VSV-G infection of HeLa cells (Supplemental Figure 3), and depletion of cellular CPSF6 with short-interfering (si) RNAs that discriminate between human and mouse CPSF6 forms did not relieve the block imposed by mCPSF6-358 in HeLa cells (Supplemental Figure 4). A consistent increase in HIV-1 infection was observed in control cells depleted for endogenous CPSF6 in these and other experiments.
To understand whether stable expression of mCPSF6-358 restricts infection by other retroviruses, control and mCPSF6-358-expressing HeLa cells were challenged with HIV-RFP/VSV-G or MLV-RFP/VSV-G (Figure 2D). HIV-1, but not MLV infection, was strongly impaired by mCPSF6-358. These data indicate that the antiviral function of mCPSF6-358 is specific for a subset of retroviruses. Similar data were obtained using HIV-1 and MLV pseudotyped with MLV amphotropic envelope glycoprotein (Supplemental Figure 5).
We next assayed whether other primate lentiviruses would be susceptible to mCPSF6-358 interference. Control and mCPSF6-358-expressing MAGI-Hi5 cells were challenged with replication competent HIV-1LAI, HIV-1YU-2, HIV-1JR-CSF, HIV-2ROD, SIVmac239, or SIVmne cl.8. An MLV vector encoding HIV-1 Tat and pseudotyped with VSV-G served as control. Although MLV-Tat/VSV-G infected both cell populations equally, the infectivities of HIV-1, HIV-2, and SIV were impaired approximately 10-fold or greater by mCPSF6-358 (Figure 2E). These effects were magnified in continuous replication assays. For three weeks or longer culture periods, HIV-1NL4-3, HIV-1NL4-AD8, HIV-1YU-2, and HIV-1JR-CSF did not replicate to detectable levels in HUT-R5 cells that expressed mCPSF6-358, although all of these viruses replicated efficiently in the parental cells (Supplemental Figure 6).
C-Terminally Truncated CPSF6 Blocks HIV-1 Trafficking to the Nucleus
Given the role of wild-type CPSF6 in pre-mRNA processing, it was conceivable that mCPSF6-358 interfered with HIV-1 expression. The presence of mCPSF6-358 in the cytoplasm could titrate essential transcription or processing factors from the nucleus, or low levels of the truncated protein might enter the nucleus and interfere with gene expression. However mCPSF6-358 had no measurable effect on HIV-1 expression (Supplemental Figure 7) or on the production of infectious virus (Supplemental Figure 8).
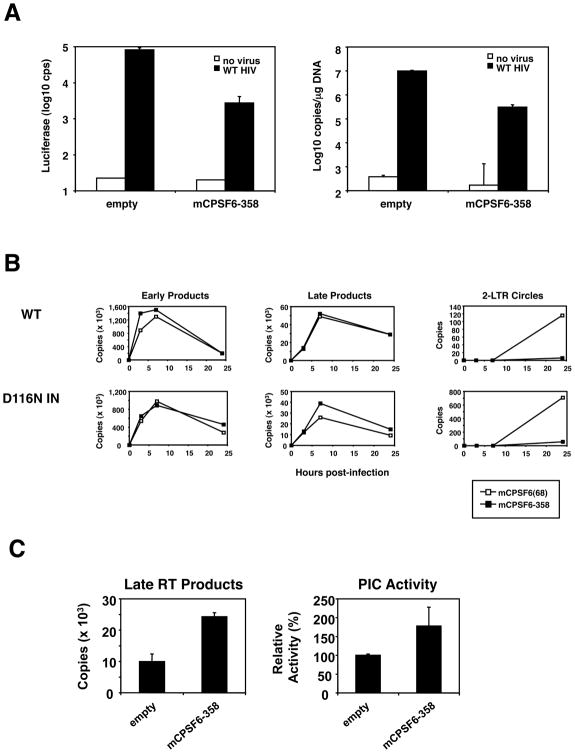
If mCPSF6-358 targeted an early replication step of HIV-1, fewer proviruses would be generated in the presence of the antiviral factor. Supporting this hypothesis, one week after infection with HIVNLdE-luc/VSV-G, NIH3T3.hCycT1 cells expressing mCPSF6-358 possessed 33-fold less vDNA than did NIH3T3.hCycT1 cells expressing an empty control vector (Figure 3A). This vDNA level was comparable with the 30-fold infection reduction measured by luciferase transduction three days after infection, presumably reflecting the number of integrated proviruses. Because virus entry pathways appeared to be intact in mCPSF6-358-positive cells, these data suggested that mCPSF6-358 might interfere with reverse transcription or integration.
Figure 3. Analysis of HIV-1 replication in the presence of truncated CPSF6.
(A) Total HIV-1 DNA is reduced in NIH3T3.hCycT1 cells expressing mCPSF6-358. Left: 3 d post-infection with HIV-luc/VSV-G, a portion of infected cells were analyzed for luciferase activity. Right: 7 d post-infection, DNA extracted from cells was analyzed by qPCR using gag-specific primers. Viral DNA copies per microgram genomic DNA are depicted on the y-axis.
(B) qPCR measurements of HIV-1 DNA synthesis. 293T cells expressing MIGR1-mCPSF6 or MIGR1-mCPSF6-358 and infected with HIV-HSA/VSV-G or HIVIN/D116N-HSA/VSV-G carrying an active site mutation in the viral IN (Engelman et al., 1995) were lysed at various times to measure the amount of viral DNA using primers for specific steps in reverse transcription. Early (minus strand), late (plus strand), and 2-LTR circle vDNA forms are labeled on the tops of the graphs.
(C) Late reverse transcripts and PIC activity from NIH3T3.hCycT1 cells expressing mCPSF6-358. NIH3T3/MIGR1 or NIH3T3/MIGR1-mCPSF6-358 cells infected with HIV-luc/VSV-G were lysed at 9 h post-infection. Late reverse transcripts were measured by qPCR as shown in the left graph, and the amount of PIC activity normalized to the levels of late reverse transcription products in the different samples is shown on the right. The activity of PICs extracted from empty vector control cells was arbitrarily set at 100%. Error bars indicate the standard errors of means derived from duplicate integration assays.
The course of reverse transcription was measured using quantitative PCR (qPCR) in 293T cells expressing wild-type mCPSF6 or mCPSF6-358 after infection with wild-type HIV-HSA/VSV-G or HIVIN/D116N-HSA/VSV-G, the latter vector encoding IN with an inactivating mutation (Figure 3B). mCPSF6-358 did not interfere with the kinetics of synthesis or the overall accumulation of early or late HIV-1 reverse transcription products. However, the accumulation of the 2-LTR circular form of vDNA, a dead-end form produced in the nucleus, was reduced in mCPSF6-358-expressing cells. Even infection with HIVIN/D116N-HSA/VSV-G, which is deficient in integration and thus produces higher levels of 2-LTR circular vDNA (Engelman et al., 1995), made far fewer circles in cells expressing mCPSF6-358 (Figure 3B).
