Summary
Many surface proteins in Gram-positive bacteria are covalently linked to the cell wall through a transpeptidation reaction catalysed by the enzyme sortase. Corynebacterium diphtheriae encodes six sortases, five of which are devoted to the assembly of three distinct types of pilus fibres – SrtA for the SpaA-type pilus, SrtB/SrtC for the SpaD-type pilus, and SrtD/SrtE for the SpaH-type pilus. We demonstrate here the function of SrtF, the so-called housekeeping sortase, in the cell wall anchoring of pili. We show that a multiple deletion mutant strain expressing only SrtA secretes a large portion of SpaA polymers into the culture medium, with concomitant decrease in the cell wall-linked pili. The same phenotype is observed with the mutant that is missing SrtF alone. By contrast, a strain that expresses only SrtF displays surface-linked pilins but no polymers. Therefore, SrtF can catalyse the cell wall anchoring of pilin monomers as well as pili, but it does not polymerize pilins. We show that SrtA and SrtF together generate wild-type levels of the SpaA-type pilus on the bacterial surface. Furthermore, by regulating the expression of SpaA in the cell, we demonstrate that the SrtF function becomes critical when the SpaA level is sufficiently high. Together, these findings provide key evidence for a two-stage model of pilus assembly: pilins are first polymerized by a pilus-specific sortase, and the resulting fibre is then attached to the cell wall by either the cognate sortase or the housekeeping sortase.
Introduction
The surface proteins displayed on the cell envelope of Gram-positive bacteria perform a variety of physiological functions. Many surface proteins interact with specific host cells directly, and thus are crucial determinants of bacterial virulence and fitness (Foster and McDevitt, 1994). A striking feature of the Gram-positive bacterial envelope is that numerous surface proteins are covalently attached to the peptidoglycan cross-bridge via transpeptidation. This reaction is catalysed by the enzyme called sortase (Mazmanian et al., 1999), which requires a C-terminus-sorting signal with an LPXTG motif that is essential for a protein to be anchored covalently to the cell wall (Navarre and Schneewind, 1999; Mazmanian et al., 2001). According to the current model, the surface protein containing the LPXTG motif is captured by sortase within the exoplasm upon translocation of the precursor polypeptide by the general secretion (Sec) system. Sortase then catalyses the cleavage of the LPXTG motif between threonine (Thr) and glycine (Gly), forming an intermediate in which the catalytic cysteine (Cys) residue of the enzyme is linked to the threonine residue of the substrate, and ultimately joins the substrate polypeptide to the peptidoglycan using the same Thr residue and an amine in lipid II precursor (Ton-That et al., 2004a; Marraffini et al., 2006). Since the discovery of the first sortase in Staphylococcus aureus (Mazmanian et al., 1999), its homologues have been characterized in various Gram-positive model organisms. Sortase mutants in different pathogens show defects in bacterial colonization, evading innate immunity and the ability to cause disease (Mazmanian et al., 2000; Bierne et al., 2002; Garandeau et al., 2002; Jonsson et al., 2002; 2003; Weiss et al., 2004).
Another striking feature of most Gram-positive bacteria is that they encode a handful of different sortase enzymes dedicated for different classes of surface proteins (Comfort and Clubb, 2004; Dramsi et al., 2005). The archetype SrtA of S. aureus, which anchors protein A to the cell wall, is referred to as the class A, or the housekeeping, sortase. The class B, C and D sortases are specifically involved in iron acquisition (Mazmanian et al., 2003), pilus assembly (Ton-That and Schneewind, 2004; Telford et al., 2006), or sporulation (Marraffini and Schneewind, 2006). Unlike the housekeeping sortase, which is encoded in a chromosomal location farther away from the genes that encode specific surface protein substrates, all other sortases are encoded in pathogenicity islands together with their substrates (Telford et al., 2006). The class C sortases belong to the largest family, and they are present in multiple copies in the genome of many significant pathogens, including Actinomyces naeslundii, Corynebacterium diphtheriae, Enterococcus faecalis and streptococci (Ton-That and Schneewind, 2003; Mora et al., 2005; Barocchi et al., 2006; Dramsi et al., 2006; Nallapareddy et al., 2006; Rosini et al., 2006; Mishra et al., 2007). These sortases are devoted to the assembly of proteinaceous filaments, named pili or fimbriae, on the bacterial surface (Ton-That and Schneewind, 2004). These surface organelles are required for bacterial adherence and biofilm formation, and they contribute to bacterial pathogenesis and modulation of the host immune system (Yeung, 1999; Barocchi et al., 2006; Nallapareddy et al., 2006; Maisey et al., 2007; Manetti et al., 2007; Mandlik et al., 2007).
In C. diphtheriae, five pilus-specific sortase genes (named srtA–E) with their respective LPXTG-containing substrates are arranged in three chromosomal clusters (Ton-That and Schneewind, 2003). srtF, encoding the designated housekeeping sortase, is found in a chromosomal region (complement, 2 363 265–2 363 330) that is not associated with pilus genes. Each gene cluster encodes a distinct heterotrimeric pilus structure (Ton-That and Schneewind, 2003; Gaspar and Ton-That, 2006; Swierczynski and Ton-That, 2006). For example, the SpaA-type pilus is composed of the major pilus shaft SpaA and two minor pilins, SpaB, decorating the shaft, and SpaC, largely found at the tip regions (Ton-That and Schneewind, 2003). A covalent linkage between the SpaA monomers in the pilus shaft is thought to be formed between threonine of the LPXTG motif and lysine (Lys) of a conserved motif, named the pilin motif, found in the SpaA protein sequence (Ton-That et al., 2004b). Ultimately, the pilus polymers are anchored to the bacterial cell wall. Located in the SpaA gene cluster, pilus-specific sortase SrtA is essential for the formation of the SpaABC pili on the bacterial surface (Ton-That and Schneewind, 2003; Mandlik et al., 2007). Similarly, sortases SrtB/SrtC are essential for the assembly of the SpaD-type pili (Gaspar and Ton-That, 2006), whereas sortases SrtD/SrtE are specific for the SpaH-type pili (Swierczynski and Ton-That, 2006).
Notably, recent work from our laboratory revealed that corynebacteria display not only distinct polymers containing the various pilins, but also individual pilins through the sortase catalysed attachment of the pilins to the cell wall (Mandlik et al., 2007). This non-fibrilar display of surface pilins seems to be biologically significant, because our work also revealed that the corynebacterial adherence to pharyngeal epithelial cells requires SpaB and SpaC minor pilins, but not the major pilin SpaA, which is essential for the formation of the pilus polymer (Mandlik et al., 2007). A key question not addressed so far is whether the housekeeping sortase SrtF is somehow involved in pilus assembly as well as the surface display of various pilins and pilus structures. That the housekeeping sortase may play some role in pilus biogenesis was first indicated from the initial analysis of the srtF-deletion mutant in C. diphtheriae, wherein significant amount of pilus polymers were found to be extracted from corynebacteria without any treatment with muramidase, a cell wall hydrolase (Ton-That and Schneewind, 2003). This suggested that in the absence of SrtF, the SpaA pili were not properly attached to the cell envelope, as would be expected if SrtF were to catalyse the cell wall anchoring of the pili. In a more recent study in Streptococcus agalactiae, the deletion of the housekeeping sortase gene was shown to cause a significant reduction in pilus polymerization as well as pilus assembly on the bacterial surface (Dramsi et al., 2006). These findings prompted us to carry out a systematic genetic and biochemical analysis of SrtF function in corynebacterial pilus assembly reported here.
