Abstract
Nonsense mediated RNA decay (NMD) has long been viewed as an important constitutive mechanism to rapidly eliminate mutated mRNAs. More recently it has been appreciated that NMD also degrades multiple non-mutated transcripts, and that NMD can be regulated by wide variety of cellular stresses. Many of the stresses that inhibit NMD, including cellular hypoxia and amino acid deprivation, are experienced in cells exposed to hostile microenvironments, and several NMD targeted transcripts promote cellular adaptation in response to these environmental stresses. Because adaptation to the microenvironment is crucial in tumorigenesis, and because NMD targets many mutated tumor suppressor gene transcripts, the regulation of NMD may have particularly important implications in cancer. This review briefly outlines the mechanisms by which transcripts are identified and targeted by NMD and reviews the evidence demonstrating NMD is a regulated process which can dynamically alter gene expression. While much of the focus in NMD research has been in identifying the proteins that play a role in NMD and identifying NMD targeted transcripts, recent data regarding the potential functional significance of NMD regulation, including the stabilization of alternatively spliced mRNA isoforms, the validation of mRNAs as bona-fide NMD targets, and NMD's role in tumorigenesis, are explored.
Cellular stress is a common feature of many physiological and pathological conditions including cancer, where tumor growth and an unorganized and faulty vascular system lead to significant hypoxia, amino acid deprivation, and reactive oxygen species (ROS) generation. The cellular response to these stresses includes dynamic alterations of gene expression which is mediated by a variety of mechanisms. For example, a plethora of research over the last two decades has emphasized the importance of transcription factors not only in cancer etiology, but also in the adaptive response of cancer cells to their microenvironment. However a gene's steady state expression level is a product not only of its rate of transcription but also of its rate of mRNA degradation. In fact, just as the complex regulation of a transcription factor is responsible for coordinating the expression of functional sets of genes, several distinct mechanisms of RNA decay are also responsible for degrading groups of transcripts with similar functions.
It is well established that one mechanism of RNA degradation, nonsense mediated RNA decay (NMD) contributes to the rapid degradation of many mutated mRNAs, including mutated tumor suppressor transcripts (1). More recently NMD has also been shown to degrade transcripts that participate in the adaptive response of cells to their microenvironment (2, 3). Furthermore alternatively spliced mRNA isoforms, which are increasingly identified in cancer, may also be regulated by NMD (4). These functions take on further biological significance with the recent observations that NMD activity is inhibited by distinct forms of cellular stress that commonly occur in the tumor microenvironment (2, 3). Here we review the regulation of NMD by the tissue microenvironment, focusing on cancer as a model.
Nonsense mediated RNA decay is carried out by several multiprotein complexes
NMD is an efficient mRNA surveillance process that selectively eliminates aberrant transcripts which contain premature termination codons (PTCs). The identification and degradation of NMD transcripts is mediated by several multi-protein complexes. Over the last decade these complexes have been at least partially characterized, and many individual proteins in these complexes have been validated as playing important roles in NMD. NMD is an evolutionarily conserved process and, for the most part, components of the NMD complex in mammalian cells have been identified by their homology to those in lower organisms and/or by their physical interaction to previously identified members of the NMD complex. Many of these proteins have been validated by knock-down, over-expression, and knock-out mouse model experiments. Experiments have also been carried out in which proteins are tethered to mRNA species to determine whether these proteins are sufficient to elicit NMD (5). These many experiments will not be detailed, as they are discussed extensively in several excellent recent reviews (6-10). Instead the current prevailing model of NMD in mammalian cells will be briefly reviewed (Fig. 1) although it should be noted that this model is undoubtedly simplistic and additional/alternative models exist. It should also be noted that many of the proteins that play important roles in NMD, including those that are not extensively detailed in this review, also have seemingly independent roles in other diverse pathways, including DNA replication, and the protection against genomic instability (as reviewed in (9)).
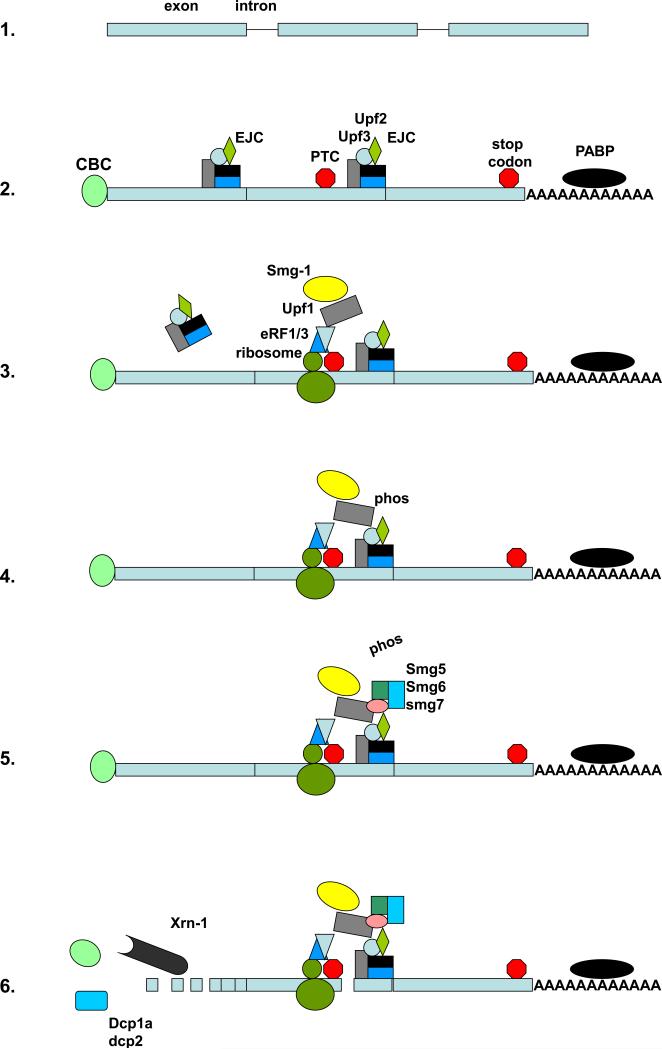
Fig 1.
NMD is carried out by several multi-protein complexes. As described in the text, after introns splicing (1) nascent mRNAs contain a unique cap binding complex (CBP) as well as an exon junctional complex (EJC), and the 3’ UTR binds the poly(A) binding protein C1 (PABPC1) (2). During the pioneer round of translation (3) the EJCs are removed. In the presence of a PTC the eRF proteins recruit Upf1/Rent1, which then binds Smg1 to form the SURF complex. Upf1/Rent1 in the SURF complex bridges the EJC, and Upf1/Rent1 is phosphorylated by SMG1 (4). Subsequently SMG 5-7 are recruited, which then dephosphorylate Upf1/Rent1 (5). The mRNA is then cleaved near the PTC by endonuclease activity of SMG-6, and degraded via decapping by the decapping proteins dcp1a and dcp2, followed by exonucleases activity of xrn-1 (6). See text for details and references, including recent reviews.
During the processing of mammalian pre mRNA, introns are excised and marked by a multi-protein complex termed the exon junction complex (EJC) (Fig. 1, step 2). This EJC contains at least 10 proteins that are deposited 20-24 nucleotides upstream of the exon-exon junction, and include the core NMD components UPF2/Rent2 and UPF3 (11-14). Newly synthesized mRNAs are also capped at the 5’ terminus by the cap-binding complex (CBC). Later the CBC will be replaced by eukaryotic initiation factor 4E (eIF4E) at which time mammalian transcripts appear to become immune to NMD (15-17) although some of the data supporting this model is indirect (as reviewed in (10). mRNAs containing the CBC are thought to be translated through a pioneering round of translation, shortly after nuclear export, by a translation complex including eIF4G, eIF3, and eIF2α (18, 19). Other members of the steady state translation process are also required, but the 4E binding protein 4EBP-1, which is commonly over-expressed in some cancers and which regulates protein translation in hypoxic and metabolically starved tumors does not affect this pioneer round of translation or NMD (18, 20, 21).
