Abstract
Plants, similar to animals, form polarized axes during embryogenesis upon which cell differentiation and organ patterning programs are orchestrated. During Arabidopsis embryogenesis, establishment of the shoot and root stem cell populations occurs at opposite ends of an apical-basal axis. Recent work has identified the PLETHORA (PLT) genes as master regulators of basal/root fate1–3, while the master regulators of apical/shoot fate have remained elusive. Here we show that the PLT1 and PLT2 genes are direct targets of the transcriptional corepressor TOPLESS (TPL) and that PLT1/2 are necessary for the homeotic conversion of shoots to roots in tpl-1 mutants. Using tpl-1 as a genetic tool, we identify the CLASS III HOMEODOMAIN-LEUCINE ZIPPER (HD-ZIP III) transcription factors as master regulators of embryonic apical fate, and show they are sufficient to drive the conversion of the embryonic root pole into a second shoot pole. Furthermore, genetic and misexpression studies reveal an antagonistic relationship between the PLT and HD-ZIP III genes in specifying the root and shoot pole.
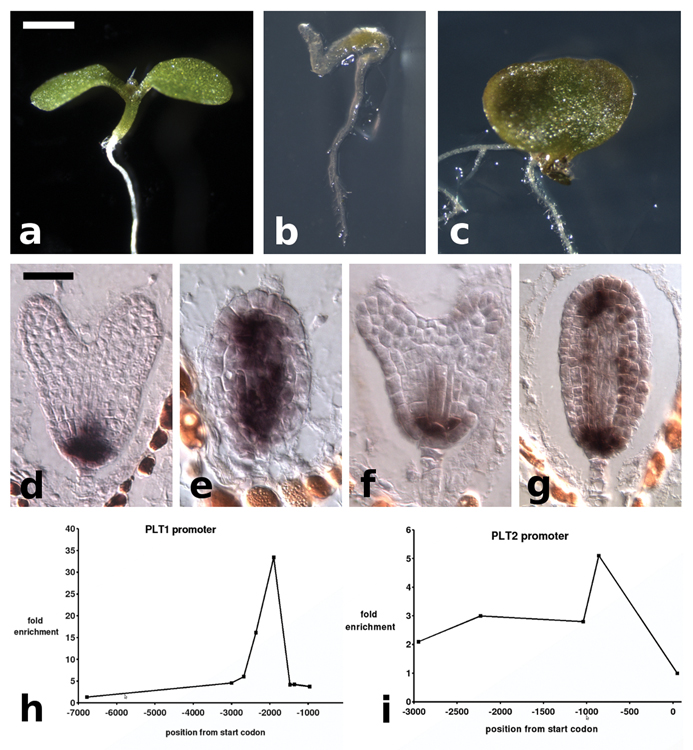
During early embryogenesis, the phytohormone auxin plays a critical role in apical-basal axis formation4. Auxin-induced gene expression involves degradation of AUX/IAA transcriptional repressors5, and IAA12/BODENLOS was shown to require the transcriptional co-repressor TOPLESS (TPL) for its function during embryonic root development6. In the dominant negative tpl-1 allele the embryonic shoot pole is transformed into a second root pole (Fig. 1a, b), indicating that root specifying genes must be actively repressed in the apical half of the embryo for normal apical/basal patterning7,8. tpl-1 is temperature sensitive and shows a high frequency of shoot to root transformation when embryos develop at 29°C8. In wild-type (WT) plants, PLT1 and PLT2 are expressed in the root meristem throughout embryo development1 (Fig. 1d, f). In tpl-1 grown at 29°C, both PLT1 and PLT2 are misexpressed in the apical domain, beginning at the heart stage (Fig. 1e, g). It has been shown that the PLT genes are sufficient to initiate ectopic roots when driven from an embryonic promoter1, suggesting that the misexpression seen in tpl-1 is causative of the double root phenotype. In agreement with this, double root formation was never observed in tpl-1 plt1-5 plt2-1 embryos grown at 29°C (n>1000) (Fig. 1c). Thus, the PLT genes are necessary for apical root formation in tpl-1. To assess whether the PLT genes are direct targets of TPL repression, we performed Chromatin Immunoprecipitation (ChIP) on TPLp::TPL-HA dissected ovules containing globular to heart stage embryos. We observed enrichment of regions in both the PLT1 and PLT2 promoters in the TPL ChIP samples (Fig.1h, i), indicating that TPL acts in the apical region of the embryo by directly repressing PLT expression.