By examining a large battery of corynebacterial mutants, we show that the housekeeping sortase SrtF is not required for pilus polymerization, per se, in agreement with previous work (Ton-That and Schneewind, 2003). By analysing the culture medium, we demonstrate that the srtF-deletion mutant secretes a large fraction of pilus polymers into the extracellular milieu, while a strain that expresses only the housekeeping sortase displays surface-linked pilins but not polymers. Thus, SrtF is not only sufficient to catalyse the cell wall-anchoring reaction, but it is also required physiologically for the efficient attachment of pilus fibres to the cell wall. We show that SrtF and the pilus-specific sortase SrtA together produce wild-type levels of the SpaA-type pili on the bacterial surface, and permit optimal corynebacterial adhesion to pharyngeal epithelial cells. Finally, by controlling the expression level of the major subunit SpaA, we demonstrate that the SrtF requirement for surface anchoring of the SpaA pilus becomes prominent when the SpaA level is high. The fact that pilus polymers are secreted in the absence of SrtF demonstrates that pilus assembly involves a polymerization step followed by the cell wall-anchoring step.
Results
The housekeeping sortase SrtF is dispensable for pilus polymerization
To determine whether the housekeeping sortase SrtF of C. diphtheriae is required for pilus polymerization, we generated a library of non-polar, in-frame deletion mutants that express individual sortases (Table S1). We then monitored pilus polymerization in the various corynebacterial strains grown in liquid culture to mid-exponential phase by Western blotting, using equivalent samples representing the bacterial cell wall (lanes indicated by W) and the bacteria-free culture medium (lanes marked M) (see Experimental procedures). It is important to note that the cell wall fraction was generated by spinning out bacterial cells and washing the cells prior to muramidase treatment to exclude medium contamination, and also removing the protoplasts from the soluble cell wall extracts after muramidase treatment, to exclude other cellular contaminants. The culture media and the cell wall fractions were then precipitated and dissolved in SDS-sample buffer for gel electrophoresis (Fig. 1). With wild-type bacteria, using an antibody against the major subunit SpaA (α-SpaA), we observed a minute amount of pilus polymers (HMW) in the culture medium, but most of the SpaA polymers were found in the cell wall fraction (Fig. 1A). By contrast, with the mutant strain expressing sortase A (SrtA+) only, the majority of pilus polymers were found in the culture medium. This shows clearly that the pilus-specific sortase SrtA by itself is insufficient for the optimal cell wall anchoring of the pilus.
Fig. 1.
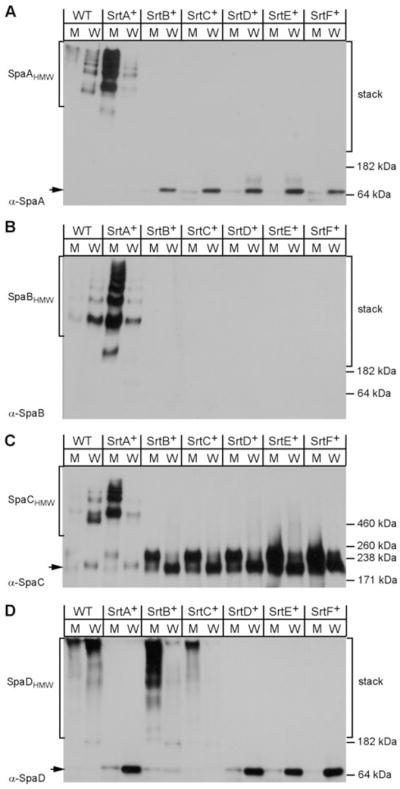
The housekeeping sortase SrtF is not required for pilus polymerization. Uniformly treated protein samples were collected from the culture medium (M) and cell wall fragments (W) of the C. diphtheriae wild-type (WT) strain or its isogenic derivatives expressing individual sortase (SrtA–F) prior to extraction with hot SDS sample buffer. Proteins were separated on SDS-PAGE and detected by immunoblotting with α-SpaA (A), α-SpaB (B), α-SpaC (C) or α-SpaD (D). The monomer (arrow) and high-molecular-weight products (e.g. SpaHMW) of pilus polymerization, the position of molecular weight markers and the stacking gel portion of SDS-PAGE (stack) are indicated.
Importantly, no SpaA polymers were detected in strains expressing only SrtB, SrtC, SrtD, SrtE or SrtF, confirming that SrtA is essential and sufficient for the polymerization reaction (Fig. 1A). This also establishes the important fact that none of the other sortases, including the housekeeping sortase, are able to catalyse SpaA polymerization. The same conclusion is reached for the two minor pilins SpaB (Fig. 1B) and SpaC (Fig. 1C), which respectively decorate the SpaA pilus shaft and the tip region (Ton-That and Schneewind, 2003). Recall that our previous studies have shown that sortases SrtB/SrtC and SrtD/SrtE are essential for polymerization of the SpaD- and SpaH-type pili respectively (Gaspar and Ton-That, 2006; Swierczynski and Ton-That, 2006; Mandlik et al., 2007). Additional experiments demonstrated that neither pilus requires SrtF for the formation of polymers (Figs 1D and S1). Thus, if the housekeeping sortase played a role in pilus polymerization, as was shown for S. agalactiae (Dramsi et al., 2006), this is likely to be a species-specific phenomenon.
SrtF is required for the efficient cell wall anchoring of SpaA-type pili
The secretion of SpaA polymers by the ΔsrtB–F mutant described above suggests that there must be a factor besides SrtA that facilitates the efficient cell wall anchoring of the SpaA pili. That SrtF may be this missing component was first suggested by the previous observation that when srtF was deleted, some pilus polymers were readily extractable from the cells without any muramidase treatment (Ton-That and Schneewind, 2003). We, therefore, analysed pilus protein samples of different corynebacterial variants collected from both the culture medium and the cell wall fragments as described above by Western blotting with α-SpaA. The result showed that the strain that expresses only SrtA and the strain that is devoid of SrtF both secreted the majority of pilus polymers in the culture medium, while the cell wall-linked pili were drastically reduced in each case as compared with that of the wild type (Fig. 2). This showed that SrtF plays a major role in the cell wall anchoring of SpaA pili. To examine a potential contribution by other sortases, if any, we generated a corynebacterial strain that expresses SrtA and SrtF but not the other sortases. This mutant strain produced the same amount of cell wall-anchored pili as the wild-type bacteria, and it secreted very little pilus polymers in the medium like the wild-type strain (Fig. 2). The same phenotype was observed when SrtF was overproduced in the strain that expresses only SrtA (Fig. 2, last two lanes). Therefore, SrtA and SrtF are both necessary and sufficient to generate wild-type levels of SpaA-type pilus on the cell surface. Note that some monomeric Spa pilins were detected in the cell wall fraction in mutant strains expressing single sortase (Figs 1 and 2); we speculate that without their cognate sortase, the pilins might be missorted.
