Abstract
Various cell-surface multisubunit protein polymers, known as pili or fimbriae, have a pivotal role in the colonization of specific host tissues by many pathogenic bacteria. In contrast to Gram-negative bacteria, Gram-positive bacteria assemble pili by a distinct mechanism involving a transpeptidase called sortase. Sortase crosslinks individual pilin monomers and ultimately joins the resulting covalent polymer to the cell-wall peptidoglycan. Here we review current knowledge of this mechanism and the roles of Gram-positive pili in the colonization of specific host tissues, modulation of host immune responses and the development of bacterial biofilms.
Sortase covalently links proteins to peptidoglycan
Surface components of a bacterium enable the organism to sense and assess its environment, and function as major virulence determinants in many pathogens. In Gram-negative bacteria, the surface proteins are embedded within the outer membrane lipid bilayer, which encapsulates the periplasm and the peptidoglycan layer. Devoid of an outer membrane, the Gram-positive bacteria use the cell wall as the scaffold for displaying a wide variety of surface molecules, which include teichoic acid, lipoteichoic acid and several protein adhesins [1]. Although some of these surface proteins are bound to the cell wall noncovalently, many other proteins are anchored covalently to the peptidoglycan [1].
The discovery of covalent linkage of a protein to the peptidoglycan emerged from the classic work on protein A of Staphylococcus aureus by Sjöquist and colleagues [2,3]. They showed that protein A is associated with the cell wall, and that it could only be released by treating bacteria with cell-wall hydrolases. Three decades later, Schneewind and colleagues discovered the sortase enzyme (SrtA, for surface protein sorting A) that catalyzes cell-wall anchoring of surface proteins in Staph. aureus [4,5]. Joining of protein A to the cell wall requires a specific motif, the cell-wall sorting signal (CWSS), within the C-terminus of the protein. The CWSS comprises the amino acid sequence LPxTG, a sequence that is conserved in all surface proteins anchored by SrtA. This motif is followed by a hydrophobic membrane-spanning domain and a positively charged tail, which are also important for sortase-catalyzed anchoring of surface proteins [6]. To anchor a surface protein to the cell wall, sortase cleaves the LPxTG motif, between threonine (T) and glycine (G), and links the threonine residue of the cleaved polypeptide to the amino group of the cross-bridge within the peptidoglycan structure [7,8].
The sortase SrtA, referred to as the housekeeping sortase, is present in all Gram-positive bacterial genomes sequenced to date, except for Mycobacterium and Microplasma [9–11]. Many pathogens harbor additional sortases, designated class B, C and D [9,11] (Figure 1), which are involved in iron acquisition [12], sporulation [13] and pilus assembly (see below). Sortases of the class C family form the largest group and are often present in multiple copies in a genome [9,11,14]. These sortases are encoded together with their substrates, which constitute the various types of pili in many pathogens. For a comprehensive description of cell-wall protein sorting, the reader is referred to an excellent recent review [15]. Here we focus on the mechanism of sortase-mediated pilus assembly and the function of pili in pathogenesis of Gram-positive bacteria.
Figure 1.
Phylogeny of sortase homologs. Clustal X [58] was used to align the protein sequences of sortase homologs of Gram-positive bacteria. The phylogenetic tree of the housekeeping sortases (green) was reconstructed with the neighbor-joining algorithm [59] using the program PAUP 4.0 10β. Numbers on the branches specify bootstrap values. Different classes of sortase are color-coded; the number of sortases in each class is indicated by dots.
Assembly and architecture of pili: lessons from Corynebacterium diphtheriae
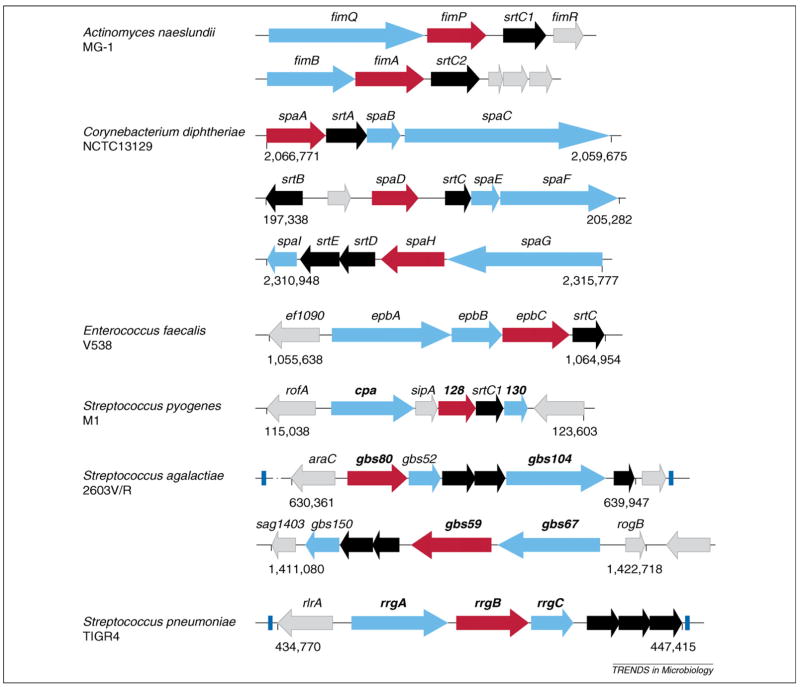
Corynebacterium diphtheriae, the causative agent of pharyngeal diphtheria [16], was one of the earliest organisms in which the presence of pili was described [17]. A major advance in our understanding of Gram-positive pili and a possible mechanism of their assembly, however, came from the characterization by Yeung and colleagues of fimbriae in Actinomyces naeslundii [18,19]. By cloning and sequencing the fimbriae-encoding genes, this work disclosed the presence of the LPxTG motif in both FimP and FimA, two proteins that, respectively, constitute the type 1 and type 2 fimbriae of Actinomyces (Figure 2). Thus, this work provided the earliest clue that pilus or fimbria assembly in Gram-positive bacteria might be catalyzed by sortase. Subsequent work in corynebacteria proved this to be true.