Our analysis of reverse transcription in mCPSF6-358 expressing cells did not distinguish whether integration-competent forms of the vDNA were synthesized but blocked from entering the nucleus, or whether the linear vDNA was unable to circularize or integrate. We thus examined whether functional PICs were produced in the cytoplasm of HIV-1 infected cells that express mCPSF6-358. PIC function in vitro requires prior trimming of dinucleotides from both 3′ ends of linear vDNA (Chen and Engelman, 2001) and thus is a measure of functional DNA end synthesis. As shown in Figure 3C, functional HIV-1 PICs were readily isolated from the cytoplasm of mCPSF6-358-expressing cells. The integration efficiency was comparable for PICs isolated from cells that did, and did not, express mCPSF6-358. These data show that mCPSF6-358 does not interfere with the completion of reverse transcription by HIV-1 and moreover suggest that mCPSF6-358 does not directly interfere with IN function. Given the presence of functional PICs in the cytoplasm of mCPSF6-358 expressing cells and the absence of 2-LTR circle forms or persisting vDNA in the same cells, we inferred that mCPSF6-358 interfered with the nuclear entry of HIV-1 PICs.
Mutation of HIV-1 CA Confers Resistance to Truncated CPSF6 and Alters the Infection of Nondividing Cells
We next asked whether we could select for resistance to mCPSF6-358. Most HIV-1 isolates we tested showed no replication in HUT-R5 cells that constitutively expressed mCPSF6-358 (Supplemental Figure 6 and not shown). An exception was HIV-1 NL4-3/BaL which replicated to detectable levels under these conditions (Figure 4A). HIV-1NL4-3/BaL that had been passaged in the presence of mCPSF6-358 importantly revealed wild-type viral replication kinetics when passed onto new HUT-R5.mCPSF6-358 cells (Supplemental Figure 9). Sequence analysis of vDNA isolated from cells freshly infected with the resistant stock revealed a AAT -> GAT mutation in codon 74 of CA, causing an N74D change in the protein. N74 and its surrounding residues are highly conserved amongst known HIV-1, HIV-2, and SIV isolates (http://www.hiv.lanl.gov).
Figure 4. A mutation in HIV-1 CA confers resistance to mCPSF6-358.
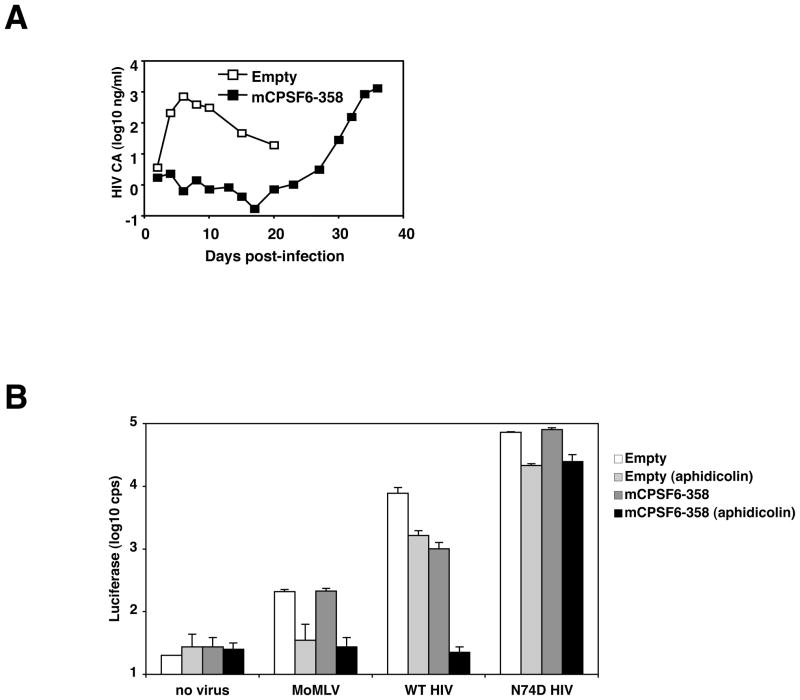
(A) Selection for HIV-1 resistance to mCPSF6-358. HUT-R5 cells that contain the empty vector or HUT-R5.mCPSF6-358 cells were infected with HIV-1NL4-3/BaL and passaged every 2–3 days. Culture supernatants were collected and analyzed for CA by HIV-1 p24 ELISA.
(B) HIV-1 with the N74D mutation in CA efficiently infects nondividing HeLa cells and infection is not blocked in nondividing HeLa cells expressing mCPSF6-358. HeLa.MIGR1 and HeLa.MIGR1-mCPSF6-358 cells were maintained in the presence or absence of aphidicolin and infected with MLV, HIVNLdE-luc/VSV-G, or HIVNLdE CA N74D-luc/VSV-G. Two days after infection, cells were lysed and assayed for luciferase activity.
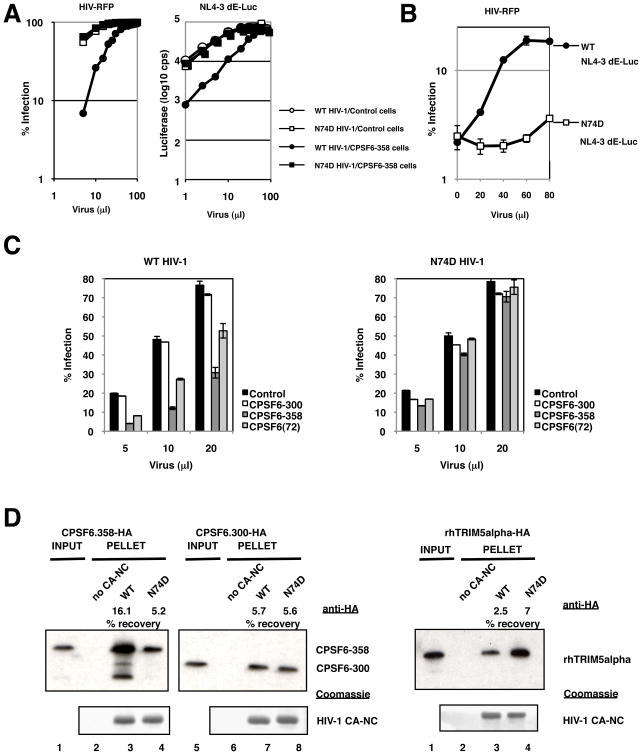
We tested the effect of this single substitution on the replication of HIV-1NL4-3/BaL and HIVNLdE-luc/VSV-G in cells expressing mCPSF6-358. Similar to the uncloned resistant isolate, HIV-1NL4-3 CA N74D/BaL was neither impaired for infection or replication in HUT-R5.mCPSF6-358 cells (not shown). The single-cycle vector carrying the N74D mutation (HIVNLdE CA N74D-luc/VSV-G) also infected cells expressing an empty vector or mCPSF6-358 equally well; whereas the wild-type virus was restricted about 10-fold by mCPSF6-358 (Figure 4B). These data demonstrated that the N74D CA mutation was necessary and sufficient for HIV-1 resistance to mCPSF6-358.