Fig. 2.
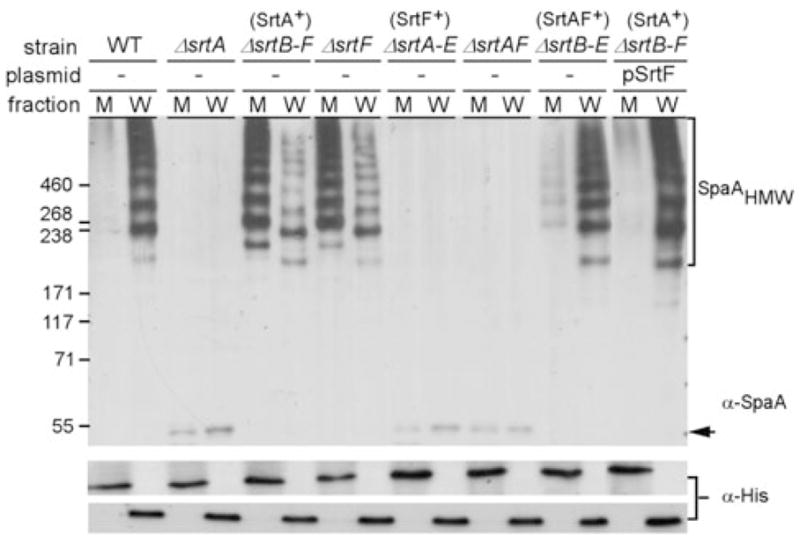
SrtF is required for the efficient cell wall anchoring of SpaA pili. Samples were treated exactly as in Fig. 1. Proteins were separated on 4–12% Tris-glycine gradient gels and detected by immunoblotting with α-SpaA. Anti-pentahistidine (α-His) was used for loading controls. The monomer (arrow) and high-molecular-weight products (e.g. SpaHMW) of pilus polymerization, and the position of molecular weight markers are indicated.
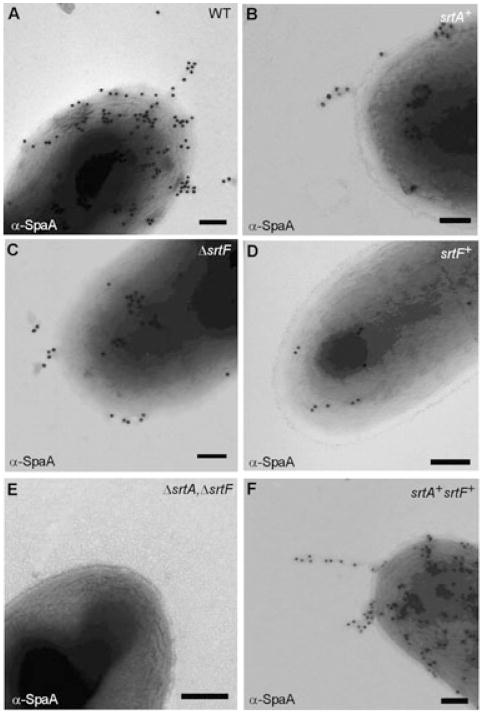
We next examined pilus assembly microscopically, whereby we treated the above strains with α-SpaA antibody, followed by staining with IgG-conjugated gold particles and viewed by a transmission electron microscope, as described previously (Mandlik et al., 2007). In agreement with previous results (Ton-That and Schneewind, 2003; Mandlik et al., 2007), SpaA-stained gold particles were found on the surface as well as pilus structures in the wild type (Fig. 3A). By comparison, the strain expressing SrtA alone and the srtF-deletion mutant each showed a significant reduction in SpaA-stained structures (Fig. 3B and C). Importantly, no pilus structures were detected on the surface of the strain expressing only SrtF or the srtA/srtF double deletion strain (Fig. 3D and E). On the other hand, the strain expressing SrtA and SrtF sortases displayed the SpaA pilus at a level comparable to wild-type bacteria (Fig. 3F).
Fig. 3.
Efficient assembly of SpaA pili on bacterial surface mediated by SrtF. Cells of the wild type (A) or its isogenic derivatives (B–F) were immobilized on carbon grids, stained with specific antiserum against SpaA (α-SpaA) and goat anti-rabbit IgG conjugated to 18 nm gold particles. Samples were viewed by transmission electron microscopy. Scale bars indicate the length of 0.2 μm.
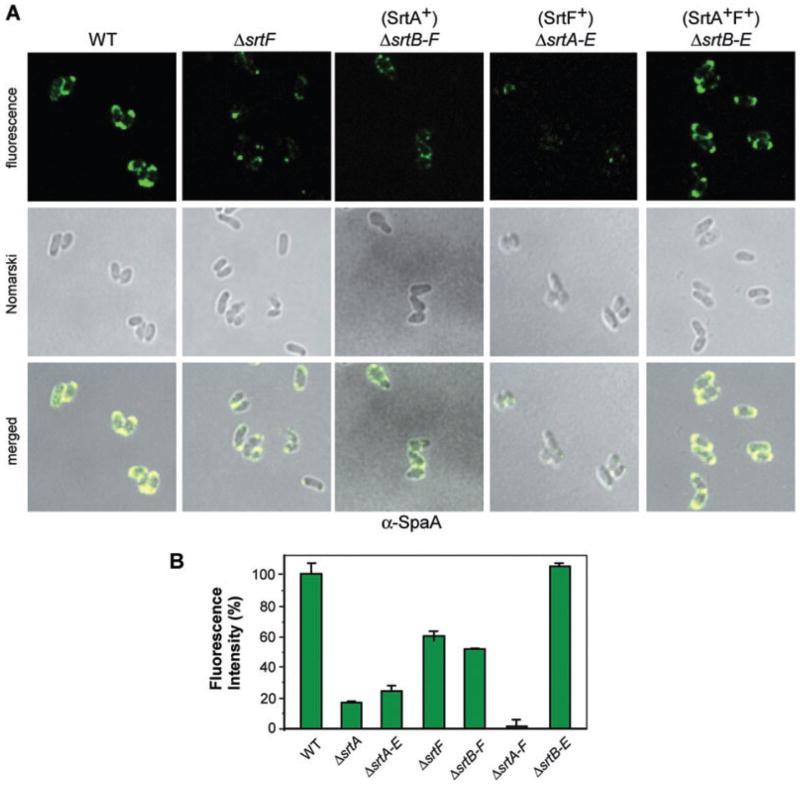
To examine SrtF function in the cell wall anchoring of the SpaA pili more quantitatively, we employed immunofluorescence microscopy, a technique we used previously to detect both pilins and pilus fibres displayed on the cell surface (Mandlik et al., 2007). Corynebacteria were treated with a specific antibody, followed by the AlexaFluor 488-stained chicken anti-rabbit IgG, and cells were observed by a confocal microscope (Fig. 4A). Quantification of fluorescence intensity over a given surface area was then carried out using the Metamorph software (see Experimental procedures). Average fluorescence intensity of each corynebacterial mutant strain was compared with that of the wild type (n = 25), which was set to 100% (Fig. 4B, first column). Upon staining corynebacteria with α-SpaA, we observed ~80% reduction of SpaA fluorescence signal in the srtA-deletion mutant compared with the wild-type signals (Fig. 4B, second column). This same level of reduced intensity of the fluorescence signal was detected in the strain that expresses only SrtF (Fig. 4B, third column). This demonstrates that the housekeeping sortase plays a physiological role in the cell wall anchoring of SpaA pilins/pili. Interestingly, about half of the SpaA signal was detected in the strain that lacks SrtF or the strain that lacks all but sortase SrtA (Fig. 4B, fourth and fifth columns). This demonstrates clearly that SrtA acts as both a polymerase and a sortase, linking the SpaA pilus to the cell wall, and that although SrtF contributes to the optimal cell wall anchoring of the SpaA pilus, SrtF is not essential for this reaction. That SrtF is the only other sortase involved in SpaA pilus assembly is evident from the fact that the wild-type level of fluorescence signal is generated in a strain that expresses only SrtA and SrtF (Fig. 4B, last column). When we labelled these strains with anti-SpaB or anti-SpaC, very similar results were observed (Fig. S2). We conclude that the housekeeping sortase SrtF is required for the efficient cell wall anchoring of the SpaA-type pili.