Figure 1. Misregulation of PLT genes is necessary for tpl-1 apical to basal transformation.
a–c, Seedlings from embryos grown at 29°C. a, WT seedling. b, tpl-1 double root. c, tpl-1 plt1-5 plt2-1 monocot. d–g in situ hybridization with PLT1 and PLT2 antisense probe, embryos grown at 29°C. d, PLT1 expression in WT. e, PLT1 expression in tpl-1. f, PLT2 expression in WT. g, PLT2 expression in tpl-1. h, graph of fold enrichment at the PLT1 locus from ChIP of TPLp::TPL-HA. i, graph of fold enrichment at the PLT2 locus from ChIP of TPLp::TPL-HA. Scale bars, 1 mm (a–c) and 50 µm (d–g).
A second-site modifier screen on tpl-1 uncovered a semi-dominant mutant that completely suppressed the formation of double-root seedlings (Supplementary Fig. 2). Map-based cloning identified a single mis-sense mutation within the HD-ZIP III transcription factor PHABULOSA (PHB) in this mutant, which we designate phb-14d. This mutation resides within a known microRNA (miR)165/166 family binding site and is predicted to result in a loss of miR165/166 mediated regulation9,10. Notably, the observed increase in PHB transcript abundance is less severe than previously described alleles (Supplementary Fig. 2).
All five HD-ZIP III genes (PHB, PHAVOLUTA (PHV), REVOLUTA (REV), INCURVATA4/CORONA (ICU4/CNA), and ARABIDOPSIS THALIANA HOMEOBOX-8 (ATHB-8)) are predicted to be regulated by miR 165/1669–12. Semi-dominant gain-of-function (GOF) mutations in the miR binding site of PHB, PHV, REV, and ICU4 have been previously characterized for their role in specifying adaxial/dorsal fate in lateral organs and vasculature11,13–16. Although neither rev-10d nor icu4-1d display obvious embryonic patterning defects (Supplementary Fig. 2), they also completely suppress the shoot to root transformation seen in tpl-1 when grown at 29°C (n>1000) (Supplementary Fig. 2). These results suggest that the HD-ZIP III genes play an important role in promoting apical fate in early embryogenesis.
Consistent with the observation that the GOF mutations in HD-ZIP III genes restore apical fate to tpl-1 embryos, PHB, PHV, REV, and ICU4 are all expressed in an apical/central domain of the globular embryo17 (Fig. 2a, Supplementary Fig. 3). By the heart stage, the expression of all four genes expands to the adaxial domain of the cotyledons and throughout the provascular tissue17 (Fig. 2b, Supplementary Fig. 3). In tpl-1 embryos grown at 29°C, PHB, PHV, REV, and ICU4 expression is identical to WT at the globular stage, but is absent from the apical domain by the heart stage (Fig. 2c, Supplementary Fig. 3). Persistent PHB expression in the apical domain in tpl-1 phb-14d embryos grown at 29°C (Fig. 2d) suggests that increasing HD-ZIP III transcript abundance is sufficient to restore apical fate in tpl-1.