Figure 2.
Pilus gene operons. Graphic presentation of pilus gene clusters identified in the chromosome of Actinomyces naeslundii MG-1, Corynebacterium diphtheriae NCTC13129, Enterococcus faecalis V538, Streptococcus pyogenes M1, Streptococcus agalactiae 2603V/R and Streptococcus pneumoniae TIGR4. Each cluster contains pilus-specific sortase gene(s) (black), genes encoding a major subunit (red) and minor pilins (aqua). Some of the clusters are flanked by transposable elements (blue). Genes encoding pilins used in vaccine studies are shown in bold. Uncharacterized genes are colored in gray. Numbers below clusters indicate the genomic location of pilus gene clusters.
When the unassembled genome sequence of C. diphtheriae was interrogated for the presence of sortase homologs that are colocalized with genes encoding LPxTG-containing proteins, three different pilus gene clusters were uncovered [14] (Figure 2). By raising antibodies against these proteins and performing biochemical, genetic and immunoelectron microscopic studies, corynebacterial pili were found to be made of a major subunit and two minor components, one decorating the pilus shaft and the other restricted to the tip region (Box 1) [14]. Moreover, each gene cluster formed a distinct pilus, designated as the SpaA-, SpaD- and SpaH-type pilus according to the major subunit that makes up each pilus (Box 1 and Figure 2). The assembly of these pili required cognate sortases that are encoded within each pilus gene cluster [14,20–22]. These sortases (SrtA, SrtB, SrtC, SrtD and SrtE) belong to the class C family, whereas the housekeeping sortase (SrtF) belongs to class A. All function by a common mechanism (Box 2).
Box 1. Assembly of heterotrimeric corynebacterial pili requires pilus-specific sortases.
The SpaA-type pili are encoded by the spaA-srtA-spaB-spaC gene cluster (Figure 2). Considered the prototype, these pili are composed of the major pilin (SpaA), forming the shaft, a minor pilin (SpaB), found dispersed along the shaft, and the tip pilin (SpaC) [14]. Studies of nonpolar, in-frame deletion mutants showed that SpaA is essential for the formation of the pilus structure, whereas the two minor pilins, SpaB and SpaC, are dispensable for pilus assembly [23]. That these pili are covalently crosslinked and anchored to the cell wall was evident from biochemical analysis. Treating cells with muramidase, a murein hydrolase, released pilus polymers into the extracellular milieu [14]. Moreover, pilus polymers remained intact when cells were treated with formic acid or boiled in sodium dodecyl sulfate (SDS), conditions that disrupt noncovalently bound polymers [14].
The assembly of the heterotrimeric SpaD- and SpaH-type pili requires the sortases that are encoded within the respective gene clusters (Figure 2). Unlike the SpaA-type pili, which are assembled by a single sortase, two sortases catalyze the assembly of the SpaD-type pili. Whereas either SrtB or SrtC is sufficient to polymerize the major pilin, SpaD, and attach the tip pilin, SpaF, crosslinking of the other minor pilin, SpaE, requires sortase SrtB [22]. The case of the SpaH-type pili is different in that although SrtD seems more important, the efficient assembly of the pilus requires both SrtD and SrtE, as indicated by the reduced length of SpaH polymers and the accumulation of SpaH monomers in either srtD or srtE single mutants [20].
Box 2. Sortase reaction mechanism.