The positional information provided by the EJC persists during export until the mRNA is translated; at least some components of the EJC are displaced by ribosomes during the pioneering round of translation ((22) and reviewed in (23-25)). When the translation complex pauses at a PTC that is upstream of an EJC, eukaryotic release factors (eRFs) physically bind to and recruit the RNA helicase UPF1/Rent1, a vital component of the NMD mechanism (12, 26-28). Subsequently the phosphatidyl inositol-3 kinase SMG-1 is recruited to complete the formation of the SMG-1, UPF1, eRF (SURF) complex (29). This SURF complex then binds to the UPF proteins in the EJC (Fig. 1, step 3), thus bridging these two complexes and promoting the phosphorylation of UPF1 by SMG1 (Fig. 1, step 4). Phosphorylated UPF1 then recruits SMG-5, SMG-6, and SMG-7 with the subsequent de-phosphorylation of UPF-1 by SMG-7 (30) (Fig. 1, step 5). SMG-1 kinase activity can be inhibited by two other members of the SURF complex, SMG-8 and SMG-9 (31). The phosphorylation and de-phosphorylation of Upf1 are thought to be necessary and crucial steps in the NMD pathway (29, 32, 33). Interestingly, Upf1 phosphorylation has also recently been shown to repress translation by binding to eIF3 subunits and preventing the formation of active ribosomes (34).
Whereas mRNA degradation by other mechanisms occurs via a variety of exo-nucleases and endo-nucleases, NMD targeted transcripts are thought to be primarily degraded via removal of the 5’ 7-methyl guanosine cap by the decapping enzymes dcp1 and dcp2 and the subsequent 5’ →3’ exonuclease activity of xrn-1 (35) (Fig. 1, step 6). Inhibition of decapping activity has been demonstrated to interfere with the stability of NMD targets but not other transcripts (35). Phosphorylated Upf1 may help recruit decapping enzymes (34). A rapid de-adenylation step and 3’ → 5’ nuclease activity may also take place (35, 36), although the exact contribution of each pathway is difficult to determine because of possible redundancies elicited when individual pathways are silenced. Recently SMG-6 has been determined to contain endonuclease activity on single stranded RNA and it has recently been proposed that NMD transcripts also undergo endo-nucleolytic cleavage in the vicinity of their PTCs by SMG-6 prior to terminal degradation by xrn-1 (37, 38).
The decapping enzymes dcp1a and dcp2 as well as xrn-1 are concentrated in cytoplasmic foci termed processing bodies (39). Processing bodies are distinct from stress granules, which form in response to a variety of cellular stresses found in tumors including hypoxia and metabolic starvation, though studies suggest that mRNAs can be transferred from stress granules to processing bodies (40, 41). In Saccharomyces cerevisiae knock-down of xrn-1 leads to the accumulation of mRNA and Upf1 in processing bodies as do mutations in Upf1 that fail to bind or hydrolyze ATP and thus cannot participate in NMD (42, 43). Furthermore, tethering of Upf1/Rent1 to an mRNAs is sufficient to both target that mRNA to processing bodies and rapidly degrade the mRNA (43). These data supporting a model in which processing bodies are the sites of NMD associated degradation, however, are primarily derived from yeast studies. Although mammalian Upf1/Rent1 accumulates in processing bodies during the inhibition of NMD (it is not yet known if this is accompanied with NMD targeted mRNAs), recent studies suggest that human ATP deficient Upf1/Rent1 accumulation in processing bodies is not accompanied by other components of the SURF complex, and that disruption of visible processing body formation does not affect the stability of NMD targets, suggesting that processing bodies may be a consequence of NMD and are not required for mammalian NMD (3, 44, 45).
As is obvious from this brief and simplified review, the mechanism by which NMD degraded mRNAs are targeted and degraded involves many multiunit complexes and an intricate process of phosphorylation and other enzymatic steps. NMD activity has been shown to differ amongst cell lines (46) and the absence or over-expression of a variety of proteins involved in the NMD pathway have been shown experimentally to either inhibit or accelerate the degradation of NMD transcripts respectively, without interfering with other forms of RNA decay. Although Upf1/Rent1 has been noted to be up-regulated in colon cancer tissue compared to adjacent normal colonic tissue (47) to date there has been no systematic examination of the expression levels of key components of NMD in various tissues or pathological conditions such as cancer. Similarly, it is not known if any members of the NMD complex are up-regulated by oncogenes.
NMD targets share distinct characteristics and include mutated transcripts commonly found in cancer
The existence of a pathway that rapidly degrades mutated mRNAs was suggested approximately 30 years ago when studies of several common mutations in human genes, including the β globin gene, determined that PTCs do not always result in a truncated protein but rather a markedly diminished amount of mRNA (48, 49). Indeed, several years later it was determined that mutations commonly responsible for thalassemia, including virtually all thalassemia in Sardinia, is caused by a PTC that results in unstable globin mRNA (50, 51). Since these early experiments many additional NMD targets have been identified and similarities between their gene structures have been characterized. Many of these characteristics support the current model of the molecular mechanism of NMD, specifically the interaction between the SURF complex recruited at a PTC and the EJC that triggers NMD. Thus several “rules” or algorithms have been derived in attempt to identify NMD targeted transcripts.
Authentic stop codons do not elicit NMD because they are typically present in a gene's last exon, and mammalian cells require at least one intron downstream of a PTC in order for that transcript to be targeted for NMD. Generally it is thought that for a transcript to be targeted by NMD the PTC must also be greater than approximately 55 nucleotides upstream of an exon-exon junction Based on this algorithm, bioinformatics studies have estimated that up to 30% of all known mutations causing human disease generate mRNAs that are degraded by NMD (reviewed in (1)). These mutations include those responsible for many forms of thalassemia, Duchene's muscular dystrophy, and cystic fibrosis. Mutations predicted to lead to NMD are also commonly found in tumor suppressor genes, and several of these will be discussed in detail below as a way to review the strategies for identifying bona-fide NMD targets, as well as an opportunity to critically evaluate several assumptions about the degradation, translation, and function of mRNAs predicted to be NMD targets.
Despite efforts to identify NMG targets with bioinformatic tools, the number of transcripts degraded by NMD may be under-estimated since not every NMD target conforms to the above algorithm. For example, T cell receptor gene rearrangements which increase receptor diversity often result in PTC containing transcripts; these transcripts may be degraded by NMD despite having a PTC located within the last 50 nucleotides of the penultimate exon (52). Alternative mechanisms also exist to target some transcripts for NMD. In both Drosophila melanogaster and Saccharomyces cerevisiae (which do not have introns) a stop codon is recognized as premature if it occurs far upstream of a poly(A) tail. Similarly, in mammalian cells a long 3’ UTR can also target a transcript for NMD; SMG-5 is one such example of a transcript regulated by NMD by virtue of its long 3’UTR (53). This mechanism is thought to involve a disruption of the poly (A)-binding protein C1's (PABPC1) normal role of inhibiting interaction between eRF3 and Upf1. In the absence of this inhibition, for example when a transcript contains a long sequence between a stop codon and the 3’ poly(A) tail, Upf1 is recruited and NMD occurs (53, 54). This has led to the model that NMD is a function of a competition between components of the EJC which stimulate the recruitment of the Upf/Rent complex to a stop codon, and PABPC1 which antagonizes the recruitment of the Upf complex to the ribosome at a stop codon (53). However, to complicate this model, there are also features of some long 3’UTRs which confer immunity to NMD, at least in Drosophila (55).
As will be discussed subsequently, a number of non-mutated transcripts generated in the cell are also targeted by NMD (2, 3). While some of these normal transcripts have recognizable features that could render them sensitive to NMD (e.g. long 3’ UTRs, upstream open reading frames and alternatively spliced isoforms as discussed later in detail) many do not (2, 3, 53). Therefore, it is possible that other, as yet unknown characteristics, render still additional mutated transcripts sensitive to NMD. Conversely, many mutant transcripts presumed to be NMD targets have not been validated, and several transcripts and reporter constructs predicted to be NMD targets do not appear to be degraded by this pathway (53, 56). Thus the full range of transcripts degraded by NMD remains largely elusive.