Prior experiments suggested that mCPSF6-358 interferes with the nuclear entry of HIV-1 PICs. If true, the mCPSF6-358 block could be intensified in nonmitotic cells where HIV-1 PICs must traverse an intact nuclear membrane to gain access to host cell chromatin. To test this prediction, HeLa cells expressing an empty vector or mCPSF6-358 were arrested in the G1/S phase of the cell cycle with aphidicolin, an inhibitor of DNA polymerase alpha and delta. Cell-cycle arrest was verified by FACS analysis of the aphidicolin-treated cells (not shown). Equal numbers of arrested and untreated HeLa cells were challenged with MLV-luc/VSV-G and HIVNLdE-luc/VSV-G (Figure 4B). Although MLV was insensitive to mCPSF6-358 expression, growth arrest of HeLa cells, as expected, abolished MLV infection. In contrast, the aphidicolin-induced growth arrest modestly impaired HIV-1 infection of HeLa cells expressing the empty vector, partly due to toxicity. The combination of growth arrest and mCPSF6-358 expression potently interfered with HIV-1 infection. The block was approximately 70-fold relative to nondividing control cells, near the limit of sensitivity of the assay. Growth arrest by etoposide, a less toxic inhibitor, revealed a similar enhancement in mCPSF6-358 restriction (Supplemental Figure 10). In nondividing cells expressing mCPSF6-358, HIV-1 infection is no more efficient than MLV infection. By contrast, infection by HIVNLdE CA N74D-luc/VSV-G was unaffected by the combination of cell-cycle arrest and mCPSF6-358 (Figure 4B).
CPSF6-358 Binds HIV-1 Core Complexes
The ability of the N74D mutant to evade mCPSF6-358 restriction suggested that the antiviral protein might directly target wild-type CA protein. Supporting such a model, challenge of 293T cells expressing human CPSF6-358 with increasing amounts of virus resulted in saturation of the block such that wild-type HIV-1 infected the restrictive cells as well as it did control cells, or as well as N74D HIV-1 infected CPSF6-358-expressing cells (Figure 5A). We thus tested whether pre-exposure of CPSF6-358-expressing cells to virions with wild-type CA could abrogate the infection restriction to a second virus. CPSF6-358-expressing cells were first infected with increasing amounts of wild-type and N74D HIV-1NLdE-luc. Soon after, cells were challenged with a fixed volume of wild-type HIV-RFP. Only cells pre-exposed to higher amounts of wild-type HIV-1NL4dE-luc were permissive to HIV-RFP infection (Figure 5B). Pre-infection with N74D HIV-1NL4dE-luc did not saturate the CPSF6-358 block to wild-type HIV-1.
Figure 5. CPSF6-358 interacts with wild-type HIV-1 CA.
(A) Different HIV-1 vectors can saturate the CPSF6-358 restriction. 293T.LPCX (control) or 293T.LPC-CPSF6-358-HA cell populations were challenged with increasing volumes of WT vs. N74D HIV-RFP/VSV-G or luciferase vectors (HIVNLdE-luc/VSV-G or HIVNLdE CA N74D-luc/VSV-G). Infectivity was measured by FACS or luciferase activity.
(B) Pre-infection with WT HIV-1 abrogates the CPSF6-358 restriction to a second virus. 293T.LPC-CPSF6-358-HA cells were challenged with increasing volumes of HIVNLdE-luc/VSV-G or HIVNLdE CA N74D-luc/VSV-G and then with a fixed amount of WT HIV-RFP/VSV-G. Infectivity by WT HIV-RFP/VSV-G was measured by FACS.
(C) CPSF6-300 does not restrict WT HIV-1. WT vs. N74D HIV-RFP/VSV-G infection of 293T cells transiently transfected with empty vector (control) or HA-tagged constructs expressing human CPSF6-300, CPSF6-358, or CPSF6 (encoding the full length 72 kD isoform). The percent of the cells that were infected was measured by FACS for RFP expression; the amounts of the virus containing supernatants is shown on the x-axis.
(D) Enhanced binding of CPSF6-358 to WT CA-NC complexes. Recombinant WT and N74D CA-NC complexes were incubated with lysates from 293T cells transfected with HA-tagged constructs expressing CPSF6-300, CPSF6-358, or rhTRIM5alpha. Mixtures were then layered on top of a 70% sucrose cushion followed by ultracentrifugation. Top panels are Western blots using anti-HA; bottom panels are Coomassie stains of pelleted material. Lanes 1 and 5 are lysate input lanes that were not subjected to ultracentrifugation. Lanes 2 and 6 are pelleted material in the absence of CA-NC complexes. Recovery of HA-tagged proteins relative to input was measured by densitometry.
These findings implied a CA-specific interaction with CPSF6-358. We sought to test whether a physical interaction between both proteins could be detected. As a control for such assays, we employed another cytoplasmic form of CPSF6, the truncation mutant CPSF6-300. Transient expression of CPSF6-300 in 293T cells did not impair wild-type HIV-1 infection; by contrast, transient transfection of constructs expressing CPSF6-358 and even a full-length splice variant, CPSF6(72), restricted wild-type but not N74D HIV-1 (Figure 5C).
To assay for binding of CPSF6-358 to CA, we prepared wild-type and N74D CA-NC complexes in vitro (Ganser et al., 1999). The pre-assembled complexes were then incubated with lysates of 293T cells transfected with CPSF6-358, CPSF6-300, or rhTRIM5alpha, and these mixtures were ultracentrifuged through a sucrose cushion. As shown previously (Stremlau et al., 2006), wild-type CA-NC complexes pellet rhTRIM5alpha under these conditions (Figure 5D, right panel). Interestingly, N74D CA-NC complexes more efficiently precipitated rhTRIM5alpha (about two-fold) than wild-type CA-NC complexes in four separate trials (data not shown). In comparison, CPSF6-300 purified with wild-type and N74D CA-NC complexes at an equal efficiency (Figure 5D, middle panel) in this and other experiments. By contrast, CPSF6-358 co-purified more efficiently with wild-type versus N74D CA-NC cores (Figure 5D, left panel), at an average difference of 3.9-fold over four trials (data not shown).