How does sortase catalyze the assembly of a covalently crosslinked heterotrimeric pilus structure? The reaction mechanism was revealed by a series of molecular genetic experiments, which showed that the major subunits provide all the necessary elements for pilus assembly. Homology analysis of the major pilins SpaA, SpaD and SpaH uncovered three key motifs involved in pilus assembly: the pilin motif, WxxxVxVYPKN with an invariant lysine, the E box with an invariant glutamate and the CWSS [23]. Conservation of these motifs led to the hypothesis that sortase-mediated pilus assembly is similar to the transpeptidation reaction defined for cell-wall anchoring of surface proteins. To join two pilin subunits covalently, sortase would cleave the LPxTG motif of a pilin monomer and form an acyl–enzyme intermediate, which is in turn resolved by the nucleophilic attack from a specific amino group provided by the second monomer (Figure 3; see Refs [25] and [15]). According to this model, the lysine of the pilin motif provides the necessary electron donor through its side-chain amino group for the transpeptidation reaction (Figure 3). Consistent with this, an alanine substitution of the lysine in the SpaA pilin motif abolishes pilus assembly, as is also true for a scrambled LPxTG motif [23]. The sufficiency of these motifs was demonstrated by creating a chimeric protein in which the staphylococcal enterotoxin B was fused to the SpaA amino terminus, which provided the pilin motif, and the SpaA carboxyl terminus, which provided the sorting signal. When expressed in corynebacteria, this fusion protein is polymerized and cell-wall-anchored, and the process is dependent on the conserved lysine of the pilin motif in addition to sortase SrtA [23]. Therefore, the pilin motif and the sorting signal are the only two elements that determine sortase specificity in pilus assembly.
Functions of a pilus-specific sortase and the housekeeping sortase in pilus biogenesis
SrtA, the single sortase encoded within the SpaA gene cluster, specifically catalyzes the covalent crosslinking of individual pilin monomers. Immunoelectron microscopy and biochemical analysis showed that a strain expressing only SrtA not only forms the SpaA-type pili, but also anchors pili to the cell wall [21,23]. However, this strain secretes significant amounts of polymerized pilins into the culture medium, indicating that an efficient cell-wall anchoring of the pili might involve one or more other sortases. Indeed, a strain that lacks only the housekeeping sortase SrtF also releases SpaA polymers into the culture medium [23]. Moreover, this strain also secretes the SpaD and SpaH pili abundantly [24]. Clearly, pilus assembly in C. diphtheriae involves two sortases, a pilus-specific sortase for pilin polymerization and the housekeeping sortase for efficient anchoring of pili to the cell wall (Figure 3) [24].
Figure 3.
Model of pilus biogenesis. Pilin precursors (SpaA, denoted by pink circles; SpaB, denoted by dark-aqua ovals; and SpaC, denoted by light-aqua ovals) are synthesized in the cytoplasm and translocated across the membrane by the Sec machinery (step 1). At the exoplasm, the precursors subsequently form acyl–enzyme intermediates with the housekeeping sortase (green) (step 2) or pilus-specific sortase (gray). These enzyme intermediates are capable of transferring these pilins to the lipid II precursor, thus anchoring monomeric pilins to the cell wall (step 3 in A). The pilus-specific sortase catalyzes pilus polymerization (step 4) by the mechanism described in Box 2. Pilus polymerization is terminated when pilus polymers are transferred to lipid II in one of two possible ways. In one pathway, the housekeeping sortase having a SpaA monomer would receive the pilus polymer from the pilus-specific sortase (step 5) and transfer the polymer to lipid II (step 6). In the alternative pathway (not shown), the pilus-specific sortase would transfer the polymer directly to lipid II. Red diamonds denote the D-diaminopimelic moiety of the cell wall pentapeptide. SecYEG stands for the three subunits of the general secretion machinery (Sec).
Crosslinking of minor pilins
A crucial question is how are the minor pilins linked to the pili. Like the major pilin, these proteins contain the LPxTG motif; however, neither an E box nor the pilin motif is present in minor pilins. Because SpaC is located at the tip of the pilus shaft, it is logical to propose that SpaC assembly occurs by the same sortase-mediated pathway, employing the SpaC sorting signal and the SpaA pilin motif (Figure 3) [23,25]. However, the mechanism by which the minor pilin SpaB is incorporated into the structure remains a mystery. One piece of evidence points to the importance of the SpaA E box in SpaB incorporation. When the invariant glutamate residue of the E box is substituted by alanine or arginine, neither SpaA polymerization nor SpaC joining at the tip is affected; however, there is no incorporation of SpaB into the structure [23]. It is conceivable that an anhydride bond might be formed between the SpaA E box glutamate and the carboxyl group of the SpaB sorting signal threonine, but there is no evidence that sortase possesses such an activity. Instead, the E box might provide some form of structural role for pilus assembly [23], which remains to be investigated. To date, the site of SpaA to which SpaB is crosslinked remains unknown.