Several screening strategies have been designed and implemented to identify transcripts as bona-fide NMD targets. A SNP2NMD database for human single nucleotide polymorphisms (SNPSs) that encode PTCs upstream of EJCs has been constructed (57) although this database suffers from the issues mentioned above, specifically that some transcripts may fit the criteria of a NMD target but still not be degraded by NMD and still other transcripts may not fit these criteria and yet in fact be degraded by NMD. Many experimental strategies to identify transcripts degraded by NMD are based on early assays which chemically inhibited NMD via the global inhibition of protein translation, and then amplified and in vitro translated stabilized NMD transcripts (58). The presence of a truncated protein generated only with the inhibition of NMD suggests that the corresponding transcript is an NMD target.
With the advent of expression array profiling, a protocol termed gene identification by NMD inhibition (GINI) has been developed to identify NMD transcripts in a high-throughput manner (59). In this assay expression arrays are used to identify transcripts up-regulated in the presence of emetine, an antibiotic that inhibits translation and thus NMD. Transcripts up-regulated in the presence of emetine are considered to be NMD targets. NMD inhibitors that may have less non-specific effects in some cell lines, including caffeine which inhibits SMG-1, have also been used (60). GINI has also been modified to treat cells with the RNA synthesis inhibitor actinomycin D, which will lead to decreased expression of unstable NMD transcripts in the absence of NMD inhibitors but not in the presence of NMD inhibitors (60).
These strategies, as well as the systematic sequencing of common tumor suppressor genes, have led to the identification of a number of NMD conferring mutations in genes that play a role in cancer. In fact, a systematic analysis of mutations in human genes revealed that while most mutations in oncogenes are missense mutations, tumor suppressor genes exhibit a disproportionate number of nonsense mutations, many of which are predicted to lead to NMD targeted transcripts (61). Such mutated tumor suppressor transcripts include Wilm's tumor 1 gene mutations which result in truncated transcripts that are stabilized with emetine treatment and encode for proteins with dominant negative properties (62). The classic tumor suppressor genes p53 and RB each contain PTC mutations in mantle cell lymphoma cell lines, and both these transcripts are stabilized in the presence of emetine suggesting that they are NMD targets (63). In fact, one mutant p53 transcript found in breast cancer not only demonstrates increased mRNA stability with the inhibition of NMD, but the resulting c-terminal truncated protein is more stable than wild-type p53 protein (64). In the familial form of colon cancer, familial adenomatous polyposis (FAP), many of the mutations found result in PTCs in early exons, including those found in nine out of ten Scottish kindreds (65, 66). Most BRCA2 mutations also result in PTCs, and expression of the transcript generated from the PTC mutated gene is decreased compared to transcript levels observed from other mutations in the gene, suggesting that this transcript is de-stabilized (64). Similarly, 80% of the PTC containing transcripts found in BRCA1 result in reduction of mRNA abundance (67). Several PTC mutations in MRE11, a partner of ATM in the DNA damage response, can only be detected from the sequencing of genomic DNA, but not from cDNA unless RNA decay is inhibited, suggesting that these mutations destabilizes the transcript (68, 69). Finally, in gastric cancer 80% of the mutations in the e-cadherin gene result in a PTC predicted to be subjected to NMD, and indeed the e-cadherin mRNA from PTC containing alleles are down-regulated compared to the non-mutated allele, and these mutated transcripts are also stabilized with emetine or with Rent1/Upf1 depletion (70).
In colon cancer cells with microsatellite instability, the mutator phenotype results in one or two base pair insertions or deletions, frame-shifts and often PTCs. Validation studies of the GINI technique demonstrated that approximately 4% of all transcripts were up-regulated in several microsatellite colon cancer cell lines with emetine (59). A significant number of these transcripts were not altered in a control fibroblast cell line, suggesting that these transcripts were up-regulated due to colon cancer specific mutations. The DNA mismatch repair enzyme MLH1, which is known to carry a PTC mutation in several colon cancer cell lines, was strongly up-regulated with NMD inhibition specifically in colon cancer cell lines, thus validating this approach. This finding led to an interesting modification and application of GINI, developed to rapidly identify mutations in the large and difficult to sequence MLH-1 gene in patients suspected of having hereditary non-polyposis colon cancer (HNPCC) (71). Blood samples from three patients who fit the criteria of HNPCC were treated with a chemical inhibitor of NMD prior to RNA isolation from white blood cells. RT-PCR for the MLH1 transcript was preformed, and the amplicon was examined for up-regulation or alternatively spliced isoforms that were stabilized in the presence of the NMD inhibitor. In two out of the three cases these analyses revealed transcripts with PTC mutations likely to cause NMD that were barely detectable in the absence of NMD inhibition.
Another study looking at several genes mutated specifically in microsatellite instable colon cancer cell lines was able to validate approximately 50% of their NMD candidate transcripts, including several genes that play a role in cancer biology, by sequencing and identifying PTC mutations in these genes (60). Out of 10 candidate genes, bi-allelic inactivating PTC mutations were found in six. Many of these genes were mutated in several colon cancer cell lines which exhibit microsatellite instability, and several were also found to be mutated in macrosatellite instable colon cancer tumor samples. In addition, the transcript generated from the EP300 gene encoding p300, the co-activator that plays an important role in the transcriptional activity of several oncogenes, has also been found to be mutated and targeted by NMD in microsatellite instable colon cancer cell lines (72). The mRNA of some, though not all transcripts identified by GINI in these studies were up-regulated when Upf1/Rent1 was down-regulated, suggesting that these were indeed bona-fide NMD targets (60).
Stable transcripts with a PTC, if translated, result in a truncated protein. Thus NMD is thought to have evolved in order to selectively eliminate truncated proteins which might otherwise serve as either activating mutants or as dominant negatives against an intact protein encoded by the non-affected allele. Because many tumor suppressor genes mutated in cancer contain PTCs and are either predicted to be or have been validated to be NMD targets, NMD is commonly thought to prevent tumorigenesis. Although this is a popular and satisfying model, validation requires documentation that in the absence of NMD both the transcript indeed encodes a truncated protein, and that this truncated protein can affect the phenotype of the cell. In many cases these criteria have not been satisfied.
For example, as discussed earlier, many mutations predicted to promote NMD do not, including multiple nonsense mutations described in the ATM gene which result in PTCs upstream of exon-exon junctions but do not de-stabilize the ATM transcript (73). In addition, even when NMD targeted transcripts are abundant, these transcripts are not necessarily translated. For example, even when a CHK2 transcript harboring an NMD provoking mutation is stabilized this does not result in a translated (truncated) CHK2 protein (64). Another study found that several transcripts with PTCs not predicted to elicit NMD (e.g. were in the last exon) were translated into truncated proteins, but several transcripts predicted to be degraded by NMD, even when stabilized, were still not translated (56). This may be related to the recently described ability of phosphorylated Upf1/Rent1 to repress translation (34).
Finally, the biological significance of truncated proteins, if translated, is often unknown. While some of these truncated proteins, (e.g. described WT1 and p53 mutations) may indeed act as dominant negatives (62) the generation of some truncated proteins can function normally (as in other p53 mutations (74)). Although several oncogenes have PTCs that are predicted and/or validated to elicit NMD, including the tyrosine kinase EPHB2 in both prostate cancer cell lines and greater than >5% of primary prostate tumors, and JAK1 mutations in prostate cancer transcripts (75, 76), it is unclear whether these transcripts (if they were not degraded by NMD) would result in truncated proteins with gain of function properties. Thus experimental validation that NMD protects against deleterious mutations in cancer is still necessary.