CA Regulates the Interactions of HIV-1 PICs with Transport and Nuclear Pore Associated Co-Factors
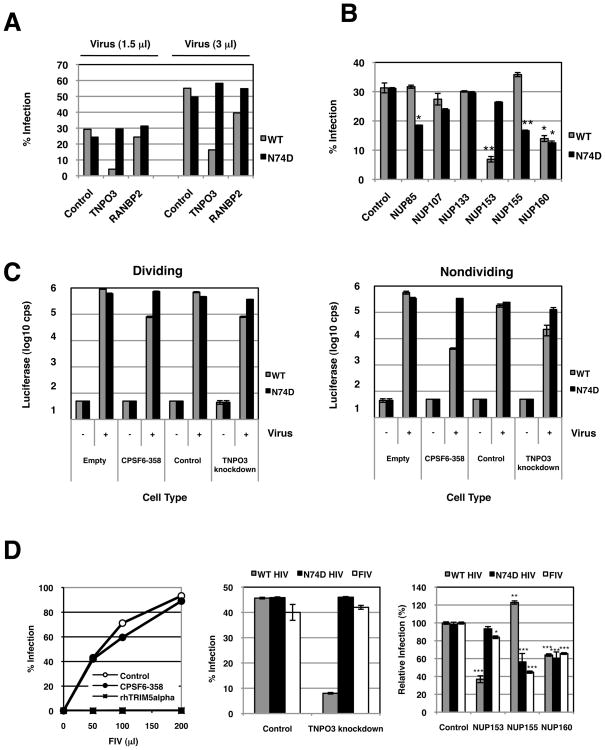
Having demonstrated that the N74D mutation allowed HIV-1 to escape the mCPSF6-358 block to nuclear entry, we asked whether any of the recently described host factors implicated in the nuclear transport of HIV-1 PICs were CA-dependent. We depleted HeLa cells of the karyopherin, TNPO3, and the nuclear pore-associated factor, RANBP2, by transient transfection of siRNAs. It has been previously shown by Brass and colleagues (Brass et al., 2008) that the knockdown of either of these proteins impaired HIV-1 infection and we observed similar results, most strongly upon TNPO3 depletion (Figure 6A). Notably, TNPO3 protein levels (Supplemental Figure 11) and RANBP2 protein levels (Supplemental Figure 12) were reduced after siRNA transfection, although RANBP2 knockdowns were more cytotoxic to cell cultures (data not shown). Infection with N74D HIV-1 was relatively unaffected by TNPO3 or RANBP2 depletion. The observation that infection with the N74D mutant was independent of TNPO3 suggested that this virus was transported to the nuclear pore by a pathway different from wild-type HIV-1, and thus might interact with different pore components.
Figure 6. WT and N74D HIV-1 require different nuclear pore factors.
(A) N74D HIV-1 vectors do not require TNPO3 or RANBP2 for infection. HeLa cells were transiently transfected with control siRNA oligos and with oligos directed at TNPO3 or RANBP2. One day after siRNA transfection, cells were replated for virus challenge. Two days after siRNA transfection, the transfected cells were infected with 1.5 and 3.0 μl of WT or N74D HIV-RFP/VSV-G and assayed 48 h later by FACS.
(B) Differential NUP requirements by WT HIV-1 and the N74D mutant. HeLa cells were transfected with siRNAs that target different NUP genes (NUP85, NUP107, NUP133, NUP153, NUP155, and NUP160). Nontargeting siRNA was also transfected as a control. Infections with WT or N74D HIV-RFP/VSV-G were performed in duplicate and assayed 48 h later by FACS. Statistical analysis was performed by Student’s t test; *p < 0.05 and **p <0.01 versus WT or N74D HIV-1 infection of respective control infected cells.
(C) Cell growth arrest does not increase the dependence of WT HIV-1 on TNPO3. HeLa cells that contain only the empty vector or cells that express mCPSF6-358 and HeLa cells transduced with control shRNA vector or TNPO3 shRNA were infected with either WT HIV-1 or N74D mutant vectors that express a luciferase reporter (HIVNLdE-luc/VSV-G or HIVNLdE CA N74D-luc/VSV-G, respectively). Cell lysates were measured for luciferase activity 48 h after infection. Luciferase values reflect averages of duplicate infections with standard deviations. The data in the right panel were from cells treated with aphidicolin.
(D) N74D HIV-1 is an FIV phenocopy. Left panel, 293T cells stably expressing BabePuro (empty vector control), BabePuro-mCPSF6-358, or LPC-rhTRIMalpha were infected with FIV-GFP(GinSin)/VSV-G (Loewen et al., 2003). The percentage of infected cells was measured by FACS for GFP expression; the amounts of the virus containing supernatants is shown on the x-axis. Middle panel, HeLa.control_shRNA or HeLa.TNPO3_shRNA cells were infected in duplicates with WT HIV-RFP/VSV-G, N74D HIV-RFP/VSV-G, or FIV-GFP/VSV-G. Infection was measured by FACS. Right panel, HeLa cells transiently transfected with nontargeting and NUP targeting siRNA were infected in duplicates with WT HIV-RFP/VSV-G, N74D HIV-RFP/VSV-G, or FIV-GFP/VSV-G. The average of four independent experimental trials is depicted as a relative infection value. Statistical analysis was performed by Student’s t test; *p = 0.0205, **p = 0.0009, and ***p < 0.0001 relative WT or N74D HIV-1 infection of respective control infected cells.
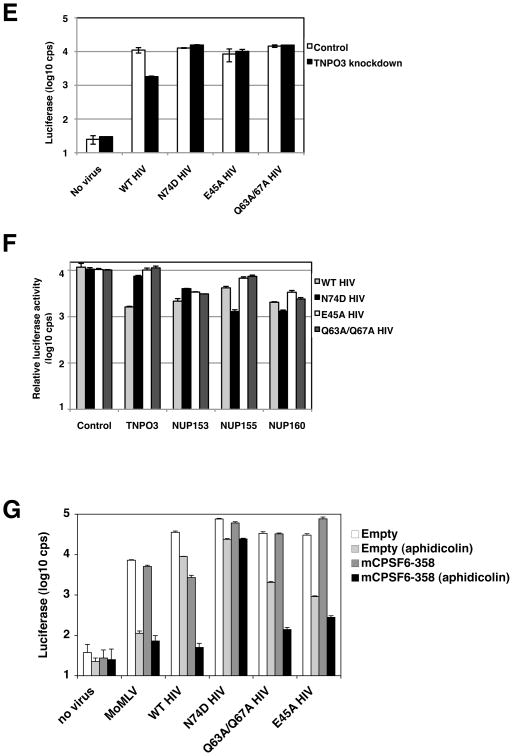
(E) E45A and Q63A/Q67A HIV-1 are TNPO3-independent. HeLa cells stably transduced with control shRNA or TNPO3 shRNA vectors were infected with WT HIV-1 or different CA mutant HIV-1 (HIVNLdE-luc/VSV-G, HIVNLdE CA N74D-luc/VSV-G, HIVNLdE CA E45A-luc/VSV-G, or HIVNLdE CA Q63A/Q67A-luc/VSV-G). After 48 h of infection, cell lysates were collected to measure luciferase activity. Luciferase values reflect averages of duplicate infections with standard deviations.