Cell-wall anchoring
The next obvious question is how does sortase catalyze the anchoring of pili to the cell wall? Surface proteins in Gram-positive bacteria are covalently attached to the cell wall by joining the sorting signal threonine to the amino group of the peptidoglycan cross-bridge. There is no reason why the same mechanism would not be used by sortase to link pilus fibers to the cell wall (Figure 3). Consistent with this, mutations in the SpaA LPxTG motif abrogate pilus polymerization and cell-wall anchoring of SpaA [21,23].
Cell-wall anchoring of monomeric pilins
The presence of the LPxTG motif in all pilins makes them substrates for sortase-catalyzed cell-wall anchoring in the monomeric form. In fact, when the SpaA pilin motif lysine is mutated, pilus polymerization is abolished, but the mutant protein is avidly anchored to the cell wall [21,23]. In the absence of SpaA, the minor pilins SpaB and SpaC are also linked to the cell wall [21]. This cell surface display of monomeric pilins is also observed for the SpaD- and SpaH-type pili (A. Swierczynski, unpublished). Display of pilins on the cell surface, both in the form of pili and as typical cell-wall-anchored proteins, is probably a general feature of pilins in various Gram-positive bacteria [26,27].
General pilus assembly mechanism
In the past few years, many laboratories have investigated pili from important pathogens using molecular genetic and biochemical approaches similar to those described for corynebacteria. Studies of Streptococcus agalactiae, Strep. pyogenes, Strep. pneumoniae, Enterococcus faecalis and A. naeslundii show typical clustering of pilus genes together with multiple pilus-specific sortases (Figure 1), often flanked by transposon elements (Figure 2), indicative of their acquisition by horizontal gene transfer [28–33]. Similar to corynebacterial pili, the streptococcal and enterococcal pili are heterotrimeric, with the pilus shaft containing two minor pilins, whose localization has not been well defined. A distinguishing feature of A. naeslundii is that the two different types of fimbriae produced by this bacterium are each made of two pilins, with the major fimbrial protein forming the shaft and the minor fimbrial protein localized at the tip and the cell surface [33]. A conserved feature of most Gram-positive pili is that their assembly requires specific sortases located within their respective loci; moreover, the major pilins harbor the pilin motif, E box and the CWSS. However, certain strains of Strep. pyogenes assemble a pilus structure made of a major subunit (T antigen) that lacks both the pilin motif and the E box [10,29]. The corresponding motif responsible for T antigen polymerization needs to be identified. Curiously, pilus and sortase genes of streptococci are flanked by genes that encode transcriptional regulators belonging to the RofA or AraC family, and these factors promote the transcription of pilus genes [27,30,31]. This regulation could be physiologically significant, and thus unraveling the cues that modulate the activity of these regulators will be important. Unlinked regulators might modulate the expression of pilus gene clusters in other Gram-positive pathogens in which no obvious regulatory gene is associated with the pilus gene cluster.
Function of pili in bacterial colonization
The role of pili in host cell adherence and tissue tropism has been well established from studies of Gram-negative bacteria, which contain distinct types of pili, namely type I pili, Pap pili, type IV pili and curli pili [16,34–36]. Gram-positive pili also have a major role in host–pathogen interaction and the eventual colonization of proper tissues by many pathogens.
Pilus-mediated adherence of C. diphtheriae to pharyngeal epithelial cells
Recent work has revealed that of the three pilus structures displayed by C. diphtheriae, only the SpaA-type pili mediate corynebacterial adherence to the human pharyngeal cells, as the SpaD- and SpaH-type pili help adhesion to the laryngeal and lung epithelial cells [21]. Evidence for this important conclusion was reached from a comprehensive analysis of host cell binding using a large battery of corynebacterial mutants. First, corynebacteria that lack all sortases bind poorly to several types of epithelial cells tested. By contrast, deletion of srtA alone abrogates binding only to the pharyngeal cells. Second, a strain that expresses only the SpaA-type pili adheres avidly to the pharyngeal cells, but not so well to other cells [21], thus demonstrating a crucial role of pili in tissue tropism.
Minor pilins as adhesins
To determine the identity of the adhesin(s) involved, individual pilin mutants were analyzed [21]. Surprisingly, the spaA deletion mutant, which does not form any pilus fibers, binds pharyngeal cells well, whereas the adherence is compromised significantly when either spaB or spaC is deleted. Consistent with the role of minor pilins in selective adherence, the spaBC double mutant shows marginal binding to pharyngeal cells. As expected, the two minor pilins are displayed on the surface in the absence of the pilus shaft, and this surface display is dependent on the sortase SrtA and the LPxTG motif of SpaB and SpaC pilins [21]. Antibodies against either SpaB or SpaC but not SpaA abrogate corynebacterial adherence to pharyngeal cells. The crucial evidence for the direct role of minor pilins in adherence comes from biochemical studies in which latex beads conjugated with recombinant minor pilins bind to the pharyngeal cells, but not to cells of lung or laryngeal origin [21].