Non-mutated transcripts targeted by NMD targets: characteristics and implications for alternatively spliced transcripts
While NMD has long been appreciated to degrade many mutated transcripts, such as nonfunctional transcripts with retroviral or transposon insertions, recently NMD has also been found to regulate many non-mutated transcripts including, as discussed later, transcripts that play an important role in the cellular response to stress (2). This unexpected finding resulted from the siRNA knock-down of Upf1/Rent1 in Hela cells followed by microarray analysis for transcripts whose expression was altered. While this study documented that NMD regulates up to 10% of the Hela transcriptome, it should be noted that this type of analysis cannot distinguish between those transcripts directly stabilized by the inhibition of NMD and those indirectly stabilized by NMD. Transcripts directly targeted by NMD demonstrate increased stability with Upf1/Rent1 knock-down, while the indirect up-regulation of a given transcript can occur via the stabilization and up-regulation of either a transcription factor or of an inhibitor of RNA degradation. Approximately half of the transcripts up-regulated with Upf1/Rent1 knock-down do not have recognizable features which could make them sensitive to NMD. Because a large proportion of transcripts altered when NMD is disabled are down-regulated, without an alteration in their stabilities (2), indirect processes clearly exist, as there is no obvious mechanism by which decreased NMD activity can directly result in the decreased expression of a transcript. Thus NMD has the potential to regulate a marked number of transcripts in addition to those directly targeted by the pathway.
The observation that many cellular transcripts are degraded by NMD raises the question of how these are recognized by the NMD apparatus. Many mRNAs have upstream open reading frames (uORFs) that can result in ribosomal pausing upstream of a stop codon, and theoretically trigger NMD (77). Bioinformatic studies have demonstrated that, in general, transcripts with uORFs are expressed to a lower degree than transcripts without uORFs, suggesting either decreased transcription or increased degradation (78). While many short uORFs are not sufficient to render a transcript sensitive to NMD, the uORF of one important NMD target discussed in detail later, ATF-4, has been shown to be sufficient to promote NMD (3, 79). As previously discussed, a second mechanism that can trigger NMD in normal transcripts requires long distances between eRFs deposited at a normal stop codon and PABPC1 bound to a distant 3’ UTR, which then perimits the recruitment of Upf1 and subsequent NMD (53). Additional potential mechanisms include functional genes with insertions of retroviral or transposon, and genes with an introns in their 3’ UTR. (2, 4, 80-82).
Yet another mechanism which may lead to a non-mutated transcript's degradation by NMD is the alternative splicing of that transcript (Fig 2). Alternatively spliced transcripts vary in different tissues, occur in cells exposed to stresses common in the tumor microenvironment, have been increasingly identified in cancer cells, and have been hypothesized to play a causal role in cellular phenotypes (83-88). Alternatively spliced isoforms which play a role in cancer, for example, include vascular endothelial growth factor (VEGF) and BCL-x, which has an isoform that inhibits apoptosis (Bcl-x(L)) and an isoform that promotes apoptosis (Bcl-x(s)) (89) Up to 30% of all human genes may be affected by alternative splicing, although many of these do not affect coding sequences (52, 90-92). Many alternative splicing events can lead to altered reading frames and a PTC upstream of an exon-exon junction, thus rendering them susceptible to NMD. As opposed to transcripts with a nonsense mutation, these alternatively spliced transcripts can result in a new reading frame and thus a novel protein. Depending on the alternatively spliced transcript, the upstream protein sequence of the original protein may be maintained, and thus a fusion protein may result. Using bioinformatics to assess expressed sequence tags (ESTs) approximately 20-35% of alternatively spliced events are predicted to lead to a PTC that would render them targets of NMD (93-96). These alternatively spliced transcripts include several of those transcripts found to be up-regulated in Hela cells when NMD is disabled (2).
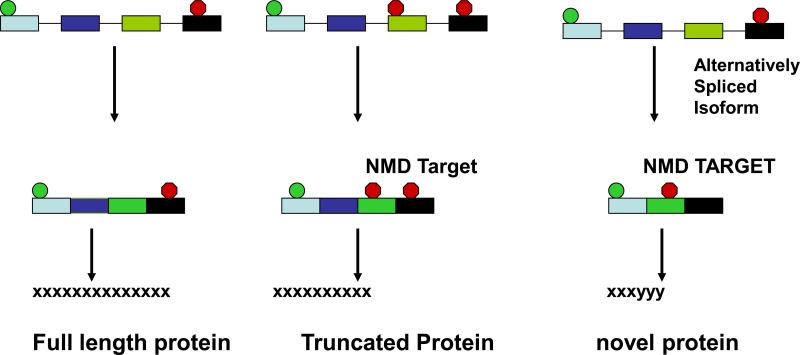
Fig 2.
Transcript degradation by NMD can lead to stabilization of truncated and alternatively spliced isoforms. During normal translation a full length protein is generated (left panel). A PTC normally triggers NMD, but if that transcript is stabilized via inhibition of NMD then a truncated protein may be generated, which may then serve as dominant negative or may function as a wild-type protein (middle panel). If an alternatively spliced isoform, e.g. due to a skipped exon, generates a PTC, in the absence of NMD this isoform may result in a protein with deletions or, if the alternatively spliced isoform results in a new reading frame, then a novel protein (right panel).
Many of alternatively spliced transcripts degraded by NMD are found in low abundance, are not conserved between mouse and human, result in frameshift mutations unlikely to reflect bona-fide genes, and are thus considered to represent genomic noise generated from an error prone splicing process (97, 98). However, when othologous exons can be found there is evidence that the NMD specific isoform is under selective pressure, suggesting that these are indeed translated under at least some conditions, and are in fact detrimental to survival (98). While alternatively spliced transcripts degraded by NMD have been identified by experimental techniques, including the MLH gene in patients with HNPCC, (71) experimental evidence to support a wide-spread role for NMD is controversial. While one study utilizing exon arrays, in which most gene exons are represented by multiple probes, found a limited effect of Rent1/Upf1 depletion, another study identified and validated over 200 exons with altered expression upon NMD inhibition (4, 99). Many of these NMD degraded isoforms contained a PTC in all three reading frames, and could be confirmed with conditional knock-out of Upf2/Rent2 in mouse hematopoietic cells (82). These studies suggest that NMD can stabilize and increase the expression of alternatively spliced mRNA isoforms, which may play an important role in cancer, but more experimental work must be done to determine the extent and biological significance of NMD regulation in this process.
Cellular transcripts targeted by NMD are involved in the cellular stress response
While the identification and validation of an individual NMD transcript may in turn suggest a specific functional role for NMD, it is important that this be validated with a formal documentation of phenotype. In published studies this documentation is often absent, as is even the evidence that with the inhibition of NMD protein expression of the targeted transcript is increased. It is clear, however that gross genetic and molecular manipulation of NMD results in dramatic organismal and cellular phenotypes. For example, components of NMD (e.g. Upf1/Rent1, Upf2) have been shown to be necessary for zebrafish embryonic differentiation and survival (100). Upf1/Rent1 is necessary for mouse viability and the knock-out of Upf2/Rent2 in mouse hematopoietic cells leads to lethality within ten days and a failure of definitive hematopoiesis through the loss of stem and progenitor populations (82, 101). Because differentiated cells are spared, it has been hypothesized that NMD is primarily essential for proliferating cells (82). NMD targeted transcripts that are responsible for these described phenotypes, however, have not yet been identified. Although several of the proteins involved in NMD appear to play a role in diverse phenotypes such as cell cycle checkpoints and telomere maintenance in mammalian and lower organisms (reviewed in (9)), it is unclear if they affect these functions through NMD or through other pathways. For example, SMG1 can respond to double strand breaks and contribute to a G2 checkpoint, not via Upf1/Rent1 phosphorylation but through p53 phosphorylation (102). In addition, the inactivation of NMD surely results in the stabilization of mutant transcripts, and results in truncated and mutant proteins which, in turn, may non-specifically elicit many of the described phenotypes. Thus these phenotypes do not necessarily help in the identification of specific functional genes or pathways that are normally regulated by NMD.