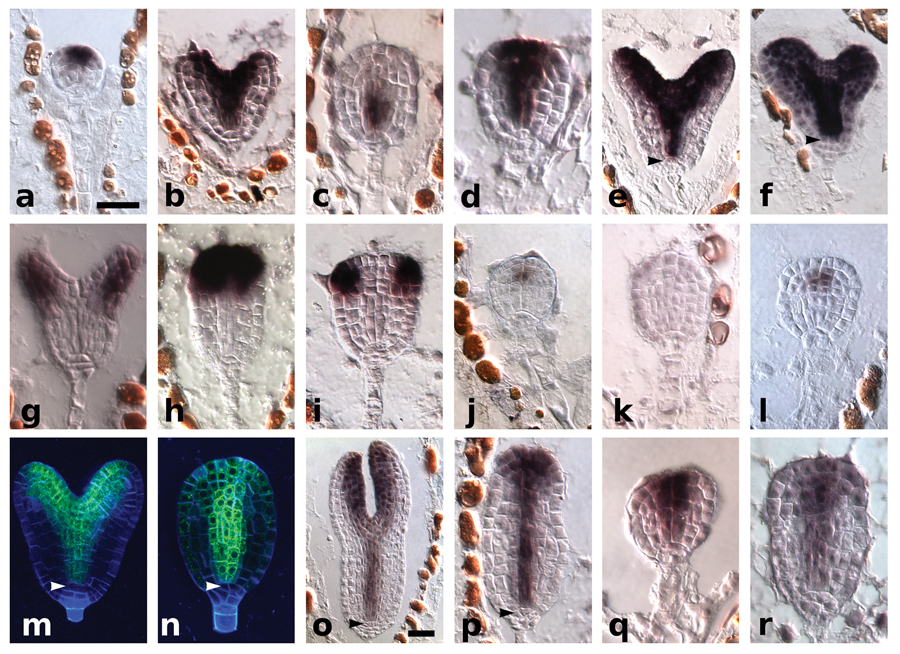
Figure 2. Molecular characterization of tpl-1, phb-14d, and tpl-1 phb-14d in embryos grown at 29°C.
a–f, PHB in situ hybridizations. WT globular (a) and early heart (b) stage, tpl-1 transition stage (c), tpl-1 phb-14d transition stage (d), phb-14d heart stage (e), and phb-1d heart stage (f). g–i, FIL in situ hybridizations. WT (g), tpl-1 (h), and tpl-1 phb-14d (i). j–l, WUS in situ hybridizations. WT (j), tpl-1 (k), and tpl-1 phb-14d. (l). m–p, miR165/166 sensor. GFP fluorescence in WT (m) and tpl-1 (n). GFP in situ hybridizations in WT (o) and tpl-1 (p). q, PHB in situ hybridizations in tpl-1 plt1-5 plt2-1. r, REV in situ hybridization in tpl-1 plt1-5 plt2-1. Arrowheads indicate the root meristem organizing center. Scale bars, 50 µm (a–r).
We then examined the radial organization of the apical domain of these embryos at 29°C by determining the expression patterns of the FILAMENTOUS FLOWER (FIL) and WUSCHEL (WUS) genes. FIL is normally expressed peripheral to the meristem in globular stage WT embryos (Supplementary Fig. 3) and becomes restricted to the abaxial/ventral side of the cotyledons in heart and later stages (Fig. 2g, Supplementary Fig. 3). In tpl-1, FIL expression expands throughout the apical domain from late globular to heart stage and is subsequently lost (Fig. 2h, Supplementary Fig. 3). WUS plays a critical role in SAM initiation and maintenance, and serves as a central-apical marker throughout embryogenesis18. In tpl-1, WUS is expressed correctly through the globular stage but is subsequently lost7 (Fig. 2j, k). These patterns of FIL and WUS misexpression in tpl-1 are identical to what is seen in the triple loss-of-function mutant rev-9 phb-6 phv-5, and show that the apical portion of tpl-1 embryos lose adaxial identity during the shoot to root transformation (Supplementary Fig. 4). FIL and WUS expression are restored to the WT pattern in tpl-1 phb-14d (Fig. 2i, l Supplementary Fig. 3). We then asked whether the loss of HD-ZIP III apical expression in tpl-1 was due to expansion of miR165/166 activity. Green Fluorescent Protein (GFP) based sensors for miR165/166 activity, in which the sensor is inactivated in all cells where miR165/166 are active, accumulate in a pattern similar to that of the HD-ZIP III mRNA and protein accumulation in the embryo (Fig. 2b, m, o, Supplementary Fig. 3, 