Independent studies of other organisms further emphasize the crucial role of minor pilins and indicate that Gram-positive pili have a general role in tissue tropism. Dramsi and coworkers showed that adhesion of Strep. agalactiae (group B streptococcus, GBS) strain NEM316 to human lung and cervial epithelial cells involves a minor pilin (GBS1478), and that the major shaft protein is dispensable for this binding [27]. Similarly, the minor pilin PilA is required for the initial attachment of another GBS strain to human brain microvascular endothelial cells (hBMEC), whereas the major protein PilB mediates intracellular invasion of brain endothelium by this organism [37]. Finally, Kehoe and colleagues demonstrated that minor pilins mediate adhesion of Strep. pyogenes (group A streptococcus, GAS) to both human tonsil epithelium and primary human keratinocytes, the two main sites of infection by this human-specific pathogen [38]. However, in this case, pilus biosynthesis is also required for efficient adhesion.
Role of minor pilins in intimate adhesion and pathogenesis: a speculative model
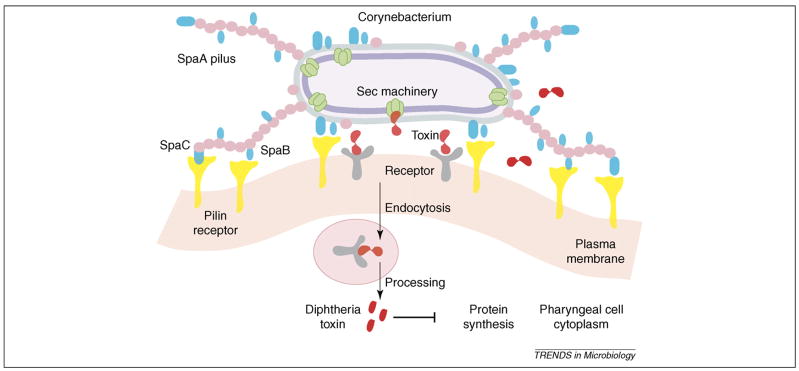
The mounting evidence that the minor pilins are crucial for tissue tropism and that they are components of both the cell wall and the pili suggests a molecular model of how pili might orchestrate bacterial adhesion during infection [21,39]. The display of adhesins as part of extended pilus fibers would aid in the initial distant contacts that bacteria must make to seek out and attach to the desired host cells. The bacteria might then undergo additional contacts with the host cells through the binding of cell-wall-anchored minor pilins, thereby forming an intimate zone of adhesion between the bacterium and the host cells (Figure 4). This intimate adhesion would serve three important functions in pathogenesis. First, the proximity of the bacterial surface to the plasma membrane of the host cell would promote additional ligand–receptor interactions involving a variety of non-pilus adhesins. Second, and most importantly, the intimate zone of adhesion would permit the efficient delivery of virulence factors, essentially recapitulating the efficiency of the direct delivery of toxins as mediated by the type III secretion systems of Gram-negative bacteria [40]. Third, the intimate adherence would facilitate the intracellular invasion of host cells by Gram-positive pathogens such as GBS and GAS.
Figure 4.
Model of pilus-mediated adhesion and pathogenesis. This general model applies to various Gram-positive pathogens. We depict Corynebacterium diphtheriae as a specific case. Adhesive fibers make initial contact with host cell receptors, whereas cell-wall-linked pilins mediate the formation of an intimate zone of adhesion. This enables additional ligand–receptor interactions, the efficient delivery of virulence factors, and the intracellular invasion of certain pathogens.
Host cell receptors for Actinomyces fimbriae
Pilus-mediated adherence to host cells must involve specific receptors for the various pilin adhesins. Although nothing is known for corynebacterial, streptococcal or enterococcal pilus receptors, important progress on this has been made in Actinomyces. A. naeslundii is one of the earliest organisms found in the oral cavity after birth and on cleaned tooth surfaces [41]. A. naeslundii has the ability to colonize the tooth and mucosal surfaces, thus providing a niche for subsequent colonization of other oral bacteria, including streptococci and Gram-negative anaerobes such as Veillonella atypica and Fusobacterium nucleatum [41,42] (see below). A. naeslundii assembles two types of fimbriae on the cell surface [43]. The type 1 fimbriae mediate bacterial adherence to salivary acidic proline-rich proteins (PRPs) and statherines that coat the tooth enamel [44,45]. By contrast, type 2 fimbriae recognize β-linked galactose and N-acetylgalactosamine-containing motifs present in host cell surface glycoconjugates and in the polysaccharides of oral streptococci [43,46]. These interactions are neutralized by fimbria-specific antibodies [43,47,48]. Each of the two fimbrial types contains, in addition to the major fimbrial subunit, a minor protein that is localized at the fimbrial tip and the bacterial surface (Figure 2) [33]. These minor fimbrial proteins might act as adhesins for the host cell receptors and consequently have important roles in the development of dental plaques [33].