The non-mutated mRNAs normally targeted by NMD, as identified by array studies, cover a wide range of ontologic categories including those involved in cell cycle, differentiation, and signaling (2). A strong argument that NMD plays an important role in regulating normal gene expression comes from the appreciation that multiple transcripts from the same functional classes of are targeted by NMD. For example, several of the alternatively spliced isoforms degraded by NMD are splicing related factors, leading to the auto-regulation, or regulated unproductive splicing and translation (RUST) hypothesis ((4, 82, 94) and reviewed in (9)). In this model, when genes which promote splicing (e.g. SR genes) are highly expressed they then promote alternative splicing of their own transcripts. This alternative splicing leads to a transcript that is degraded by NMD, and thus down-regulation of mRNA expression. Conversely, genes that inhibit splicing (e.g. hnRNP) repress the inclusion of the coding exon that creates a frameshift and triggers NMD.
In addition to splicing factors, a disproportionate number of reported NMD regulated transcripts are involved in stress response and nutrient homeostasis pathways (2, 3, 82). Multiple studies in yeast have also demonstrated an important role of NMD in regulating transcripts involved in amino acid transport and synthesis and oxidative stress (103, 104), suggesting that the targeting of mRNAs which can promote the cellular adaptation to hostile environments is a conserved feature of NMD. This, along with the assumption that when multiple transcripts with similar functions are predicted to be regulated by NMD then these transcripts are likely bona-fide NMD targets with important physiological roles, it makes it probable that the regulation of stress response/nutrient homeostasis is an important function of NMD in mammalian cells.
Specific mammalian stress/nutrient related transcripts up-regulated with the inhibition of NMD include those of two intimately related pathways: amino acid transport and synthesis (including asparagine synthetase, cystathione-g-lyase, cysteinyl-tRNA synthetase, seryl-tRNA synthetase, and glutamate/neutral, and dibasic/neutral amino acid transporters) and the endoplasmic reticulum (ER) stress response pathway known as the unfolded protein response (UPR) (2, 3). Unfolded proteins accumulate in the ER when there is a marked increase in the synthesis of secreted or membrane bound proteins, a decrease in the availability of ER chaperones to fold their client proteins, and/or in the setting of ER micro-environmental pertubations including cellular hypoxia and/or generation of ROS ((105-108) and reviewed in (109)). Activation of the UPR leads to the coordinated degradation of ER-associated proteins, the processing of the transcript for the xbp-1 transcription factor, and the phosphorylation of the α subunit of eukaryotic initiation factor 2 (eIF2α) by the PKR-like ER kinase (PERK) (110). In a mechanism termed the integrated stress response, eIF2α can also be phosphorylated by a variety of cytoplasmic kinases that are activated by diverse forms of stress, including ROS generation and amino acid deprivation (105, 111).
Phosphorylation of eIF2α leads to both a general suppression of protein translation as well as the paradoxical translational induction of the transcription factor ATF-4. NMD targeted transcripts include ATF-4 and the ATF-4 targets CHOP (gadd153) and ATF-3 (2, 3, 82). The UPR plays a role in numerous phenotypes, including proliferation and differentiation (as reviewed in (112)). However the best delineated role for the up-regulation of ATF-4 is the cellular adaptation to ER stress via the transcriptional up-regulation of protein chaperones and other stress-response genes. The UPR also protects against oxidative stress, which can be generated under conditions of ER stress, transformation with the Ras oncogene, hypoxia, as well as by by-products of O2-utilizing cellular processes (105, 111, 113-116).
There are several links between the UPR and the amino acid metabolism pathways. The UPR is activated by amino acid starvation (111). ATF-4 targets include many genes involved with amino acid import and metabolism, particularly ones which contain thios, and UPR deficient cells are more sensitive to amino acid deprivation (111, 117, 118). Regulation of both the UPR and amino acid metabolism also plays a particularly important role in cancer progression. Just as glucose metabolism is known to be altered in many cancer cells, it is increasingly appreciated that the transport and metabolism of amino acids play an important role in tumor cell proliferation and survival (119, 120). Studies have also demonstrated that the unfolded protein response is also activated in hypoxic areas of xenografted tumors and human tumors and the generation of ATF-4 promotes the survival and growth of hypoxic tumors, and induced angiogenesis (108, 121).
Many of the transcripts involved in amino acid transport and the unfolded protein response are directly targeted by NMD (2, 3). Importantly, transcripts involved in the unfolded protein response are not up-regulated when NMD is inhibited simply because of the accumulation of mutant truncated unfolded proteins in the ER (3). Thus while many stress response and amino acid transporter genes are up-regulated by the NMD targeted ATF-4, transcripts for these genes are also independently degraded by NMD. Specifically, these transcripts are 1. Down-regulated by the over-expression of Upf1/Rent1 (which is sufficient to drive NMD); 2. Up-regulated by the knock-down of Upf1/Rent1; and 3. Stabilized by the knock-down of Upf1-Rent1 (2, 3). Many of the transcripts of the unfolded protein response (e.g. ATF-4 and CHOP), while necessary for survival to stress, are detrimental to cellular proliferation and survival when highly expressed in non-stressed cells, thus suggesting a potential reason for their rapid degradation by NMD. Thus further work is required to determine the role of NMD in both normal physiology and cancer biology.
NMD is regulated by stresses commonly found in the tumor micro-environment
The appreciation that NMD plays a role in degrading non-mutated, functional transcripts raises the question whether NMD might be regulated and thus stabilize both transcripts and alternatively spliced isoforms during physiological or patho-physiological conditions. Indeed, NMD activity is known to vary between different cell lines and even amongst distinct tissues (46). In patients with Schmid metaphyseal chondrodysplasia, collagen X PTC mutations leads to complete NMD in cartilage, but the mutant mRNA is not subjected to NMD in non-cartilage (lymphoblasts and bone) cells (122). Experimentally in mice, when the prevalence of PTC mutated Men1 mRNA is compared to mRNA from the non-mutated allele, there is a decrease in PTC vs. wild-type mRNA in testis, but not in lung, intestine and thymus (123).
Because NMD has been found to degrade transcripts vital in amino acid transportation and transcripts necessary for the cellular response to ER stress, it is logical that amino acid starvation and ER stress might be two conditions that suppress NMD activity to up-regulate these transcripts and allow a robust cellular response to these stimuli. Both amino acid deprivation and hypoxia (which activates the UPR) were found to inhibit the degradation of NMD reporter constructs (i.e. genomic sequences with PTCs) but not control constructs (2, 3). The inhibition of NMD in hypoxic cells and amino acid deprived cells was found to increase the stabilities, and in many cases the steady state expression level, of a variety of endogenous NMD transcripts (2, 3).
Insights into the mechanism by which NMD is inhibited are provided by the observation that a common link between amino acid starvation and hypoxia is that they both result in eIF2α phosphorylation. As discussed, eIF2α can also be phosphorylated by ROS and intriguingly in Schizosaccharomyces pombe the up-regulation of mRNAs in response to ROS also requires Rent1/Upf1 (104). To test whether eIF2α phosphorylation was necessary for the hypoxic inhibition of NMD, cells which could not phosphorylate eIF2α due to the genetic knock-in of an eIF2α mutant gene were rendered hypoxic and NMD activity was tested (3). In the absence of eIF2α phosphorylation neither a PTC reporter construct nor endogenous NMD targets were stabilized in hypoxic cells, indicating that eIF2α phosphorylation is necessary for at least hypoxic inhibition of NMD. As discussed, ribosomal scanning during the pioneer round of protein translation is necessary for NMD, and although eIF2α phosphorylation attenuates protein translation this is unlikely to be the mechanism by which eIF2α phosphorylation inhibits NMD for a variety of reasons, including the fact that many of the transcripts stabilized by the inhibition of NMD are actually robustly translated during this inhibition (3).