(F) E45A and Q63A/Q67A HIV-1 are less sensitive to NUP depletions. HeLa cells were transiently transfected with nontargeting siRNA or siRNAs that target different host factors (TNPO3, NUP153, NUP155, and NUP160). Cells were infected with HIVNLdE-luc/VSV-G, HIVNLdE CA N74D-luc/VSV-G, HIVNLdE CA E45A-luc/VSV-G, or HIVNLdE CA Q63A/Q67A-luc/VSV-G and lucifease activity from lysates was measured 48 h later. Control cell virus infections are within 10-fold of their actual activities and were used to normalize infection values of other samples.
(G) CPSF6-358 restriction of E45A or Q63A/Q67A HIV-1 is cell-cycle dependent. HeLa.MIGR1 and HeLa.MIGR1-mCPSF6-358 cells maintained in the presence or absence of aphidicolin were infected in duplicate with MLV, HIVNLdE-luc/VSV-G, HIVNLdE CA N74D-luc/VSV-G, HIVNLdE CA E45A-luc/VSV-G, or HIVNLdE CA Q63A/67A-luc/VSV-G. Cells were lysed and assayed for luciferase activity two days after infection. Error bars represent standard deviations of duplicates.
We next treated cells with siRNAs directed against NUP85, NUP107, NUP133, NUP153, NUP155, and NUP160, also identified as potential HIV-1 dependency factors (Brass et al., 2008). Knockdowns of NUP85, NUP107, NUP133, NUP153, and NUP155 were verified by Western blotting (Supplemental Figure 12). Although we did not have antibodies to detect NUP160 levels, siRNA targeting of NUP160 did not significantly impair infection by MLV (data not shown). Reductions in NUP153 or NUP160 led to a reproducible reduction of HIV-1 infection (Figure 6B). By contrast, N74D HIV-1 infection was more strongly diminished in cells transfected with siRNAs for NUP85 or NUP155, and it was also impaired in cells transfected with siRNA to NUP160.
We wanted to understand whether growth arrest would enhance the effects of the siRNAs for nuclear pore factors on HIV-1 infection. However, under the conditions that we employed, the combination of the siRNAs to NUPs and the different types of growth arrest was cytotoxic (data not shown). By contrast, cells transfected with siRNA for TNPO3 were able to survive growth arrest.
To minimize manipulation of cells, we developed HeLa cells stably depleted of TNPO3 (Supplemental Figure 13). The TNPO3 knockdown cells were similar to mCPSF6-358 expressing cells in that both types of cells were about 10-fold less susceptible to infection by wild-type HIV-1 than were control cell lines during normal growth (Figure 6C). As expected, infection by the N74D mutant was essentially unimpaired by mCPSF6-358 expression or TNPO3 depletion. When such cells were growth arrested, a different infection pattern emerged. As shown before, nondividing cells expressing mCPSF6-358 strongly restricted infection by wild-type HIV-1 but not N74D HIV-1. By contrast, the reduction of HIV-1 infection in TNPO3-depleted cells relative to control cells did not increase when the cells were growth arrested. Thus, the requirement for TNPO3 in HIV-1 infection was the same in dividing and nondividing cells. Together, these data indicate that the karyopherin and nuclear pore requirements for wild-type and N74D HIV-1 vary due to a single residue change in CA, and karyopherin use by HIV-1 may be cell-cycle independent.
Although primate lentiviruses were susceptible to mouse and human CPSF6-358 restriction, we observed feline immunodeficiency virus (FIV) to be insensitive to the antiviral factor (Figure 6D, left panel). By contrast, FIV is susceptible to CA-mediated restriction by rhTRIM5alpha (Saenz et al., 2005). To understand whether CPSF6-358 sensitivity predicted lentivirus interactions with nuclear transport or pore proteins, we examined the infection of FIV in different knockdown cells. Similar to N74D HIV-1, FIV was unimpaired in the infection of cells depleted of TNPO3 (Figure 6D, middle panel). We next tested cells transiently depleted of NUP153, NUP155, and NUP160, factors whose depletion more significantly affected either wild-type or N74D HIV-1 infection. Like N74D HIV-1, depletion of NUP155 or NUP160 impaired FIV infection whereas depletion of NUP153 was tolerated (Figure 6D, right panel). We did not observe effects on FIV infection of cells depleted for NUP85, NUP107, or NUP133 (data not shown).
Cell-Cycle Dependent HIV-1 Mutants Are TNPO3-Independent and Restricted by CPSF6-358
E45A or Q63A/Q67A mutation of CA impairs HIV-1 infection of cell-cycle arrested cells (Yamashita et al., 2007). We thus examined whether these changes in CA could alter HIV-1 interactions with transport factors or mCPSF6-358. Unlike wild-type HIV-1, E45A or Q63A/Q67A HIV-1 infection was unaffected by reduced levels of TNPO3 in stable knockdown cells (Figure 6E).
TNPO3-independence by E45A and Q63A/Q67A HIV-1 prompted further examination of their NUP requirements in siRNA-treated cells. As in previous experiments, wild-type HIV-1 infection was more greatly diminished after transient depletion of TNPO3, NUP153, or NUP160 (Figure 6F). N74D HIV-1 infection was again decreased in cells depleted of NUP155 and NUP160. Under these experimental conditions (see Supplemental Text for Figure 6), NUP153 depletion of HeLa cells also modestly impaired N74D HIV-1 infection. The infection profiles of E45A and Q63A/Q67A HIV-1 were distinct. These CA mutants viruses were typically more resistant to the depletion of NUPs in target cells. Not only were they TNPO3-independent, E45A and Q63A/Q67A HIV-1 were less affected by siRNA targeting of NUP155 than N74D HIV-1.
These data collectively implied a reduced dependence of E45A and Q63A/Q67A HIV-1 on the nuclear pore for infection, or, stated differently, an inefficient interaction by these HIV-1 mutants with the nuclear pore complex. If these viruses were less dependent on CA interactions with pore components, then mCPSF6-358 might not interfere with their infection. Indeed E45A or Q63A/Q67A HIV-1 was not restricted in dividing cells expressing mCPSF6-358 (Figure 6G). Consistent with past reports, both E45A and Q63A/Q67A HIV-1 were impaired approximately 30-fold and 15-fold, respectively, in the infection of aphidicolin-treated control HeLa cells (Figure 6G). Because these viruses retained the N74 residue that appears critical for CPSF6-358 interactions, we next tested infection interference in nondividing cells, when transport through the nuclear pore would be required to access cellular chromatin. In marked contrast to N74D HIV-1, mCPSF6-358 expression in the growth arrested HeLa cells strongly restricted infection by either E45A or Q63A/Q67A HIV-1. E45A HIV-1 infection was reduced 275-fold relative to dividing cells expressing mCPSF6-358, and Q63A/Q67A HIV-1 infection was decreased nearly 230-fold compared to cycling counterparts (Figure 6G).