Pili and biofilm formation
Biofilms are differentiated bacterial communities encased in a protective and adhesive matrix that typically resists antibiotic therapy and poses significant health problems. A striking example of a bacterial biofilm is dental plaque, which harbors more than 500 bacterial species, including Actinomyces and oral streptococci [49]. The oral actinomycetes and streptococci coaggregate together to colonize the tooth surface, and this promotes subsequent colonization by other oral bacteria. That coaggregation is essential for the formation of oral biofilms has been demonstrated in a flow-cell system for biofilm development [50,51]. In this experimental setting, Strep. oralis and A. naeslundii T14V, when grown together, exhibit a mutualistic interaction characterized by ‘luxuriant interdigitated growth’. When grown individually, however, neither organism displays growth characteristic of a single-species biofilm, although the bacteria do adhere to the flow-cell surface. Binding of Actinomyces with the tooth enamel and other oral bacteria are fimbria-mediated. Whether fimbriae are required for biofilm formation in vivo has not been determined, and neither has the contribution of streptococcal pili in this process.
Two recent studies further highlight the importance of pili in biofilm development in other Gram-positive pathogens [32,52]. Genetic studies of E. faecalis suggest that pili could be directly involved in this process [32]. The E. faecalis pili (named Ebp, for endocarditis and biofilm-associated pili) are heterotrimeric, and their assembly is sortase-mediated (Figure 2). Non-piliated deletion mutants of E. faecalis show a prominent defect in biofilm formation, an essential step for successful infection by this pathogen. These mutants are also attenuated for virulence in a rat endocarditis model [32]. It is not clear whether the major subunit contributes to optimal biofilm formation, although a minor component seems sufficient for this process. Similar studies of GAS demonstrate the role of pili in both bacterial adherence to pharyngeal cells and biofilm formation, and the data suggest that the major pilin is involved in both cases [52].
Pili and the host immune response
The fact that pili promote bacterial adherence to host epithelium suggests that pili might also contribute to the virulence of pathogens. A recent study of Strep. pneumoniae has uncovered important functions of pili in pathogenesis and host immune responses [31]. By engineering a streptococcal mutant that lacks sortases and pilins encoded by a single pilus gene cluster, the authors demonstrated that this mutant produces no pili and is defective in binding to epithelial cells grown in cell culture. This nonpiliated mutant is attenuated in a mouse infection model and outcompeted by the wild type in the upper airways, lungs and blood. Importantly, piliated pneumococci evoke host inflammatory responses, as evident from the elevated levels of tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) release compared with the deletion mutant [31]. This inflammation might promote subsequent bacterial invasion of the tissue consequent on damage to the mucosal barrier. Thus, both initial bacterial binding and subsequent invasion are enhanced by pili in pneumococci. A key problem that remains to be solved is how do pili evoke the inflammatory response. If pilins are not the direct modulators of immune signaling, the intimate adherence promoted by pilins could certainly enhance the required signaling, as proposed above (Figure 4).
Pili as vaccine candidates
The presence of pili on bacterial cell surfaces and their demonstrated role in bacterial adherence make them ideal candidates for vaccines. The immunogenicity of pilins is well documented [29,31,53]. In addition, there is evidence for a natural antibody response to the enterococcal pilus, because sera from infected patients contain antibodies against the various pilins of this organism [54]. To design a universal protein-based vaccine against GBS strains, Maione and coworkers immunized pregnant mice with several hundred individual recombinant proteins [53]. The resulting offspring were then challenged with a dose of wild-type streptococci that killed 80% to 90% of the pups. Four antigens tested in this study conferred significant protection on the infected infant mice [53]. Moreover, when used in combination, these antigens were more effective and induced complement-dependent opsonophagocytic killing [53]. Remarkably, three of these four protective antigens were pilins, which are encoded by the two pilus islands present in many strains of GBS [27,28,55]. The protective function of the minor pilin is consistent with its documented role in adherence [27]. Pili might have more functions in pathogenesis and the modulation of the host immune response than currently known. Certainly, the major and minor pilins might trigger independent signaling events in the infected cells, thus culminating in a more robust immune response.
The immunoprotective effect of the pilin-based vaccine has been reported for additional pathogens. In the case of GAS, immunization of mice with a combination of recombinant pilus proteins of serotype M1 conferred protection against mucosal challenge with virulent GAS [29]. Similarly, in a mouse model of intraperitoneal infection, both active and passive immunization with recombinant pilus subunits afforded protection against lethal challenge with the Strep. pneumoniae serotype 4 strain [56]. Although these reports are encouraging for efforts to develop pilus-based multivalent vaccines against potent streptococcal pathogens, such vaccines could not have broad use for every pathogen because many clinical isolates of these pathogens do not contain pilus genes [57].