It is also possible that NMD is dynamically regulated by other mechanisms in addition to eIF2α phosphorylation. For example, radiation exposure has been reported to trigger Upf1/Rent1 phosphorylation by SMG-1 and by ATM (102) and the fact that NMD requires multiple protein complexes and phosphorylation and de-phosphorylation events suggests multiple potential steps for physiological regulation. In addition, as discussed previously, experimentally the over-expression of several of these components can drive or inactivate NMD experimentally and it is possible that physiological alterations of endogenous expression of these proteins (in different tissues, in cancer, or in other pathological states) can alter NMD activity, though this has not yet been convincingly demonstrated.
Thus a model can be formed in which hypoxia, amino acid starvation, ROS generation, which are all stresses commonly generated by the microenvironment, activate the UPR, phosphorylate eIF2α, and inhibit NMD. This inhibition then stabilizes transcripts necessary for amino acid transport and the cellular response to stress, many of which are also up-regulated independently via other arms of the UPR. Thus, for example, eIF2α phosphorylation not only induces the translation of ATF-4, but also stabilizes the ATF-4 transcript. While the ATF-4 transcript is not greatly induced in cells rendered hypoxic, studies have suggested that without the inhibition of NMD the ATF-4 transcript is actually repressed in hypoxic cells (3, 107, 108). This redundant control of ATF-4 expression, together with the recent suggestion that ATF-4 protein stability is also increased in hypoxic cells in a mechanism similar to that of HIF-1α and HIF-1β (124) not only suggests that the tight up-regulation of this protein is an important stress response mechanism, but that the down-regulation of ATF-4 in non-stressed cells is also crucial. The example of ATF-4 also highlights that the UPR truly invokes a wide range of responses ranging from transcription, to mRNA processing, to translation and, with the inhibition of NMD, the stabilization of transcripts.
In light of the relevance of the UPR and amino acid metabolism in cancer, the potential biological significance of this model can be examined by considering how NMD regulation by the tumor micoroenvironment may affect tumorgienesis, (108, 119, 121) (Fig 3). During tumorigenesis, tumor growth and a leaky, disorganized vascular system lead to significant regions of hypoxia and amino acid deprivation. ROS are generated in these hypoxic cells, in addition to ROS generated via oncogenic Ras activity. All of these lead to eIF2α phosphorylation and activation of the integrated stress response, which in turn promote tumor cell survival and angiogenesis. eIF2α phosphorylation also inhibits NMD, which augments the integrated stress response and thus promotes tumorigenesis. NMD inhibition also leads to the stabilization of distinct alternatively spliced isoforms, normally degraded by NMD, encoding for both truncated proteins and novel proteins which may also play a role in tumorigenesis. Finally, inhibition of NMD by the tumor microenvironment promotes the stabilization and up-regulation of mutated tumor suppressor transcripts that would normally be degraded by NMD, leading to truncated dominant negative proteins that can inhibit the remaining un-mutated allele and promote tumorigenesis.
Fig 3.
Stresses common in the tumor microenvironment can inhibit NMD via eIF2α phosphorylation. During tumor growth hypoxia and amino acid deprivation occur (top). Both of these stresses lead to eIF2a phosphorylation through the PERK and GCN2 kinase, respectively, which then inhibits NMD. The inhibition of NMD can then not only stabilize mutated tumor suppressor genes, but up-regulate a variety of cellular transcripts, including those important for the cellular response to amino acid starvation and ER stress, which then promote tumor adaptation and growth. The inhibition of NMD may also disversify the transcriptome through the stabilization of alternatively spliced isoforms (see Fig 2).
However to date the experimental evidence indicating that NMD activity is actually diminished in hypoxic and metabolically starved tumors in vivo is lacking. It is well documented that several other RNA degradative pathways are de-regulated in cancer. For example, hypoxia is common in the tumor microenvironment and the VEGF mRNA, which plays an important role in angiogenesis, is stabilized in hypoxic cells via the RNA binding protein HuR's interaction with sequences contained in VEGF's 3’ UTR (125-127). HuR has also been shown to bind to p53 transcripts, in a manner dependent on the Von Hippel-Lindau tumor suppressor, to increase p53 mRNA stability and translation (128, 129). There is strong circumstantial evidence that NMD regulation by the tumor mricroenvironment may occur in vivo and be important in tumorigenesis. While it is clear that an active UPR, and specifically up-regulation of ATF-4, is necessary for tumor growth (108, 121) it is unknown whether one contributing factor for this is the inhibition of NMD by eIF2α phosphorylation. And while several studies have recently suggested that alternatively spliced isoforms may be increased in cancer cell lines, studies so far have focused on cis-mutations and the aberrant expression of splicing proteins found in cancer, not on the potential role of NMD in generating these isoforms (88, 130). Finally, it has recently been described that H-Ras has an alternatively spliced form that is degraded by NMD (131), but it has not yet been shown whether this alternatively spliced Ras transcript is translated and whether it serves a biological function. However, the knowledge that NMD is a regulated mechanism, and plays an important role in stress response, will undoubtedly provided the impetus for investigators to explore NMD's role in tumorigenesis.
Summary and Conclusion
Thus NMD, a process originally considered to be responsible only for degrading mutant transcripts, is a process regulated by stresses common in the microenvironment and is responsible for dynamically altering gene expression. The regulation of NMD has implications for the stabilization of mutated transcripts, the expression of alternatively spliced transcripts, and the augmentation of stress response pathways including the UPR. However, while eIF2α phosphorylation, necessary for the inhibition of NMD, and activation of the UPR clearly play important roles in many biological processes including tumor survival and growth, the documentation that NMD and/or the regulation of NMD alters cellular phenotype has not yet been clearly demonstrated (106-108). While conditional knock-out animals for key components of the NMD pathway have been generated, studies to assess the role of NMD in the initiation and/or progression of tumors have been hampered by the observation that complete cessation of NMD appears to be incompatible with the survival of proliferating cells (82, 101). Future studies will need to explore the role of more moderate modulation of NMD activity on the tumorigenic role of this important RNA degradative pathway..
The very fact that NMD can be physiologically inhibited, has activity dependent on phosphorylation events, and may play a role in regulating many transcripts including those relevant in cancer, does suggest that manipulation of NMD activity may be an achievable and novel target for the pharmacological treatment of many diseases in addition to those genetic diseases caused by NMD triggering mutations. Gentamicin and other compounds can promote ribosomal bypass of PTCs. Preclinical and clinical trials have shown such treatments can result in expression of full length proteins and ameliorate genetic diseases caused by NMD provoking mutations, including cystic fibrosis and muscular dystrophy, although inexplicably at least some of these drugs do not to appear to increase the stability of these transcripts (132). Other inhibitors of NMD have also been identified, including the natural product pateamine, which interacts with eIF4AIII but inhibits NMD independently of translation initiation inhibition (133).
Previous studies on NMD have primarily emphasized the determination of the complicated NMD mechanism and the identification of NMD targets. Little work has been done on validating and characterizing these targets. Future experiments will have to better define in vivo the role that NMD inhibition plays in biology, including tumorigenesis and other conditions (e.g. development) which are marked by hypoxia and other cellular stresses. Better insight into the mechanism by which eIF2α phosphorylation and potentially other events inhibit NMD will aid in the identification and testing of pharmacological agents that can modulate NMD activity and perhaps be clinically useful in diseases caused by PTC mutated transcripts and/or marked by cellular stress.
Acknowledgments
I apologize for the many studies that could not be cited due to space constraints. Gregory David and Heather Harding provided helpful critiques in the preparation of this manuscript, and funding was provided by the New York Community Trust and the National Institutes of Health (RO1DK08164).