Discussion
Nondividing cells provide an important reservoir for HIV-1 infection in vivo; thus the viral determinants and cellular factors required for HIV-1 nuclear entry have been subjects of considerable interest. A mechanistic understanding of this process has been limited by a failure to link viral regulators of nuclear entry to host factors involved in nuclear entry. Here we have identified an antiviral protein that restricts HIV-1 nuclear entry, selected a CA mutant HIV-1 that overcomes this block, and shown that CA regulates HIV-1 dependence on nuclear transport and pore proteins.
While our study primarily focused on CPSF6-358 restriction of HIV-1, depletion of endogenously expressed CPSF6 enhanced HIV-1 susceptibility, and reciprocally, ectopic expression of full length forms of CPSF6 impaired virus infection. In vivo expression of CPSF6 could thus restrict primate lentiviruses, especially under circumstances where the cytoplasmic concentration of the protein is elevated. The interaction of CA with the host cell cytoplasm after entry is of interest in HIV-1 antiviral development. TRIM5 and CPSF6 derived restrictions indicate significant vulnerability of primate lentiviruses during these early replication steps and suggest the feasibility of developing specific inhibitors targeting CA. In turn, antiviral drugs often provide important probes to study the replication of viruses. With this model in mind, we have used CPSF6-358 to examine early replication step requirements of HIV-1 and illuminated the role of CA in regulating nuclear entry.
Expression Screen and Restriction by mCPSF6-358
Wild-type CPSF6 is a component of CF Im, a complex that participates in the polyadenylation of pre-mRNA and is enriched in the nucleus (Dettwiler et al., 2004; Ruegsegger et al., 1998). We showed that a C-terminally truncated version of mCPSF6, mCPSF6-358, that is localized in the cytoplasm, potently restricts HIV-1 infection. HIV-2 and other SIV isolates are also restricted by mCPSF6-358, but MLV is not. HIV-1 is restricted in primary CD4+ T cells expressing mCPSF6-358, indicating the pathway targeted is important in cells that are infected by HIV-1 in patients.
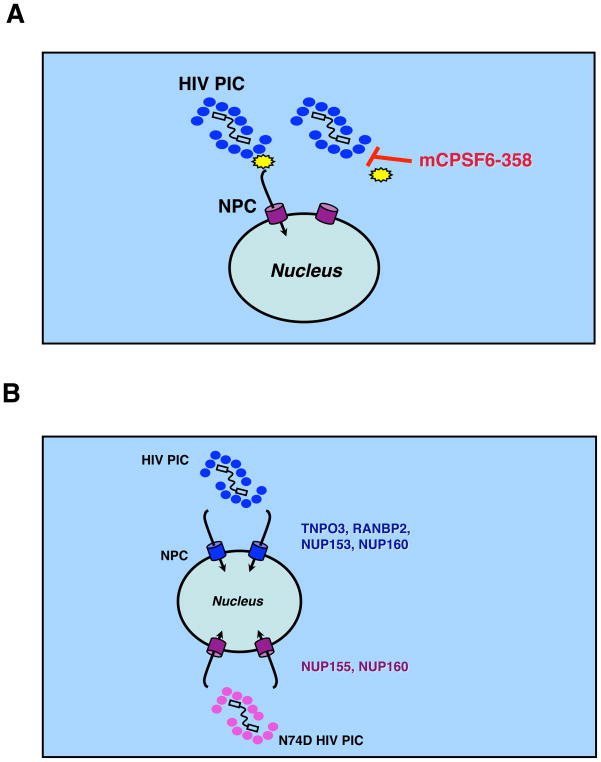
The finding that human CPSF6-358 physically interacts with wild-type HIV-1 in virus saturation experiments and with recombinant CA-NC complexes in precipitation assays suggests that CPSF6-358 targets a core-associated co-factor or directly binds to core complexes. Because there is no a priori evidence of CPSF6 aiding HIV-1 infection, we favor the hypothesis that CPSF6-358 interaction with incoming HIV-1 cores impairs productive interactions with uncoating or transport factors (Figure 7A). Indeed TRIM5 or CypA-mediated restrictions can also be enhanced in nondividing cells and have been hypothesized, at least in the case of rhTRIM5alpha, to accelerate HIV-1 CA dissociation (Yamashita and Emerman, 2009).
Figure 7. Models for HIV-1 nuclear entry.
(A) HIV-1 nuclear entry is restricted by CPSF6-358 interaction with the CA core (blue ovals), thus interfering with HIV-1 core disassembly or interaction with a host transport factor (yellow polygon).
(B) HIV-1 CA regulates nuclear transport and pore protein requirements for infection. Depletion of TNPO3, RANBP2, and NUP153 diminishes WT but not N74D HIV-1 infection. By contrast, depletion of NUP155 limits N74D HIV-1 and FIV infection. Knockdown of NUP160 impaired infection by different lentiviruses.
Does CPSF6-358 target the CA directly or through an intermediate protein? Given that CPSF6 interacts with the polyadenylation machinery (Ruegsegger et al., 1998), spliceosomes (Rappsilber et al., 2002; Zhou et al., 2002), and localizes with the nuclear paraspeckles (Dettwiler et al., 2004), the number of potential interacting factors is large. CA interaction with a co-factor could also explain the conservation of N74 and flanking residues among primate lentiviruses. N74D CA has so far been observed in one primate lentiviral isolate, SIVcpz MT145 (Keele et al., 2006), but is otherwise rare in known HIV-1/HIV-2/SIV isolates. Because N74D mutation of HIV-1 CA enhanced rhTRIM5alpha binding, it is conceivable that TRIM5alpha in vivo additionally counterselects an N74D core in primate lentiviruses.
Cytoplasmic CPSF6, rhTRIM5alpha, and TRIMCyp proteins target HIV-1 CA through different residues, and in the case of the latter two restriction factors, provide formidable cross-species transmission barriers. As a focal point for post-entry restriction factors, the retention of CA with the viral PIC while in the cytoplasm must confer a benefit during early replication. CA-associated with vDNA could cloak the viral genome from host immune sensors (Stetson et al., 2008) or could enable transport via cytoplasmic macromolecular machinery (McDonald et al., 2002). Our data further suggests that CA plays an active role in the PIC interaction with the nuclear pore complex. Understanding how CPSF6-358 physical interaction with CA impairs interaction with the nuclear pore complex is a point of ongoing investigation.
NUP Requirements for HIV-1 Infection
The enhanced restriction of wild-type but not N74D HIV-1 by CPSF6-358 in nondividing cells prompted further examination of the transport and NUPs recently identified as potential HIV-1 co-factors in siRNA screens. Because both viruses are isogenic except for a single nucleotide mutation in gag, reduced efficiency of infection by one virus but not the other would strongly suggest diminished infection was not due to cytotoxicity caused by depletion of the protein in question.