Fig. 4.
Immunofluorescent detection of SpaA pili displayed on the bacterial surface. Corynebacteria were stained with a specific antibody against SpaA (α-SpaA) and AlexaFluor 488 chicken anti-rabbit IgG. Shown are the fluorescent, the nomarski DIC and the merged images (A). The samples were observed on a Zeiss LSM 510 confocal microscope. The results are shown as representatives of at least three independent experiments.
B. Quantification of fluorescence intensity in (A) was carried out using the Metamorph software. Intensity values shown with arbitrary units were averaged over 25 cells in each strain and expressed relative to the average fluorescence intensity of the wild type (WT).
The efficient cell wall anchoring of SpaD- and SpaH-type pili depends upon SrtF
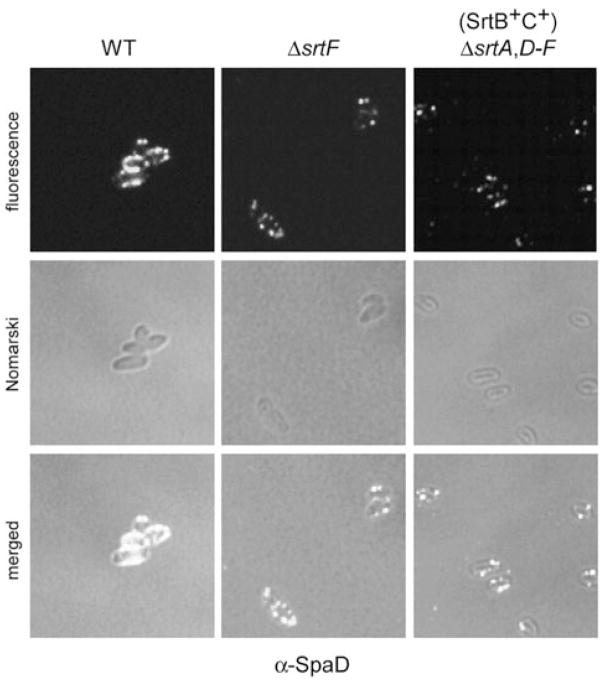
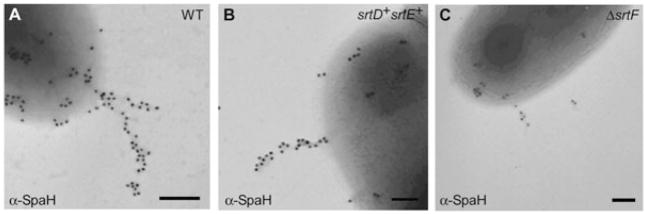
The role of the housekeeping sortase in cell wall anchoring might be a general feature of pilus assembly. To determine whether SrtF plays a role in assembly of the other pilus types, we went on to examine the surface assembly of these pili by biochemical analysis, immunofluorescence and immunoelectron microscopy. As was found for SpaA pilus, SpaD- and SpaH-pilus polymers were found predominantly in the culture medium with the ΔsrtF strain (Fig. S1). This strain also showed a significant reduction of SpaD signals detected by fluorescence (Fig. 5). Similar lower intensity of the SpaD signal was also observed in the strain that expresses only SrtB and SrtC, which are the sortases specific for the SpaD-type pili (Gaspar and Ton-That, 2006). Thus, the housekeeping sortase appears to play a broad role in cell wall anchoring of distinct pilus structures (Fig. 5). Further evidence for this conclusion came from the analysis of the SpaH pili (Figs 6 and S3). By immunoelectron microscopy, we observed SpaH pilus structures of comparable length in the wild type and a strain that expresses only SrtD and SrtE (Fig. 6A and B). Consistent with a role of SrtF in the cell wall anchoring of SpaH pili, however, fewer pili and pilins were observed in the strain that lacks SrtF (Fig. 6C).
Fig. 5.
SrtF is required for the efficient cell wall anchoring of the SpaD-type pili. Corynebacteria were stained with a specific antibody against SpaD (α-SpaD) and AlexaFluor 488 chicken anti-rabbit IgG. Shown are the fluorescent, the nomarski DIC and the merged images. The samples were observed on a Zeiss LSM 510 confocal microscope. The results are shown as representatives of at least three independent experiments.
Fig. 6.
SrtF is required for the efficient cell wall anchoring of the SpaH-type pili. Wild-type C. diphtheriae (A) and its isogenic derivatives ΔsrtA-C, F (designated as srtD+srtE+) (B), or ΔsrtF (C) were immobilized on carbon grids, stained with specific antiserum against SpaH and goat anti-rabbit IgG conjugated to 18 nm gold particles. Samples were viewed by transmission electron microscopy. Scale bars indicate the length of 0.2 μm.