References
- 1.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet. 1999;8(10):1893–900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 2.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36(10):1073–8. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 3.Gardner LB. Hypoxic inhibition of nonsense-mediated RNA decay regulates gene expression and the integrated stress response. Mol Cell Biol. 2008;28(11):3729–41. doi: 10.1128/MCB.02284-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ni JZ, Grate L, Donohue JP, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21(6):708–18. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clement SL, Lykke-Andersen J. A tethering approach to study proteins that activate mRNA turnover in human cells. Methods Mol Biol. 2008;419:121–33. doi: 10.1007/978-1-59745-033-1_8. [DOI] [PubMed] [Google Scholar]
- 6.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36(8):801–8. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 7.Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581(15):2845–53. doi: 10.1016/j.febslet.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 8.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 9.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008 doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebbapragada I, Lykke-Andersen J. Execution of nonsense-mediated mRNA decay: what defines a substrate? Curr Opin Cell Biol. 2009;21(3):394–402. doi: 10.1016/j.ceb.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293(5536):1836–9. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- 12.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103(7):1121–31. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 13.Mendell JT, ap Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298(5592):419–22. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 14.Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol. 2000;20(23):8944–57. doi: 10.1128/mcb.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12(10):893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 16.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106(5):607–17. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 17.Woeller CF, Gaspari M, Isken O, Maquat LE. NMD resulting from encephalomyocarditis virus IRES-directed translation initiation seems to be restricted to CBP80/20-bound mRNA. EMBO Rep. 2008;9(5):446–51. doi: 10.1038/embor.2008.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18(7):745–54. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lejeune F, Ranganathan AC, Maquat LE. eIF4G is required for the pioneer round of translation in mammalian cells. Nat Struct Mol Biol. 2004;11(10):992–1000. doi: 10.1038/nsmb824. [DOI] [PubMed] [Google Scholar]
- 20.Connolly E, Braunstein S, Formenti S, Schneider RJ. Hypoxia inhibits protein synthesis through a 4E-BP1 and elongation factor 2 kinase pathway controlled by mTOR and uncoupled in breast cancer cells. Mol Cell Biol. 2006;26(10):3955–65. doi: 10.1128/MCB.26.10.3955-3965.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braunstein S, Karpisheva K, Pola C, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28(3):501–12. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Dostie J, Dreyfuss G. Translation is required to remove Y14 from mRNAs in the cytoplasm. Curr Biol. 2002;12(13):1060–7. doi: 10.1016/s0960-9822(02)00902-8. [DOI] [PubMed] [Google Scholar]
- 23.Tange TO, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16(3):279–84. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3(3):195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 25.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309(5740):1514–8. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 26.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11(4):939–49. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 27.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4). Mol Cell Biol. 2001;21(1):209–23. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, et al. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12(11):1665–77. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashima I, Yamashita A, Izumi N, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20(3):355–67. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14-3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17(4):537–47. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 31.Yamashita A, Izumi N, Kashima I, et al. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23(9):1091–105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15(17):2215–28. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohnishi T, Yamashita A, Kashima I, et al. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12(5):1187–200. doi: 10.1016/s1097-2765(03)00443-x. [DOI] [PubMed] [Google Scholar]
- 34.Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell. 2008;133(2):314–27. doi: 10.1016/j.cell.2008.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12(3):675–87. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 36.Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23(14):4805–13. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14(12):2609–17. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16(1):49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 39.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172(6):803–8. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kedersha N, Stoecklin G, Ayodele M, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169(6):871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10(6):430–6. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 42.Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26(1):253–64. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125(6):1095–109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stalder L, Muhlemann O. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA. 2009 doi: 10.1261/rna.1672509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27(11):3970–81. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linde L, Boelz S, Neu-Yilik G, Kulozik AE, Kerem B. The efficiency of nonsense-mediated mRNA decay is an inherent character and varies among different cells. Eur J Hum Genet. 2007;15(11):1156–62. doi: 10.1038/sj.ejhg.5201889. [DOI] [PubMed] [Google Scholar]
- 47.El-Bchiri J, Guilloux A, Dartigues P, et al. Nonsense-mediated mRNA decay impacts MSI-driven carcinogenesis and anti-tumor immunity in colorectal cancers. PLoS ONE. 2008;3(7):e2583. doi: 10.1371/journal.pone.0002583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang JC, Kan YW. beta 0 thalassemia, a nonsense mutation in man. Proc Natl Acad Sci U S A. 1979;76(6):2886–9. doi: 10.1073/pnas.76.6.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci U S A. 1979;76(10):5134–7. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981;27(3 Pt 2):543–53. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 51.Kan YW, Lee KY, Furbetta M, Angius A, Cao A. Polymorphism of DNA sequence in the beta-globin gene region. Application to prenatal diagnosis of beta 0 thalassemia in Sardinia. N Engl J Med. 1980;302(4):185–8. doi: 10.1056/NEJM198001243020401. [DOI] [PubMed] [Google Scholar]
- 52.Okazaki Y, Furuno M, Kasukawa T, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–73. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 53.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6(4):e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14(3):563–76. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26(6):1591–601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.You KT, Li LS, Kim NG, et al. Selective translational repression of truncated proteins from frameshift mutation-derived mRNAs in tumors. PLoS Biol. 2007;5(5):e109. doi: 10.1371/journal.pbio.0050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han A, Kim WY, Park SM. SNP2NMD: a database of human single nucleotide polymorphisms causing nonsense-mediated mRNA decay. Bioinformatics. 2007;23(3):397–9. doi: 10.1093/bioinformatics/btl593. [DOI] [PubMed] [Google Scholar]
- 58.Bateman JF, Freddi S, Lamande SR, et al. Reliable and sensitive detection of premature termination mutations using a protein truncation test designed to overcome problems of nonsense-mediated mRNA instability. Hum Mutat. 1999;13(4):311–7. doi: 10.1002/(SICI)1098-1004(1999)13:4<311::AID-HUMU8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 59.Noensie EN, Dietz HC. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat Biotechnol. 2001;19(5):434–9. doi: 10.1038/88099. [DOI] [PubMed] [Google Scholar]
- 60.Ivanov I, Lo KC, Hawthorn L, Cowell JK, Ionov Y. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene. 2007;26(20):2873–84. doi: 10.1038/sj.onc.1210098. [DOI] [PubMed] [Google Scholar]
- 61.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat. 2008;29(8):1037–47. doi: 10.1002/humu.20763. [DOI] [PubMed] [Google Scholar]
- 62.Reddy JC, Morris JC, Wang J, et al. WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J Biol Chem. 1995;270(18):10878–84. doi: 10.1074/jbc.270.18.10878. [DOI] [PubMed] [Google Scholar]
- 63.Pinyol M, Bea S, Pla L, et al. Inactivation of RB1 in mantle-cell lymphoma detected by nonsense-mediated mRNA decay pathway inhibition and microarray analysis. Blood. 2007;109(12):5422–9. doi: 10.1182/blood-2006-11-057208. [DOI] [PubMed] [Google Scholar]
- 64.Anczukow O, Ware MD, Buisson M, et al. Does the nonsense-mediated mRNA decay mechanism prevent the synthesis of truncated BRCA1, CHK2, and p53 proteins? Hum Mutat. 2008;29(1):65–73. doi: 10.1002/humu.20590. [DOI] [PubMed] [Google Scholar]
- 65.Bala S, Kraus C, Wijnen J, Meera Khan P, Ballhausen WG. Multiple products in the protein truncation test due to alternative splicing in the adenomatous polyposis coli (APC) gene. Hum Genet. 1996;98(5):528–33. doi: 10.1007/s004390050254. [DOI] [PubMed] [Google Scholar]