We confirmed that wild-type HIV-1 infection was impaired by TNPO3, RANBP2, NUP153, or NUP160 knockdown (Figure 7B). In contrast, infection by the N74D mutant was less dependent on TNPO3, RANBP2, or NUP153, suggesting that these proteins interacted, directly or indirectly, with wild-type CA during infection. The N74D mutant had an increased dependency on NUP155. NUP85 knockdowns also appeared to diminish N74D HIV-1 infection but to a lesser extent, whereas wild-type HIV-1 and the N74D mutant shared infection sensitivity to NUP160 depletion. Collectively, these data show that CA associated with the RTC or PIC modulates interactions with nuclear transport machinery.
A more distantly related lentivirus, FIV, behaved remarkably similar to N74D HIV-1 in infection assays. It was not susceptible to CPSF6-358 restriction, or sensitive to depletion of cellular TNPO3 or NUP153. By contrast, FIV infection was impaired in cells depleted of NUP155 or NUP160. These data indicate that N74D HIV-1 and FIV use similar strategies to access the nucleus and that avoiding interaction with CPSF6-358 profoundly alters the nuclear entry pathway (Figure 7B). Why distinct mechanisms of nuclear transport are favored by HIV-1 and FIV is currently unclear, but may reflect in vivo tissue requirements of the viruses, or species-specific differences in cofactors or their expression patterns.
Underscoring the pivotal role of HIV-1 CA in cytoplasmic-nuclear trafficking, other CA mutations, E45A and Q63A/Q67A, enable mCPSF6-358 evasion by HIV-1, but only in dividing cells. By contrast, E45A or Q63A/Q67A HIV-1 are impaired in the infection of nondividing cells and potently restricted when these cells express mCPSF6-358. The combined effect of cellular growth arrest and siRNA depletion of NUPs on E45A and Q63A/Q67A HIV-1 infection could not be evaluated due to cytotoxicity. Nonetheless, in dividing cells E45A and Q63A/Q67A HIV-1 were generally less affected by depletion of TNPO3 or NUPs relative to wild-type and N74D HIV-1, albeit they were not as insensitive as MLV (data not shown). Inefficient interaction with the nuclear pore complex could be why these viruses are highly susceptible to mCPSF6-358 under growth arrest conditions. The presence of N74 in both mutant viruses would enable CPSF6-358 binding and further undermine an already inefficient transport through the nuclear pore.
Consistent with our findings with HIV-1 CA mutants, we have recently shown that HIV-1 with an MLV CA is no longer dependent on TNPO3 for infection (Krishnan et al., 2009). Because the MLV/HIV-1 chimera virus is relatively impaired for infection and unable to infect nondividing cell types (Yamashita and Emerman, 2004), this study did not assess whether a loss of TNPO3 interaction was directly responsible for the impaired nuclear entry. Experiments with N74D HIV-1 indicate that TNPO3 is not required for nuclear entry. Moreover, we observed that the wild-type HIV-1 requirement for TNPO3 was the same in dividing and nondividing cells.
The roles of other nuclear transport factors and pore proteins in HIV-1 infection await elucidation. The mammalian nuclear pore complex is comprised of approximately 30 different proteins (Lim and Fahrenkrog, 2006). Subunit complexes assemble into pore structures, so disruption of one component often affects another within the same subunit. One example is depletion of NUP107 or NUP133, both part of the NUP107-160 subunit, destabilizes the reciprocal molecule (Boehmer et al., 2008; Walther et al., 2003). Although we also observed this interdependence, neither NUP107 nor NUP133 depletions, to the extent that we achieved in viable cells, impacted virus infection. Dependence on NUP155 expression correlated with resistance to mCPSF6-358, raising the possibility that this factor directly interacts with N74D HIV-1 or FIV PICs. While this type of mechanism is tempting to consider, knockdowns of essential nuclear pore components via siRNA are likely to be partial in surviving cells and with a consequence of altering overall nuclear pore structure, complicating interpretation of the roles of these proteins in wild-type and mutant virus infection. Nonetheless, if the depletion of a NUP impairs HIV-1 infection, then the nuclear pore architecture favored by the virus would appear to require the missing cofactor. Moreover, because single amino acid changes in CA can enable infection of nuclear pore factor knockdown cells, it demonstrates that CA regulates HIV-1 interaction with the nuclear pore or transport pathways.
The mechanism of HIV-1 transport through the nuclear pore will be further illuminated as host factor requirements are defined. The core of herpes simplex virus type 1 is thought to dock with the nuclear pore during infection and inject the viral genome into the nucleus (Smith and Helenius, 2004). Intact HIV-1 cores have also been found near nuclear pores (Arhel et al., 2007). It is conceivable that the HIV-1 RTC/PIC undergoes essential modifications at the nuclear pore that enables the transfer of the vDNA into the nucleus. A recent study suggested that HIV-1 interactions with NUPs may not be required for nuclear entry but could affect viral integration (König et al., 2008). Whether HIV-1 PICs are modified during their transit through the nuclear membrane so that they can interact more efficiently with co-factors in the nucleus or are coupled to these co-factors at the nuclear pore will require additional analysis. Given the essential role of CA in regulating HIV-1 nuclear entry, defining the successive interactions between CA and host factors that are necessary for this process will help to unravel the complexities of the HIV-1 cytoplasmic-nuclear journey.
Experimental Procedures
Detailed information regarding materials and methods are found in the Supplemental Text.