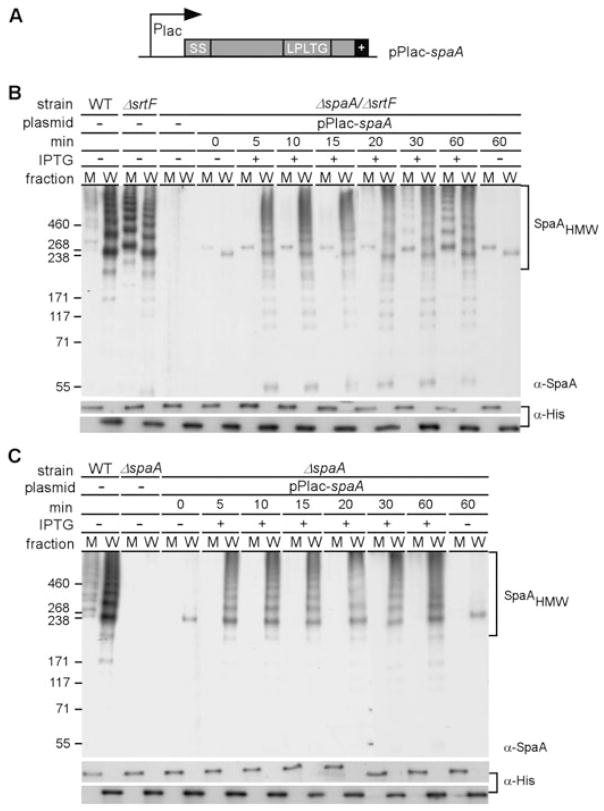
SrtF dependence is pronounced at high levels of SpaA pilin
Previous work has shown that the extent of pilus polymerization is governed by the amount of the major pilin subunit expressed in the cell (Swierczynski and Ton-That, 2006). We, therefore, wondered whether the level of the major pilin subunit also influences the SrtF-catalysed reaction. To explore this possibility, we engineered a construct that expresses the SpaA pilin under the control of Plac promoter (see Experimental procedures) (Fig. 7A). This expression vector was introduced into strains that lack chromosomal spaA or both spaA and srtF. Corynebacteria grown to mid-log phase were induced with IPTG, aliquots of equal number of cells were taken at various intervals, and samples were fractionated into culture medium and cell wall and analysed by Western blot as described above. As a control for quantification, an anti-histidine antibody was used to detect a corynebacterial poly-histidine-containing protein that appears to be cell wall associated and also found in the culture medium (our unpublished data). With the strain that lacks srtF, pilus polymers (high molecular weight, HMW) were observed exclusively in the cell wall compartment after 10 min of induction (Fig. 7B). As the amount of the SpaA pilin increased over time, polymers were detected in the culture medium as well (Fig. 7B). When SrtF was present, however, no pilus polymers were detected in the culture medium under the same time-course of SpaA induction (Fig. 7C), even though comparable amounts of SpaA accumulated in the two strains. Thus, at low levels of SpaA pilin produced in the cell, SrtA is sufficient for both pilus polymerization and cell wall anchoring. When the pilin level reaches a certain concentration in the cell, however, SrtF becomes critical for cell wall anchoring of the SpaA pili. Evidently, wild-type corynebacteria express sufficiently high amounts of SpaA that necessitates the involvement of the housekeeping sortase.
Fig. 7.
Inducible expression of SpaA pilins.
A. Plasmid pPlac-spaA was generated by cloning the coding sequence of spaA under the control of an IPTG-inducible promoter of the E. coli/C. diphtheriae shuttle vector pEKEx2.
B. Cells of the isogenic ΔsrtF or ΔsrtF-ΔspaA strain harbouring pPlac-spaA were induced by the addition of 1 mM IPTG, and samples with an equal number of cells were taken at timed intervals for isolating medium (M) and cell wall fragments (W) as described in Fig. 1. Samples of the wild-type and ΔsrtF bacteria were collected at the end of induction. Solubilized pilins were boiled in SDS sample buffer and were separated on 4–12% Tris-glycine gradient gels and detected by immunoblotting with the specific antiserum α-SpaA. Anti-his (α-His) and no IPTG (−) were used as controls.
C. The same treatment as described in (B) was employed for cells of the wild type (WT), its isogenic ΔspaA or ΔspaA strain harbouring pPlac-spaA.
SrtF facilitates the efficient binding of C. diphtheriae to pharyngeal epithelial cells
Human nasopharynx is the major site of C. diphtheriae infection (Love and Murphy, 2006). Recent studies have demonstrated that corynebacterial adherence to this tissue is mediated by the SpaA-type pili (Mandlik et al., 2007). To examine whether SrtF contributes to this process, we employed ex vivo adhesion assays, whereby pharyngeal epithelial cells were grown in multi-well culture plates to confluence and infected with the desired corynebacterial variants. The infected pharyngeal cells were washed to remove the unbound bacteria, detached from wells, and then plated on agar plates to enumerate absorbed bacteria (see Experimental procedures). Relative association of various corynebacterial strains to pharyngeal cells was then compared with that of the wild-type strain, which was set to 100% (Fig. 8, first column). Consistent with previous findings, deletion of all sortases or only srtA drastically reduced corynebacterial adherence to pharyngeal cells (Fig. 8, second and third columns). The same is true for a strain that lacks both SrtA and SrtF (Fig. 8, fourth column). The strain that expresses SrtF alone adhered to pharyngeal cells somewhat more efficiently than the strain that lacks all six sortases (Fig. 8, fifth and second columns). Moreover, the deletion of srtF alone showed a small (~20%) but reproducible reduction in host cell adhesion, a phenotype similar to that of the strain expressing only SrtA (Fig. 8, sixth and seventh columns). Finally, wild-type level adhesion was observed with the strain that expresses both SrtA and SrtF (Fig. 8, last column). Evidently, SrtF function somehow facilitates an optimal corynebacterial adhesion to the pharyngeal cells.
Fig. 8.
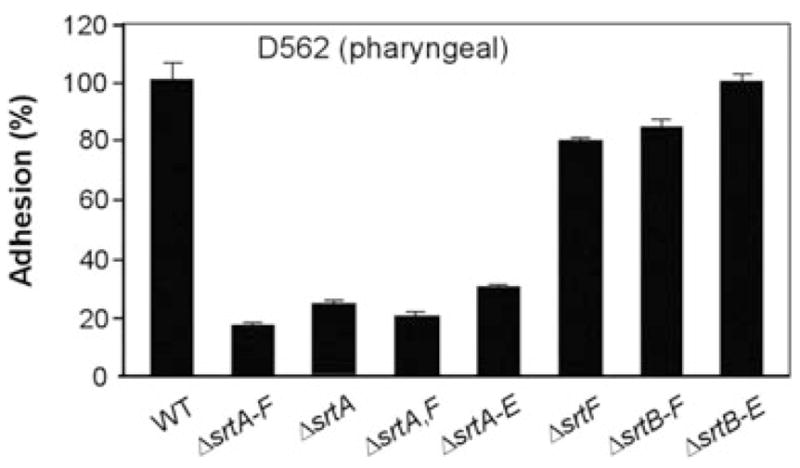
SrtF is required for efficient corynebacterial adherence to pharyngeal epithelial cells. The wild type (WT) and its isogenic deletion mutants were subjected to adhesion assays using pharyngeal epithelial cells (D562). Data are presented as percentage of adhesion relative to that of the WT. The results are presented as averages (±SD) from three independent experiments performed in triplicates.
Discussion
Gram-positive bacteria utilize a highly conserved family of sortase enzymes to anchor on the cell wall of many surface proteins that are important for bacterial virulence and fitness. The so-called housekeeping sortase (such as SrtA in S. aureus) anchors a number of LPXTG-containing proteins to the cell wall, while a handful of other sortases (such as SrtABCDE in C. diphtheriae) are involved in the assembly of various pili (Ton-That and Schneewind, 2004). In this study, we demonstrated that although the housekeeping sortase of C. diphtheriae does not participate in pilus polymerization, the enzyme clearly plays a physiological role in facilitating the efficient cell wall anchoring of pilus polymers. The cellular requirement for the housekeeping sortase for pilus assembly is most pronounced at higher levels of the major pilin subunit, which drives the extent of pilus polymerization (Swierczynski and Ton-That, 2006). Our studies showed conclusively that a pilus-specific sortase and the housekeeping sortase together produce wild-type levels of the different pili on bacterial surface, and they suffice for optimal binding of corynebacteria to pharyngeal epithelial cells, a specific adhesion that is mediated by the SpaA-type pili (Mandlik et al., 2007). Together, these findings establish for the first time a function of the housekeeping sortase in the final step of pilus assembly. Our findings also provide important evidence for the hypothesis that pilus polymerization precedes the cell wall-anchoring step during pilus biogenesis.