5). Notably, the sensor is cleared from the root meristem organizing center from the globular stage on (Fig. 2m, o, Supplementary Fig. 5). If miR165/166 were misexpressed in tpl-1, we would expect to observe clearance of the sensor similar to that of the mRNA of HD-ZIP III genes. However, the sensor continues to accumulate in the apical domain of tpl-1 embryos grown at 29°C (Fig. 2n, p, Supplementary Fig. 5). Therefore, the loss of apical HD-ZIP III expression in tpl-1 is due to a mechanism independent of mir165/166 action and is likely at the level of transcriptional control. This represents a novel aspect of the control of HD-ZIP III gene expression and suggests that HD-ZIP III genes are excluded from the root by both transcriptional and post-transcriptional mechanisms. miR165/166 independent loss of HD-ZIP III expression in tpl-1 may be caused by PLT1/PLT2 misexpression in apical tissues. This idea is supported by the finding that PHB and REV expression is maintained in the apical domain of tpl-1 plt1-5 plt2-1 triple mutants (Fig. 2q, r), suggesting PLT1/PLT2 act as negative regulators of HD-ZIP III expression during embryogenesis. This is also consistent with what is observed in mutants where PHB mRNA is thought to be completely uncoupled from miR165/166 regulation, such as phb1-d and serrate (se). PHB mRNA accumulates throughout a wide pattern in phb1-d and se-5 embryos, and can lead to root defects and embryonic lethality. However, it is still restricted from the lenticular cell in these mutant backgrounds15,19 (Fig. 2f), an area of high PLT1/PLT2 expression (Fig. 1d, f).
Double root formation in tpl-1 requires PLT1/PLT2 misexpression and is suppressed by GOF HD-ZIP III mutations. Therefore, we examined PLT1/PLT2 gene expression in tpl-1 phb-14d and tpl-1 rev-10d double mutants grown at 29°C. PLT1 and PLT2 are still misexpressed in the vascular tissue and abaxial regions of developing cotyledons (Fig. 3a–d, Supplementary Fig. 4). However, we never observed PLT gene misexpression in the cells that would give rise to the shoot meristem. This suggests that the ability of PLT genes to promote root meristem formation in tpl-1 is dependent on misexpression in the meristem, and the GOF HD-ZIP III alleles are able to repress the PLT pathway in these cells.
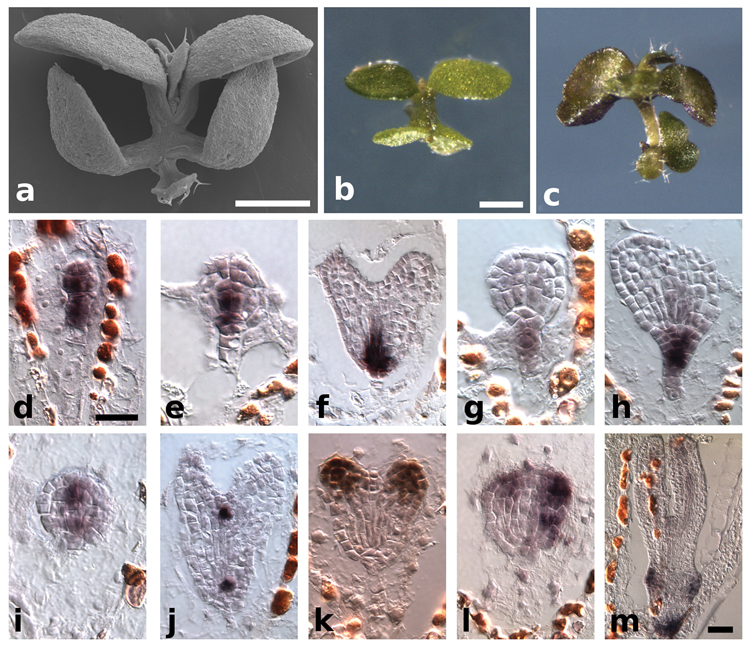
Figure 3. HD-ZIP III genes antagonize PLT function.