Supplementary Material
Acknowledgments
We thank Eric Freed for manuscript comments; Paul Bieniasz, Michael Emerman, Eric Freed, and Masahiro Yamashita for discussions; Julie Tang for assistance; Sabine Dettwiler, Michael Emerman, Eric Freed, John Kappes, Walter Keller, Ned Landau, Eric Poeschla, Wes Sundquist, and Uta von Schwedler for reagents. This work was supported by the National Cancer Institute’s intramural Center for Cancer Research, which supports the HIV Drug Resistance Program (SHH, VNK); NIH grants to AE (AI52014), DRL (AI033303; AI033856), DU (AI49131), JMC (CA089441), and JS (AI063987; AI076094); and an American Foundation for AIDS Research postdoctoral fellowship to ZA (106404-33-RFMC). JMC was a Research Professor of the American Cancer Society, with support from the George Kirby Foundation. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arhel NJ, Souquere-Besse S, Munier S, Souque P, Guadagnini S, Rutherford S, Prevost MC, Allen TD, Charneau P. HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 2007;26:3025–3037. doi: 10.1038/sj.emboj.7601740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmer T, Jeudy S, Berke IC, Schwartz TU. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouyac-Bertoia M, Dvorin JD, Fouchier RA, Jenkins Y, Meyer BE, Wu LI, Emerman M, Malim MH. HIV-1 infection requires a functional integrase NLS. Mol Cell. 2001;7:1025–1035. doi: 10.1016/s1097-2765(01)00240-4. [DOI] [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Bukrinsky M. A hard way to the nucleus. Mol Med. 2004;10:1–5. [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky MI, Haggerty S, Dempsey MP, Sharova N, Adzhubel A, Spitz L, Lewis P, Goldfarb D, Emerman M, Stevenson M. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature. 1993;365:666–669. doi: 10.1038/365666a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Engelman A. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol Cell Biol. 2001;21:6758–6767. doi: 10.1128/MCB.21.20.6758-6767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ F, Thys W, De Rijck J, Gijsbers R, Albanese A, Arosio D, Emiliani S, Rain JC, Benarous R, Cereseto A, et al. Transportin-SR2 imports HIV into the nucleus. Curr Biol. 2008;18:1192–1202. doi: 10.1016/j.cub.2008.07.079. [DOI] [PubMed] [Google Scholar]
- de Noronha CM, Sherman MP, Lin HW, Cavrois MV, Moir RD, Goldman RD, Greene WC. Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science. 2001;294:1105–1108. doi: 10.1126/science.1063957. [DOI] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Dismuke DJ, Aiken C. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J Virol. 2006;80:3712–3720. doi: 10.1128/JVI.80.8.3712-3720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorin JD, Bell P, Maul GG, Yamashita M, Emerman M, Malim MH. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J Virol. 2002;76:12087–12096. doi: 10.1128/JVI.76.23.12087-12096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A. HIV infection of non-dividing cells: a divisive problem. Retrovirology. 2006;3:74. doi: 10.1186/1742-4690-3-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A, Goff SP. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol. 2001;75:3626–3635. doi: 10.1128/JVI.75.8.3626-3635.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Englund G, Martin MA. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol. 1995;69:3949–3954. doi: 10.1128/jvi.69.6.3949-3954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Martin MA. HIV-1 infection of non-dividing cells. Nature. 1994;369:107–108. doi: 10.1038/369107b0. [DOI] [PubMed] [Google Scholar]
- Gallay P, Hope T, Chin D, Trono D. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc Natl Acad Sci U S A. 1997;94:9825–9830. doi: 10.1073/pnas.94.18.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallay P, Swingler S, Aiken C, Trono D. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell. 1995a;80:379–388. doi: 10.1016/0092-8674(95)90488-3. [DOI] [PubMed] [Google Scholar]
- Gallay P, Swingler S, Song J, Bushman F, Trono D. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell. 1995b;83:569–576. doi: 10.1016/0092-8674(95)90097-7. [DOI] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- Haffar OK, Popov S, Dubrovsky L, Agostini I, Tang H, Pushkarsky T, Nadler SG, Bukrinsky M. Two nuclear localization signals in the HIV-1 matrix protein regulate nuclear import of the HIV-1 pre-integration complex. J Mol Biol. 2000;299:359–368. doi: 10.1006/jmbi.2000.3768. [DOI] [PubMed] [Google Scholar]
- Heinzinger NK, Bukinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci U S A. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrosson-Wuilleme L, Goujon C, Bernaud J, Rigal D, Darlix JL, Cimarelli A. Transduction of nondividing human macrophages with gammaretrovirus-derived vectors. J Virol. 2006;80:1152–1159. doi: 10.1128/JVI.80.3.1152-1159.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Bibollet-Ruche F, Chen Y, Wain LV, Liegeois F, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan L, Matreyek K, Oztop I, Lee K, Tipper CH, Li X, Dar MJ, Kewalramani VN, Engelman A. The requirement for cellular transportin-3 (TNPO3/TRN-SR2) during infection maps to human immunodeficiency virus type 1 capsid and not integrase. J Virol. 2009 doi: 10.1128/JVI.01899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RY, Fahrenkrog B. The nuclear pore complex up close. Curr Opin Cell Biol. 2006;18:342–347. doi: 10.1016/j.ceb.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Limon A, Nakajima N, Lu R, Ghory HZ, Engelman A. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J Virol. 2002;76:12078–12086. doi: 10.1128/JVI.76.23.12078-12086.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen N, Barraza R, Whitwam T, Saenz DT, Kemler I, Poeschla EM. FIV Vectors. Methods Mol Biol. 2003;229:251–271. doi: 10.1385/1-59259-393-3:251. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov S, Rexach M, Zybarth G, Reiling N, Lee MA, Ratner L, Lane CM, Moore MS, Blobel G, Bukrinsky M. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 1998;17:909–917. doi: 10.1093/emboj/17.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Yang R, Aiken C. Cyclophilin A-dependent restriction of human immunodeficiency virus type 1 capsid mutants for infection of nondividing cells. J Virol. 2008;82:12001–12008. doi: 10.1128/JVI.01518-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe T, Reynolds TC, Yu G, Brown PO. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegsegger U, Blank D, Keller W. Human pre-mRNA cleavage factor Im is related to spliceosomal SR proteins and can be reconstituted in vitro from recombinant subunits. Mol Cell. 1998;1:243–253. doi: 10.1016/s1097-2765(00)80025-8. [DOI] [PubMed] [Google Scholar]
- Saenz DT, Teo W, Olsen JC, Poeschla EM. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5alpha proteins. J Virol. 2005;79:15175–15188. doi: 10.1128/JVI.79.24.15175-15188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Helenius A. How viruses enter animal cells. Science. 2004;304:237–242. doi: 10.1126/science.1094823. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A. 2006;103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unutmaz D, KewalRamani VN, Marmon S, Littman DR. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J Exp Med. 1999;189:1735–1746. doi: 10.1084/jem.189.11.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Alves A, Pickersgill H, Loiodice I, Hetzer M, Galy V, Hulsmann BB, Kocher T, Wilm M, Allen T, et al. The conserved Nup107–160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. The cell cycle independence of HIV infections is not determined by known karyophilic viral elements. PLoS Pathog. 2005;1:e18. doi: 10.1371/journal.ppat.0010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Retroviral infection of non-dividing cells: old and new perspectives. Virology. 2006;344:88–93. doi: 10.1016/j.virol.2005.09.012. [DOI] [PubMed] [Google Scholar]
- Yamashita M, Emerman M. Cellular restriction targeting viral capsids perturbs human immunodeficiency virus type 1 infection of nondividing cells. J Virol. 2009;83:9835–9843. doi: 10.1128/JVI.01084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Perez O, Hope TJ, Emerman M. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 2007;3:1502–1510. doi: 10.1371/journal.ppat.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaitseva L, Myers R, Fassati A. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 2006;4:e332. doi: 10.1371/journal.pbio.0040332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ, et al. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495–504. doi: 10.1016/j.chom.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Sim J, Griffith J, Reed R. Purification and electron microscopic visualization of functional human spliceosomes. Proc Natl Acad Sci U S A. 2002;99:12203–12207. doi: 10.1073/pnas.182427099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.