That the housekeeping sortase SrtF of C. diphtheriae is not required for pilus polymerization is evident from genetic and biochemical analysis of mutant strains, which express individual sortases. The strain expressing SrtA only (SrtA+) is able to catalyse not only pilus polymerization, but also the cell wall anchoring of some of the resulting pilus fibres (Fig. 1). Thus, the pilus-specific sortase SrtA is both necessary and sufficient for pilus assembly (see also Figs 2 and 3). Furthermore, we have shown that none of the other sortases, including the housekeeping SrtF, is able to catalyse SpaA polymerization.
The vital role of the housekeeping sortase in pilus biogenesis is clearly evident from the striking fact that, in its absence, only a small fraction of the SpaA polymers are anchored to the cell wall. Inspection of the SrtA-only strain revealed that although the efficiency of SpaA polymerization in this strain is comparable to the wild type, most of the polymers made in the mutant strain are secreted out in the culture medium, and little is anchored to the cell wall (Figs 1 and 2). Clearly, there must be a factor missing in this strain that is normally required for the efficient cell wall anchoring of the SpaA pili. That the housekeeping sortase is the missing component is established by the fact that an identical phenotype is observed with the strain that is devoid of SrtF alone (Fig. 2). Importantly, we have demonstrated that in the strain that expresses both SrtA and SrtF, but none of the other sortases, the assembly and cell wall anchoring of the SpaA pili is comparable to the wild type (Figs 2–4). This requirement of both the pilus-specific sortase and the housekeeping sortase for optimal cell wall anchoring of the SpaA pili is further underscored by our demonstration that the two sortases are indeed sufficient to allow corynebacterial adherence to human pharyngeal epithelial cells to a level that is comparable to the wild-type bacteria (Fig. 8). Finally, the role of the housekeeping sortase in pilus biogenesis is not restricted to the SpaA pili. The deletion of srtF alone significantly reduces the cell wall anchoring of both SpaD- and SpaH-type pili (Figs 5 and 6, and S1 and S3). Therefore, the housekeeping sortase plays a prominent physiological role in the pilus biogenesis in corynebacteria.
The critical question that now arises is precisely how the housekeeping sortase facilitates pilus biogenesis. An obvious function of this sortase is that it directly participates in the catalysis of cell wall anchoring of the pilus, as was pointed out in a recent review article (Scott and Zahner, 2006). An alternate possibility is that the housekeeping sortase acts indirectly, by catalysing the surface localization of an accessory component that is required for pilus anchoring to the cell wall. Fortunately, we obtained evidence that helps to distinguish these possibilities. Immunofluorescence experiments, which measured surface display of pilin antigens quantitatively, revealed clearly that a corynebacterial mutant that expresses SrtF only displays an appreciable amount of the SpaA pilin on the cell surface (see Fig. 4). Immunofluorescence analysis showed that SpaB/SpaC is also linked to the cell wall in this strain (Fig. S2). These results demonstrate the catalytic competence of the housekeeping sortase in anchoring of specific pilus proteins to the cell wall. We, therefore, favour the model that the housekeeping sortase directly catalyses the cell wall anchoring of both pilins and pilus fibres (Fig. 9).
Fig. 9.

The two-stage mechanism of pilus biogenesis: function of the housekeeping sortase. This working model, built on the most significant findings reported here, depicts the two stages of pilus polymerization and cell wall anchoring catalysed by a pilus-specific sortase and the housekeeping sortase in corynebacteria. As detailed in the text, pilus assembly requires two sortase molecules properly juxtaposed to capture the pilin precursors in the exoplasm and catalyse cycles of covalent linkage between pilin monomers. This is carried out by a pair of the pilus-specific sortases (black), the SrtA sortase for the heterotrimeric SpaABC pilus, each of which must form an acyl enzyme intermediate with the pilin precursors via the catalytic cysteine residue (Cys) and the threonine (Thr) residue of the pilin LPXTG motif. Pilus polymerization is catalysed by cross-linking Thr with the lysine of the pilin motif. Pilus polymerization is terminated when the resulting polymer is joined to the cross-bridge amino group of the lipid II precursor, which may be catalysed by the pilus-specific sortase. The main feature of the current model is the involvement of the housekeeping sortase enzyme (yellow). The key data in this paper show that this enzyme acts predominantly to terminate pilus polymerization (see text). In essence, when the housekeeping sortase captures a pilin precursor, it is in turn poised to receive and secure a pilus polymer assembled on a pilus-specific sortase (centre). Now, the pilus residing on the housekeeping sortase (not drawn in the figure) can only be transferred to the lipid II precursor and incorporated into the cell wall via transglycosylation reaction. Individual pilins are shown with letters A (SpaA), B (SpaB) and C (SpaC). Arrows indicate the direction of transfer of pilin and pilus polymers. Note that the tip pilin SpaC is believed to act as the normal nucleator for polymerization of the pilus shaft made of SpaA, on which the SpaB minor pilin is cross-linked by the SrtA sortase. How this is achieved remains one of the significant puzzles of pilus biogenesis.
Our finding that pilus polymers are secreted in the culture medium in the absence of the housekeeping sortase has another important implication. If pilus polymerization were carried out on a cell wall-anchored pilin, polymers would never be free to leak out into the medium. Thus, our data provide critical evidence for a biphasic pathway of pilus biogenesis in which cell wall anchoring is the terminal step (Fig. 9). We envision that the pathway of pilus assembly involves two discrete stages – one allowing the joining of pilin monomers to form an extended pilus fibre, and the other terminating this process by cell wall anchoring of the fibre (Mandlik et al., 2007). Upon synthesis in the cytoplasm, pilin precursors are translocated across the membrane by the Sec machinery. The translocated pilin precursor is captured within exoplasm by a sortase embedded in the nearby membrane, which cleaves the LPXTG motif between the Thr and Gly, and forms an acyl-enzyme substrate intermediate involving the catalytic Cys residue and the Thr residue of the sorting signal. Importantly, the pilus polymerization reaction requires appropriate juxtaposition of two of these intermediates, one of which must be the pilus-specific sortase. The pilin protein moiety from this intermediate is then transferred to the other sortase intermediate through the formation of a Thr–Lys linkage, which is triggered by the nucleophilic attack by an invariant Lys residue that characterizes the conserved pilin motif (Fig. 9). Additional cycles of this reaction can continue pilus growth so long as pilins are available and the sessile Thr–Cys bond in the last subunit added in the polymer is presented by the pilus-specific sortase for nucleophilic attack by another pilin subunit. This is obviously ensured when each of the two sortases participating in the reaction are both pilus-specific, namely SrtA for the SpaA pilus (Fig. 9).