a–c, in situ hybridizations with PLT1/PLT2 in 29°C grown embryos. a, PLT1 in tpl-1 phb-14d. b, PLT1 in tpl-1 rev- 10d. c, PLT2 in tpl-1 phb-14d. d, PLT2 in tpl-1 rev-10d. e–h, PLT1/PLT2 in situ hybridizations in 24°C grown embryos. e, PLT1 in tpl-1. f, PLT2 in tpl-1. g, PLT1 in tpl-1 rev-9. h, PLT2 in tpl-1 rev-9. i, phb-14d plt1-5 plt2-1 seedling. j, Scanning electron micrograph (SEM) of rev-10d plt1-5 plt2-1 seedling. k, REV in situ hybridization in rev-10d plt1-2 plt2-1. Scale bars, 50 µm (a–h, k) and 1 mm (i, j).
Our genetic studies with the GOF HD-ZIP III alleles and tpl-1 implicate the HD-ZIP III genes in promotion of apical fate and antagonism of basal fate. phb-6 phv-5 rev-9 triple loss-of-function mutants produce pin-shaped seedlings11 similar to what is observed at low frequency in tpl-18 (Supplementary Fig. 4). tpl-1 seedlings rarely form double roots at 24°C (2%, n=682), whereas tpl-1 rev-9 significantly enhances the double root phenotype (48%, n=355). Additionally, loss of PHB and PHV in the tpl-1 rev-9 background further increases the frequency of the double-root phenotype (Table S1). At 24°C PLT1 and PLT2 are not broadly misexpressed in the apical domain of tpl-1 embryos (Fig. 3e, f). However, in tpl-1 rev-9 embryos grown at 24°C, PLT1 and PLT2 are misexpressed similar to tpl-1 at 29°C (Fig. 3g, h). These results show that at lower temperatures, the HD-ZIP III genes act to prevent the misexpression of PLT1/PLT2 in a tpl-1 background.
Given that GOF HD-ZIP III alleles suppress PLT1/PLT2-dependent apical root formation in tpl-1, we investigated the genetic interactions of GOF HD-ZIP III mutants with PLT loss-of-function mutants. phb-14d and rev-10d have no discernible embryonic root defects (Supplementary Fig. 2) and plt1 plt2 have only a minor defect, resulting in a properly organized seedling1. However, phb-14d plt1-5 plt2-1 triple mutant seedlings completely lacked a root and displayed only a rudimentary hypocotyl structure (Fig. 3i, Table S2), similar to plt1 plt2 plt3 plt4/bbm segregants3. rev-10d plt-5 plt2-1 seedlings showed an even more severe loss of both root and hypocotyl tissues (Fig. 3j, Table S2). In addition, rev-10d plt1 plt2 show expansion of REV transcript into the root meristem region (Fig. 3k) further suggesting that the PLT genes play an active role in repression of the HD-ZIP III genes, in addition to negative regulation by miR165/166.
To investigate whether the HD-ZIP III genes could impart apical polarity to basal tissue if misexpressed in the basal pole of the early embryo, we expressed miR resistant cDNAs of the HD-ZIP III genes fused to the glucocorticoid receptor domain (GR) under the control of the PLT2 promoter. When induced with dexamethasone during early embryogenesis, plants harboring either PLT2p::REVΔmiR-GR, PLT2p::PHBΔmiR-GR, or PLT2p::ICU4ΔmiR-GR transgenes produce seedlings that show a complete transformation of the root pole into a second shoot pole (Fig. 4a–c, Table S2, Supplementary Fig. 6).