Concluding remarks and future directions
The past few years have seen dramatic advances in our knowledge of the biology of Gram-positive pili. The evidence obtained so far establishes the prominent roles of pili in pathogenesis by mediating bacterial adhesion, invasion, aggregation, biofilm formation and modulation of immunity. It is also clear that combinations of pilus proteins should serve as broad-coverage vaccines against some life-threatening infections, particularly with antibiotic-resistant organisms.
Less clear, however, are many aspects of the molecular mechanism of pilus assembly, and the way this process is regulated temporally and spatially (see Box 3). An equally important gap in our knowledge is the identity of the receptors that are recognized by the various pili and the function of these receptors in signaling and host defense mechanisms. In addition, important aspects of immune responses that are modulated by the various pili are far from being understood. Thus, we can look forward to an exciting era of further research addressing these problems, and hope that unique therapeutic approaches for combating some of these deadly infections will emerge.
Box 3. Outstanding questions.
How is secretion of proteins through the Sec machinery coupled to the processes of cell wall sorting and pilus assembly?
What governs the decision between pilus polymerization and cell wall anchoring?
What determines the substrate specificity of a sortase?
How does the housekeeping sortase modulate pilus assembly?
What are the other factors that take part in the assembly process?
Are pilus gene expression and assembly regulated by host cues during infection?
Are pilus gene expression and pilus assembly regulated by environmental cues?
What roles do pili have during in vivo infection?
How do pilins trigger inflammatory responses?
Acknowledgments
We are indebted to our colleagues Andrew Gasper, Arunima Mishra and Anu Swaminathan for valuable contributions that enhanced our overall understanding of pilus assembly and function. The authors’ work has been supported by grants from the Charles H. Hood Foundation and the National Institute of Allergy and Infectious Diseases, NIH grant AI061381 to H.T.T. and in part by intramural graduate fellowships to A.M. and A.S.
References
- 1.Navarre WW, Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol Mol Biol Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sjöquist J, et al. Protein A isolated from Staphylococcus aureus after digestion with lysostaphin. Eur J Biochem. 1972;29:572–578. doi: 10.1111/j.1432-1033.1972.tb02023.x. [DOI] [PubMed] [Google Scholar]
- 3.Sjöquist J, et al. Localization of protein A in the bacteria. Eur J Biochem. 1972;30:190–194. doi: 10.1111/j.1432-1033.1972.tb02086.x. [DOI] [PubMed] [Google Scholar]
- 4.Schneewind O, et al. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 5.Mazmanian SK, et al. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 6.Schneewind O, et al. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazmanian SK, et al. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol. 2001;40:1049–1057. doi: 10.1046/j.1365-2958.2001.02411.x. [DOI] [PubMed] [Google Scholar]
- 8.Ton-That H, et al. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:269–278. doi: 10.1016/j.bbamcr.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 9.Comfort D, Clubb RT. A comparative genome analysis identifies distinct sorting pathways in Gram-positive bacteria. Infect Immun. 2004;72:2710–2722. doi: 10.1128/IAI.72.5.2710-2722.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- 11.Dramsi S, et al. Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Mazmanian SK, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 13.Marraffini LA, Schneewind O. Targeting proteins to the cell wall of sporulating Bacillus anthracis. Mol Microbiol. 2006;62:1402–1417. doi: 10.1111/j.1365-2958.2006.05469.x. [DOI] [PubMed] [Google Scholar]
- 14.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 15.Marraffini LA, et al. Sortases and the art of anchoring proteins to the envelopes of Gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 17.Yanagawa R, Honda E. Presence of pili in species of human and animal parasites and pathogens of the genus Corynebacterium. Infect Immun. 1976;13:1293–1295. doi: 10.1128/iai.13.4.1293-1295.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeung MK, Ragsdale PA. Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect Immun. 1997;65:2629–2639. doi: 10.1128/iai.65.7.2629-2639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeung MK, et al. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. Infect Immun. 1998;66:1482–1491. doi: 10.1128/iai.66.4.1482-1491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swierczynski A, Ton-That H. Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188:6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandlik A, et al. Corynebacterium diphtheriae employs specific minor pilins to target human pharyngeal epithelial cells. Mol Microbiol. 2007;64:111–124. doi: 10.1111/j.1365-2958.2007.05630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ton-That H, et al. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 24.Swaminathan A, et al. Housekeeping sortase facilitates the cell wall anchoring of pilus polymers in Corynebacterium diphtheriae. Mol Microbiol. 2007;66:961–974. doi: 10.1111/j.1365-2958.2007.05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Nelson AL, et al. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol Microbiol. 2007;66:329–340. doi: 10.1111/j.1365-2958.2007.05908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dramsi S, et al. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 28.Lauer P, et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 29.Mora M, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci U S A. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LeMieux J, et al. RrgA and RrgB are components of a multisubunit pilus encoded by the Streptococcus pneumoniae rlrA pathogenicity islet. Infect Immun. 2006;74:2453–2456. doi: 10.1128/IAI.74.4.2453-2456.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barocchi MA, et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nallapareddy SR, et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest. 2006;116:2799–2807. doi: 10.1172/JCI29021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra A, et al. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol. 2007;189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanassi DG, Hultgren SJ. Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system. Methods. 2000;20:111–126. doi: 10.1006/meth.1999.0910. [DOI] [PubMed] [Google Scholar]
- 35.Sauer FG, et al. Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 36.Burrows LL. Weapons of mass retraction. Mol Microbiol. 2005;57:878–888. doi: 10.1111/j.1365-2958.2005.04703.x. [DOI] [PubMed] [Google Scholar]
- 37.Maisey HC, et al. Group B streptococcal pilus proteins contribute to adherence to and invasion of brain microvascular endothelial cells. J Bacteriol. 2007;189:1464–1467. doi: 10.1128/JB.01153-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abbot EL, et al. Pili mediate specific adhesion of Streptococcus pyogenes to human tonsil and skin. Cell Microbiol. 2007;9:1822–1833. doi: 10.1111/j.1462-5822.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 39.Telford JL, et al. Pili in Gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 40.Cornelis GR. The type III secretion injectisome. Nat Rev Microbiol. 2006;4:811–825. doi: 10.1038/nrmicro1526. [DOI] [PubMed] [Google Scholar]
- 41.Jost BH, Billington SJ. Actinomyces and Arcanobacterium spp. In: Fischetti VA, et al., editors. Gram-Positive Pathogens. ASM Press; 2006. pp. 736–749. [Google Scholar]
- 42.Nyvad B, Kilian M. Microbiology of the early colonization of human enamel and root surfaces in vivo. Scand J Dent Res. 1987;95:369–380. doi: 10.1111/j.1600-0722.1987.tb01627.x. [DOI] [PubMed] [Google Scholar]
- 43.Yeung MK. Molecular and genetic analyses of Actinomyces spp. Crit Rev Oral Biol Med. 1999;10:120–138. doi: 10.1177/10454411990100020101. [DOI] [PubMed] [Google Scholar]
- 44.Gibbons RJ, Hay DI. Human salivary acidic proline-rich proteins and statherin promote the attachment of Actinomyces viscosus LY7 to apatitic surfaces. Infect Immun. 1988;56:439–445. doi: 10.1128/iai.56.2.439-445.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibbons RJ, et al. Adsorbed salivary proline-rich protein 1 and statherin: receptors for type 1 fimbriae of Actinomyces viscosus T14V-J1 on apatitic surfaces. Infect Immun. 1988;56:2990–2993. doi: 10.1128/iai.56.11.2990-2993.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandberg AL, et al. Putative glycoprotein and glycolipid polymorphonuclear leukocyte receptors for the Actinomyces naeslundii WVU45 fimbrial lectin. Infect Immun. 1995;63:2625–2631. doi: 10.1128/iai.63.7.2625-2631.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clark WB, et al. Specific inhibition of adsorption of Actinomyces viscosus T14V to saliva-treated hydroxyapatite by antibody against type 1 fimbriae. Infect Immun. 1984;43:497–501. doi: 10.1128/iai.43.2.497-501.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Revis GJ, et al. Antibodies against the Ag2 fimbriae of Actinomyces viscosus T14V inhibit lactose-sensitive bacterial adherence. Infect Immun. 1982;36:1217–1222. doi: 10.1128/iai.36.3.1217-1222.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–437. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 50.Kolenbrander PE, et al. Communication among oral bacteria. Microbiol Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmer RJ, Jr, et al. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect Immun. 2001;69:5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manetti AG, et al. Streptococcus pyogenes pili promote pharyngeal cell adhesion and biofilm formation. Mol Microbiol. 2007;64:968–983. doi: 10.1111/j.1365-2958.2007.05704.x. [DOI] [PubMed] [Google Scholar]
- 53.Maione D, et al. Identification of a universal Group B Streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sillanpaa J, et al. A family of putative MSCRAMMs from Enterococcus faecalis. Microbiology. 2004;150:2069–2078. doi: 10.1099/mic.0.27074-0. [DOI] [PubMed] [Google Scholar]
- 55.Rosini R, et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 56.Gianfaldoni C, et al. Streptococcus pneumoniae pilus subunits protect mice against lethal challenge. Infect Immun. 2007;75:1059–1062. doi: 10.1128/IAI.01400-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Basset A, et al. Association of the pneumococcal pilus with certain capsular serotypes but not with increased virulence. J Clin Microbiol. 2007;45:1684–1689. doi: 10.1128/JCM.00265-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson JD, et al. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]