The pilus polymerization stage is terminated when the polymer is transferred to the lipid II precursor, which is in turn cross-linked to the growing cell wall. Based on the data presented here, we suggest that the housekeeping sortase performs this reaction predominantly. For that to happen, the housekeeping sortase must be located in the site of pilus assembly, and it must first form an acyl-enzyme intermediate and provide the Lys nucleophile to receive and secure the pilus polymer (Fig. 9). Once this has occurred, further growth of the pilus is irreversibly blocked because the housekeeping sortase is not a pilin polymerase. This ensures the subsequent transfer of the pilus to lipid II and in turn the cross-linking of the product to the cell wall. Our data suggest that the pilin-specific sortase also carries out this last step physiologically, albeit inefficiently (Fig. 9). Unless there is some form of an organized localization of the different machineries involved, for which there is no evidence, the pilus assembly centre could not always harbour the housekeeping sortase. This necessitates that the pilus-specific sortases perform dual function catalysing polymerization as well as surface anchoring, potentially a fail-safe mechanism evolved to optimize pilus assembly in Gram-positive bacteria.
The participation of the housekeeping sortase in the cell wall-anchoring step of pilus biogenesis helps to explain how pilins are anchored to the cell wall in a pilus-independent fashion. When a pilin precursor is captured by the housekeeping sortase, there is a definite probability that the pilin monomer is incorporated to the cell wall before another subunit or an oligomer is joined by a pilus-specific sortase (Fig. 9). This provides the cell an efficient mechanism to display both pilins and pili on the envelope, a scenario that has important implication in pathogenesis. While pili must aid in the tethering of the pathogen to the appropriate host cells, the surface-displayed pilins would enable the bacterial cell to create intimate zones of adhesion with the host cells. This tight adhesion may trigger not only appropriate host cell signalling but also an efficient toxin delivery (see Mandlik et al., 2007).
During infection, many virulence factors are likely to be abundantly produced by corynebacteria for efficient colonization and invasion of the human nasopharynx, and the expression of SpaA pili required for specific adhesion of corynebacteria to the pharyngeal cells may also be increased. Because the requirement of housekeeping sortase in cell wall anchoring is most pronounced when SpaA level is sufficiently high (see Fig. 7), it is tempting to suggest that the function of the housekeeping sortase might be most crucial in the infection setting. In this regard, it is noteworthy that a binding site for the diphtheria toxin repressor, DtxR, has been identified upstream of the srtF gene (Yellaboina et al., 2004). DtxR is an iron-activated transcription regulator that controls the expression of diphtheria toxin, and possibly many additional genes encoding virulence factors. Work is now underway to test whether DtxR regulates pilus gene expression as well as pilus assembly during corynebacterial infection. Considering the fact that three distinct classes of pili are assembled in corynebacteria, and the apparent specificity of the sortases involved in each of these systems, the findings of the present study underscore how little is known so far about the mechanisms by which pilus assembly is co-ordinated in the cell envelope and the factors that must regulate this process temporally and spatially.
Experimental procedures
Bacterial strains, plasmids and media
Corynebacteria (Table S1) were grown on heart infusion broth (HIB), heart infusion agar (HIA) or trypticase soy agar supplemented with 5% sheep blood (Hemostat) (TSASB). Escherichia coli strains were grown on Luria broth. Kanamycin was added at 50 μg ml−1 as needed. Polyclonal antibodies used in this study were previously obtained (Ton-That and Schneewind, 2003; Gaspar and Ton-That, 2006; Swierczynski and Ton-That, 2006). Reagents were purchased from Sigma unless otherwise indicated.
Gene deletions in C. diphtheriae
Non-polar, in-frame deletion mutants were obtained via homologous recombination as previously described (Ton-That and Schneewind, 2003). Briefly, E. coli S17-1 carrying a gene-deletion construct was used for mating with appropriate C. diptheriae strains (Tables S1 and S2). Co-integrates resulting from this conjugation were identified on HIA plates supplemented with 35 μg ml−1 nalidixic acid and 50 μg ml−1 kanamycin. The deletion mutants were then selected by plating co-integrates on HIA plates containing 10% sucrose and 35 μg ml−1 nalidixic acid. Gene deletions in these bacteria were confirmed by polymerase chain reaction (PCR) and Western blotting. Corynebacterial strains containing overexpression and induction plasmids were generated by electroporation.
Plasmid construction
Construction of pSrtF
Primers SrtF-promoter-5′ and SrtF-promoter-3′ or primers SrtF-5′ and SrtF-3′ (Table S2) were used to amplify, while appending BglII and EcoRI or BamHI and EcoRI sites to the fragments, the 5′ promoter sequence and UTR of the spaA- or the srtF-coding sequence respectively, from C. diphtheriae NCTC13129 chromosomal DNA. The PCR-amplified fragments were digested with BglII and EcoRI and ligated with the cleaved BglII sites of the E. coli/Corynebacterium shuttle vector pCGL0243 (Ton-That and Schneewind, 2003) to generated pSrtF. The recombinant plasmid was then electroporated into C. diphtheriae by a standard protocol (Ton-That and Schneewind, 2003).
Construction of an IPTG-inducible plasmid pPlac-SpaA
Primers SpaA-rbs-5′ and SpaA-3′ (Table S2) were used to PCR-amplify the coding sequence of spaA with chromosomal template DNA of C. diphtheriae NCTC13129. This fragment was digested with PstI/KpnI and ligated into PstI/KpnI-cut E. coli/Corynebacterium shuttle vector pEKEx2, which contains an IPTG-inducible promoter (Swierczynski and Ton-That, 2006). The recombinant plasmid was then electroporated into C. diphtheriae by a standard protocol (Ton-That and Schneewind, 2003).
Adhesion assays
Cultures of the pharyngeal cell line were carried out as described before (Mandlik et al., 2007). Briefly, human pharynx carcinoma cells (Detroit 562) were cultured in minimum essential medium supplemented with a penicillin-streptomycin solution (ATCC), non-essential amino acids, 1500 mg l−1 sodium bicarbonate, 1 mM sodium pyruvate, 2 mM L-glutamine and 10% fetal bovine serum. All cultures were maintained in a 5% CO2/95% air atmosphere at 37°C.
For adhesion assays, pharyngeal cells were grown to confluence in 12-well plates. Prior to infection with bacteria, the cells were washed with sterile phosphate-buffered saline (PBS), and fresh medium without antibiotics and serum were added. The pharyngeal cells were infected with mid-exponential phase corynebacteria with a multiplicity of infection of 10 (~107 cfu ml−1). Infection was allowed to proceed for 1 h at 37°C in a 5% CO2/95% air atmosphere. After infection, washed cells were detached and lysed by treating with diluted Trypsin-versene mixture (BioWhittaker) and 0.025% Triton X-100. To enumerate adherent bacteria, appropriate dilutions were plated on TSASB. The titre of the adherent bacteria for each strain was compared with input titre, and the percentage of adherent bacteria was determined. Each assay was performed in quadruplicates and repeated at least three times. Statistical analysis was performed by using the Student’s t-test.