Figure 4. HD-ZIP III gene misexpression can initiate apical fate and acts antagonistically to PLT gene function.
a, SEM image of PLT2p:REVΔmiR-GR seedling. b, PLT2p:PHBΔmiR-GR seedling. c, PLT2p::ICU4ΔmiR-GR seedling. d–h, in situ hybridizations with anti-sense PIN4 probe. d–f, WT 16-cell (d), globular (e), and heart (f) stage. g–h, PLT2p:REVΔmiR-GR globular stage (g) and late heart stage (h) embryos after dexamethasone induction. i–j, in situ hybridizations with anti-sense WUS probe in PLT2p:REVΔmiR-GR globular (i) and heart (j) stage embryos after induction. k–m, in situ hybridizations with anti-sense ANT probe in WT transition stage (k) and PLT2p:REVΔmiR-GR transition stage (l) and torpedo stage (m) embryos after induction. Scale bars, 1 mm (a–c) and 50 µm (d–m).
To further characterize these homeotic transformations we examined the expression of genes specifically associated with the apical and basal poles. PINFORMED4 (PIN4) is initially expressed in the embryo proper, as well as the upper cells of the suspensor (Fig. 4d). It is then restricted from the suspensor and expressed in the developing root meristem and provascular cells, and this expression has been shown to be dependent on the activity of PLT1 and PLT220 (Fig. 4e, f). In globular through heart stage PLT2p::REVΔmiR-GR embryos, PIN4 mRNA is only detectable in aberrantly dividing suspensor cells (Fig. 4g, h). The loss of PLT1/PLT2 dependant PIN4 expression in cells ectopically expressing REV again illustrates the antagonistic action of these two classes of genes.
In induced globular stage PLT2p::REVΔmiR-GR embryos, WUS is misexpressed in the basal region coincident with the presumptive second shoot position, indicating that an additional shoot organizing center has formed (Fig. 4i, j). Likewise, AINTEGUMENTA (ANT) expression, which marks the cotyledon primordia in the WT21 (Fig. 4k), is misexpressed in the basal region of transition stage embryos (Fig. 4l). In older embryos, multiple basal foci of ANT misexpression accumulate (Fig. 4m). These results show that the alteration in embryo polarity begins during the early globular stage of induced embryos. Furthermore, these results indicate that establishment of apical fate by the HD-ZIP III genes precedes WUS and ANT expression (and therefore meristem and cotyledon formation).
Our results show the HD-ZIP III genes are master regulators of apical fate in early embryogenesis. In addition, there is a clear antagonism between the HD-ZIP III and PLT gene families, both of which are under multiple modes of regulation that ensures proper spatial distribution and apical-basal patterning (Supplementary Figure 1). Whether this antagonism is direct or is a more downstream consequence of fate change will require further investigation.
Methods Summary
Plants were grown on either soil or petri dishes containing Linsmaier and Skoog salts medium. Percival growth chambers were used for controlled temperature experiments. All other plants were grown under greenhouse conditions on a 16 hour light/8 hour dark cycle. in situ hybridizations were detected with digoxigenin-labeled riboprobes using the method found at http://www.its.caltech.edu/~plantlab/protocols/insitu.htm. ChIP was performed as described24 with minor modifications. The BIO-RAD MyiQ, single color, Real-Time PCR Detection System was used with the MyiQ Optical System Software for analysis of SYBR Green I stained amplification products. Embryos were cleared with Hoyer's solution and DIC images were collected using a Leica DM5000B microscope. Confocal images were collected using a Leica DM IRE2 laser scanning confocal microscope.
Materials and Methods
Plant growth and mutant alleles
Plants were grown on either soil or petri dishes containing Linsmaier and Skoog salts medium. Percival growth chambers were used for controlled temperature experiments. All other plants were grown under greenhouse conditions on a 16 hour light/8 hour dark cycle. All mutants, with the exception of icu4-1d are in the Landsberg erecta (Ler) ecotype. Germplasm used were as follows: plt1-5 and plt2-11, rev-10d11, phb-1d16, phb-6 phv-5 rev-911, icu4-1d13. icu4-1d was isolated in the Enkheim-2 (En-2) and back crossed to tpl-1 four times.
In situ hybridization
In situ hybridizations were detected with digoxigenin-labelled riboprobes using the method found at http://www.its.caltech.edu/~plantlab/protocols/insitu.htm. PHB, PHV, REV, and FIL, probes were made generated using 300–700bp regions of coding sequence using the primers listed in Supplementary Table 3. PLT1, PLT2, ICU4, WUS, ANT, PIN4 and GFP probes were generated using full length cDNAs.
Chromatin Immunoprecipitation
ChIP was performed as described22 with the following modifications. Ovules were dissected from siliques containing globular to heart stage embryos. Tissue was fixed in 2% formaldehyde/PBS under vacuum for 2 hrs, replacing vacuum every 30 min. 500mg of starting material was used for each ChIP sample. The anti-HA monoclonal antibody HA.11 (Covance) and M-280 sheep anti-mouse IgG Dynabeads (Invitrogen) were used to immunoprecipitate TPL-HA fusion. Two negative controls were performed, including a no antibody sample and a ChIP reaction performed on wild type (no transgene) tissue.
Real-time PCR
The BIO-RAD MyiQ, single color, Real-Time PCR Detection System was used with the MyiQ Optical System Software for analysis. SYBR Green I was used as an intercalating fluorescent dye. The standard curve method was used to determine reaction efficiency for each primer pair and determine fold enrichment by comparing the CT (threshold cycle) values of IP and negative control which were normalized normalize by calculating input(IP)/input(control) when appropriate.
Plasmid Construction
The miR165/166 sensor was generated using complementary 42 base pair primers encompassing the miR165/166 recognition sequence in PHB and REV, which were annealed to generate double stranded fragments with EcoRI compatible sites at each end. These fragments were then treated with T4 polynucleotide kinase in T4 DNA ligase buffer and cloned into a unique EcoRI site in the mERGFP5 sequence that lies between the endoplasmic reticulum (ER) localization signal and mGFP5. The modified mERGFP5 were then cloned as a BamHI fragments into a pBJ36 construct, 3’ to a 925bp promoter fragment from the potato UBI3 gene23. For the negative control, three silent mutations were introduced within the 3’ end of the miR recognition sequence. Primers used are listed in Supplementary Table 3.
For construction of the PLT2p::HD-ZIPIII-GR constructs, a 4380 bp genomic fragment 5’ to the PLT2 start codon was cloned as a XhoI/SalI fragment into a SalI site of a pBJ36 vector containing the hormone binding domain of the rat glucocorticoid receptor24. HD-ZIP III miR resistant cDNAs were generated by inducing three silent mutations within the 3’ end of the miR recognition sequence by site directed mutagenesis using the primers listed in Supplementary Table 3.
Microscopy
Excised ovules were mounted in Hoyer’s solution for analysis of embryonic morphology. Embryos were imaged using a Leica DM5000B microscope, seedlings using a Leica MZ FLIII microscope. For GFP analysis, ovules were dissected into 0.5X LS media (Caisson Laboratories, Inc.; Rexburg, ID), vacuum infiltrated in 4% paraformaldehyde, rinsed with water, vacuum infiltrated with 2% SCRI Renaissance 2200 (Renaissance Chemicals Ltd.; North Yorkshire, UK) and 4% DMSO, then washed 2X and mounted in 20% glycerol. Embryos were imaged using a Leica DM IRE2 laser scanning confocal microscope. SR2200 was excited with the UV diode 405nm line, and emission was measured between at 420–470nm. GFP was excited with a 488nm argon laser line and emission was measured at 500–535nm.