Immunofluorescence microscopy
Corynebacteria grown overnight at 37°C on TSAB plates or HIB were washed in sterile PBS. Cells were blocked in PBS with 1% bovine serum albumin (BSA) for 1 h, incubated with a primary antibody diluted 1:100 in PBS with 0.1% BSA for 1 h, and followed by washing and blocking for 30 min. Subsequently, the cells were then stained for 1 h with AlexaFluor 488 chicken anti-rabbit IgG (Molecular Probes, Invitrogen) diluted 1:100 in PBS with 0.1% BSA. After washing and final suspension in PBS, samples were mounted on agarose-coated slides for observation under a Zeiss LSM 510 Meta confocal microscope using the 100× objective. Fluorescence intensity was quantified by using the Metamorph software, which computes average integrated intensity over a given surface area (n = 25). The average fluorescence of each mutant strain was compared with that of the wild type, which was set to 100%. Standard deviation was calculated after normalization.
Immunoelectron microscopy
Corynebacteria grown on blood plates were washed in 0.1 M sodium chloride. For immunogold labelling, a drop of bacterial suspension was placed onto carbon grids, washed three times with PBS containing 2% BSA, and blocked for 1 h in PBS with 0.1% gelatin. Corynebacteria were stained with a pilus specific antibody (1:100 dilution) for 1 h, followed by washing and blocking. Subsequently, the cells were treated with 18 nm gold-goat antirabbit IgG (Jackson ImmunoResearch) diluted 1:20 in PBS with 2% BSA for 1 h. Samples were washed five times with water before staining with 1% uranyl acetate. Samples were viewed in a Jeol 100CX transmission electron microscope.
Cell fractionation and Western blotting
Overnight cultures of corynebacteria were used to inoculate mid-log phage bacteria (1:50 dilution) at 37°C in HIB. Kanamycin was added to a final concentration of 50 μg ml−1 when necessary. For induction assays, a corynebacterial culture was spiked with 1 mM IPTG, and an aliquot of the same number of cells was taken at timed intervals (0, 5, 10, 15, 20, 30 and 60 min) after induction. All normalized aliquots were fractionated into medium and cell pellets by centrifugation. The washed cell pellets were treated with muramidase in sucrose buffer SMM (0.5 M sucrose, 10 mM MgCl2, and 10 mM maleate, pH 6.8) containing 0.2 μM PMSF for 6 h or overnight at 37°C. After the muramidase treatment, the soluble cell wall fraction was separated from the protoplasts by centrifugation. The culture supernatant and cell wall fractions were subjected to TCA precipitation and acetone wash. Samples were then boiled in SDS containing sample buffer, separated by 4–12% Tris-glycine gradient gels (Invitrogen) or 12% SDS-PAGE high stack gels, subjected to immunoblotting with rabbit antisera (1:20 000 for α-SpaA, 1:5000 for α-SpaB and 1:10 000 for α-SpaC), and detected with chemiluminescence. Penta-His HRP Conjugate (Qiagen) was used for loading control. After the treatment, samples were dried under vacuum and subjected to TCA precipitation and acetone wash prior to gel electrophoresis.
Supplementary Material
Acknowledgments
We thank Ann Cowan (Center for Cell Analysis and Modeling, University of Connecticut Health Center) and Vedakumar Tatvarty (University of Connecticut Health Center) for advice and help in immunofluorescence microscopy. This work was supported by the US Public Health Service Grants AI061381 from the National Institute of Allergy and Infectious Diseases to H.T.-T.
Footnotes
Supplementary material
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05968.x (This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci USA. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Mazmanian SK, Trost M, Pucciarelli MG, Liu G, Dehoux P, et al. Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol Microbiol. 2002;43:869–881. doi: 10.1046/j.1365-2958.2002.02798.x. [DOI] [PubMed] [Google Scholar]
- Comfort D, Clubb RT. A comparative genome analysis identifies distinct sorting pathways in gram-positive bacteria. Infect Immun. 2004;72:2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dramsi S, Trieu-Cuot P, Bierne H. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, et al. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- Foster TJ, McDevitt D. Surface-associated proteins of Staphylococcus aureus: their possible roles in virulence. FEMS Microbiol Lett. 1994;118:199–205. doi: 10.1111/j.1574-6968.1994.tb06828.x. [DOI] [PubMed] [Google Scholar]
- Garandeau C, Reglier-Poupet H, Dubail I, Beretti JL, Berche P, Charbit A. The sortase SrtA of Listeria monocytogenes is involved in processing of inter-nalin and in virulence. Infect Immun. 2002;70:1382–1390. doi: 10.1128/IAI.70.3.1382-1390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson IM, Mazmanian SK, Schneewind O, Verdrengh M, Bremell T, Tarkowski A. On the role of Staphylococcus aureus sortase and sortase-catalyzed surface protein anchoring in murine septic arthritis. J Infect Dis. 2002;185:1417–1424. doi: 10.1086/340503. [DOI] [PubMed] [Google Scholar]
- Jonsson IM, Mazmanian SK, Schneewind O, Bremell T, Tarkowski A. The role of Staphylococcus aureus sortase A and sortase B in murine arthritis. Microbes Infect. 2003;5:775–780. doi: 10.1016/s1286-4579(03)00143-6. [DOI] [PubMed] [Google Scholar]
- Love JF, Murphy JR. Corynebacterium diphtheriae: iron-mediated activation of DtxR and regulation of diphtheria toxin expression. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-Positive Pathogens. Washing, DC: American Society for Microbiology Press; 2006. pp. 726–737. [Google Scholar]
- Maisey HC, Hensler M, Nizet V, Doran KS. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J Bacteriol. 2007;189:1464–1467. doi: 10.1128/JB.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandlik A, Swierczynski A, Das A, Ton-That H. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti AG, Zingaretti C, Falugi F, Capo S, Bombaci M, Bagnoli F, et al. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol. 2007;64:968–983. doi: 10.1111/j.1365-2958.2007.05704.x. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Schneewind O. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol Microbiol. 2006;62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Jensen ER, Lenoy E, Schneewind O. Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. Proc Natl Acad Sci USA. 2000;97:5510–5515. doi: 10.1073/pnas.080520697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian SK, Ton-That H, Schneewind O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- Mishra A, Das A, Cisar JO, Ton-That H. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol. 2007;189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci USA. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallapareddy SR, Singh KV, Sillanpaa J, Garsin DA, Hook M, Erlandsen SL, Murray BE. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- Scott JR, Zahner D. Pili with strong attachments: gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- Swierczynski A, Ton-That H. Type III pilus of corynebacteria: pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188:6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in Gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004a;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- Ton-That H, Marraffini LA, Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004b;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- Weiss WJ, Lenoy E, Murphy T, Tardio L, Burgio P, Projan SJ, et al. Effect of srtA and srtB gene expression on the virulence of Staphylococcus aureus in animal models of infection. J Antimicrob Chemother. 2004;53:480–486. doi: 10.1093/jac/dkh078. [DOI] [PubMed] [Google Scholar]
- Yellaboina S, Ranjan S, Chakhaiyar P, Hasnain SE, Ranjan A. Prediction of DtxR regulon: identification of binding sites and operons controlled by Diphtheria toxin repressor in Corynebacterium diphtheriae. BMC Microbiol. 2004;4:38. doi: 10.1186/1471-2180-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung MK. Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med. 1999;10:120–138. doi: 10.1177/10454411990100020101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.