Supplementary Material
Figure 5.
Figure 6.
Acknowledgements
We thank J. Sewell for technical assistance with plasmid construction and microscopy; R. Biddick, B. Crawford, D.B. Jaillais, N. Krogan, J. Posakony, B. VanSchooten, and D. Kelley for critical discussions and reading of the manuscript; E. York and SIO Unified Laboratory Facility for SEM analysis; This work was supported by a Ray Thomas Edwards Foundation Career Development Award, the National Institute of Health, NIGMS (J.A.L.), Z.R.S. is a USCD Plant Systems Biology NSF-IGERT Fellow.
Footnotes
Author Contributions Z.R.S. collected the data. Z.R.S. and J.A.L. designed the study and wrote the manuscript.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
References
- 1.Aida M, et al. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119(1):109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Blilou I, et al. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature. 2005;433(7021):39–44. doi: 10.1038/nature03184. [DOI] [PubMed] [Google Scholar]
- 3.Galinha C, et al. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449(7165):1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- 4.Friml J, et al. Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature. 2003;426(6963):147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 5.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCF(TIR1)- dependent degradation of AUX/IAA proteins. Nature. 2001;414(6861):271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 6.Szemenyei H, Hannon M, Long J. TOPLESS mediates auxin dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008 doi: 10.1126/science.1151461. In Press. [DOI] [PubMed] [Google Scholar]
- 7.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312(5779):1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 8.Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK. Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development. 2002;129(12):2797–2806. doi: 10.1242/dev.129.12.2797. [DOI] [PubMed] [Google Scholar]
- 9.Tang G, Reinhart BJ, Bartel DP, Zamore PD. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17(1):49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mallory AC, et al. MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5' region. Embo J. 2004;23(16):3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emery JF, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr Biol. 2003;13(20):1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 12.Floyd SK, Bowman JL. Gene regulation: ancient microRNA target sequences in plants. Nature. 2004;428(6982):485–486. doi: 10.1038/428485a. [DOI] [PubMed] [Google Scholar]
- 13.Ochando I, et al. Mutations in the microRNA complementarity site of the INCURVATA4 gene perturb meristem function and adaxialize lateral organs in arabidopsis. Plant Physiol. 2006;141(2):607–619. doi: 10.1104/pp.106.077149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong R, Taylor JJ, Ye ZH. Transformation of the collateral vascular bundles into amphivasal vascular bundles in an Arabidopsis mutant. Plant Physiol. 1999;120(1):53–64. doi: 10.1104/pp.120.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McConnell JR, et al. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature. 2001;411(6838):709–713. doi: 10.1038/35079635. [DOI] [PubMed] [Google Scholar]
- 16.McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125(15):2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- 17.Prigge MJ, et al. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17(1):61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer KF, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95(6):805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 19.Grigg SP, et al. Repression of Apical Homeobox Genes Is Required for Embryonic Root Development in Arabidopsis. Curr Biol. 2009 doi: 10.1016/j.cub.2009.06.070. [DOI] [PubMed] [Google Scholar]
- 20.Friml J, et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell. 2002;108(5):661–673. doi: 10.1016/s0092-8674(02)00656-6. [DOI] [PubMed] [Google Scholar]
- 21.Elliott RC, et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8(2):155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowler C, et al. Chromatin techniques for plant cells. Plant J. 2004;39(5):776–789. doi: 10.1111/j.1365-313X.2004.02169.x. [DOI] [PubMed] [Google Scholar]
- 23.Garbarino JE, Belknap WR. Isolation of a ubiquitin-ribosomal protein gene (ubi3) from potato and expression of its promoter in transgenic plants. Plant Mol Biol. 1994;24(1):119–127. doi: 10.1007/BF00040579. [DOI] [PubMed] [Google Scholar]
- 24.Aoyama T, Chua NH. A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 1997;11(3):605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.