- 66.Prosser J, Condie A, Wright M, et al. APC mutation analysis by chemical cleavage of mismatch and a protein truncation assay in familial adenomatous polyposis. Br J Cancer. 1994;70(5):841–6. doi: 10.1038/bjc.1994.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11(23):2805–14. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 68.Stewart GS, Maser RS, Stankovic T, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell. 1999;99(6):577–87. doi: 10.1016/s0092-8674(00)81547-0. [DOI] [PubMed] [Google Scholar]
- 69.Pitts SA, Kullar HS, Stankovic T, et al. hMRE11: genomic structure and a null mutation identified in a transcript protected from nonsense-mediated mRNA decay. Hum Mol Genet. 2001;10(11):1155–62. doi: 10.1093/hmg/10.11.1155. [DOI] [PubMed] [Google Scholar]
- 70.Karam R, Carvalho J, Bruno I, et al. The NMD mRNA surveillance pathway downregulates aberrant E-cadherin transcripts in gastric cancer cells and in CDH1 mutation carriers. Oncogene. 2008;27(30):4255–60. doi: 10.1038/onc.2008.62. [DOI] [PubMed] [Google Scholar]
- 71.Sumitsuji I, Sugano K, Matsui T, et al. Frequent genomic disorganisation of MLH1 in hereditary non-polyposis colorectal cancer (HNPCC) screened by RT-PCR on puromycin treated samples. J Med Genet. 2003;40(3):e30. doi: 10.1136/jmg.40.3.e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene. 2004;23(3):639–45. doi: 10.1038/sj.onc.1207178. [DOI] [PubMed] [Google Scholar]
- 73.Du L, Lai CH, Concannon P, Gatti RA. Rapid screen for truncating ATM mutations by PTT-ELISA. Mutat Res. 2008;640(12):139–44. doi: 10.1016/j.mrfmmm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 74.Cardinali M, Kratochvil FJ, Ensley JF, Robbins KC, Yeudall WA. Functional characterization in vivo of mutant p53 molecules derived from squamous cell carcinomas of the head and neck. Mol Carcinog. 1997;18(2):78–88. doi: 10.1002/(sici)1098-2744(199702)18:2<78::aid-mc3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 75.Wolf M, Edgren H, Muggerud A, et al. NMD microarray analysis for rapid genome-wide screen of mutated genes in cancer. Cell Oncol. 2005;27(3):169–73. doi: 10.1155/2005/478316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rossi MR, Hawthorn L, Platt J, Burkhardt T, Cowell JK, Ionov Y. Identification of inactivating mutations in the JAK1, SYNJ2, and CLPTM1 genes in prostate cancer cells using inhibition of nonsense-mediated decay and microarray analysis. Cancer Genet Cytogenet. 2005;161(2):97–103. doi: 10.1016/j.cancergencyto.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 77.Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20(23):8635–42. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsui M, Yachie N, Okada Y, Saito R, Tomita M. Bioinformatic analysis of post-transcriptional regulation by uORF in human and mouse. FEBS Lett. 2007;581(22):4184–8. doi: 10.1016/j.febslet.2007.07.057. [DOI] [PubMed] [Google Scholar]
- 79.Stockklausner C, Breit S, Neu-Yilik G, et al. The uORF-containing thrombopoietin mRNA escapes nonsense-mediated decay (NMD). Nucleic Acids Res. 2006;34(8):2355–63. doi: 10.1093/nar/gkl277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chan WK, Huang L, Gudikote JP, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26(7):1820–30. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11(10):1530–44. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Weischenfeldt J, Damgaard I, Bryder D, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22(10):1381–96. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hang X, Li P, Li Z, et al. Transcription and splicing regulation in human umbilical vein endothelial cells under hypoxic stress conditions by exon array. BMC Genomics. 2009;10:126. doi: 10.1186/1471-2164-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Clark TA, Schweitzer AC, Chen TX, et al. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8(4):R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ladomery MR, Harper SJ, Bates DO. Alternative splicing in angiogenesis: the vascular endothelial growth factor paradigm. Cancer Lett. 2007;249(2):133–42. doi: 10.1016/j.canlet.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 86.Gardina PJ, Clark TA, Shimada B, et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thorsen K, Sorensen KD, Brems-Eskildsen AS, et al. Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol Cell Proteomics. 2008;7(7):1214–24. doi: 10.1074/mcp.M700590-MCP200. [DOI] [PubMed] [Google Scholar]
- 88.Ghigna C, Valacca C, Biamonti G. Alternative splicing and tumor progression. Curr Genomics. 2008;9(8):556–70. doi: 10.2174/138920208786847971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boise LH, Gonzalez-Garcia M, Postema CE, et al. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74(4):597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 90.Modrek B, Lee CJ. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet. 2003;34(2):177–80. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 91.Modrek B, Lee C. A genomic view of alternative splicing. Nat Genet. 2002;30(1):13–9. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 92.Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 93.Baek D, Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc Natl Acad Sci U S A. 2005;102(36):12813–8. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100(1):189–92. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hillman RT, Green RE, Brenner SE. An unappreciated role for RNA surveillance. Genome Biol. 2004;5(2):R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Green RE, Lewis BP, Hillman RT, et al. Widespread predicted nonsense-mediated mRNA decay of alternatively-spliced transcripts of human normal and disease genes. Bioinformatics. 2003;19(Suppl 1):i118–21. doi: 10.1093/bioinformatics/btg1015. [DOI] [PubMed] [Google Scholar]
- 97.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6(5):386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 98.Zhang Z, Xin D, Wang P, et al. Noisy splicing, more than expression regulation, explains why some exons are subject to nonsense-mediated mRNA decay. BMC Biol. 2009;7(1):23. doi: 10.1186/1741-7007-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pan Q, Saltzman AL, Kim YK, et al. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20(2):153–8. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wittkopp N, Huntzinger E, Weiler C, et al. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29(13):3517–28. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10(2):99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 102.Brumbaugh KM, Otterness DM, Geisen C, et al. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14(5):585–98. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 103.Gaba A, Jacobson A, Sachs MS. Ribosome occupancy of the yeast CPA1 upstream open reading frame termination codon modulates nonsense-mediated mRNA decay. Mol Cell. 2005;20(3):449–60. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- 104.Rodriguez-Gabriel MA, Watt S, Bahler J, Russell P. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol Cell Biol. 2006;26(17):6347–56. doi: 10.1128/MCB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu L, Wise DR, Diehl JA, Simon MC. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J Biol Chem. 2008;283(45):31153–62. doi: 10.1074/jbc.M805056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nemetski SM, Gardner LB. Hypoxic regulation of Id-1 and activation of the unfolded protein response are aberrant in neuroblastoma. J Biol Chem. 2007;282(1):240–8. doi: 10.1074/jbc.M607275200. [DOI] [PubMed] [Google Scholar]
- 107.Blais JD, Filipenko V, Bi M, et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24(17):7469–82. doi: 10.1128/MCB.24.17.7469-7482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bi M, Naczki C, Koritzinsky M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24(19):3470–81. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 110.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–99. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 111.Harding HP, Zhang Y, Zeng H, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–33. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 112.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40(1):14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 113.Finkel T. Reactive oxygen species and signal transduction. IUBMB Life. 2001;52(12):3–6. doi: 10.1080/15216540252774694. [DOI] [PubMed] [Google Scholar]
- 114.Genestra M. Oxyl radicals, redox-sensitive signalling cascades and antioxidants. Cell Signal. 2007;19(9):1807–19. doi: 10.1016/j.cellsig.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 115.Irani K, Xia Y, Zweier JL, et al. Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts. Science. 1997;275(5306):1649–52. doi: 10.1126/science.275.5306.1649. [DOI] [PubMed] [Google Scholar]
- 116.Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci U S A. 1998;95(20):11715–20. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Okada T, Yoshida H, Akazawa R, Negishi M, Mori K. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem J. 2002;366(Pt 2):585–94. doi: 10.1042/BJ20020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Barbosa-Tessmann IP, Chen C, Zhong C, Schuster SM, Nick HS, Kilberg MS. Activation of the unfolded protein response pathway induces human asparagine synthetase gene expression. J Biol Chem. 1999;274(44):31139–44. doi: 10.1074/jbc.274.44.31139. [DOI] [PubMed] [Google Scholar]
- 119.Ganapathy V, Thangaraju M, Prasad PD. Nutrient transporters in cancer: relevance to Warburg hypothesis and beyond. Pharmacol Ther. 2009;121(1):29–40. doi: 10.1016/j.pharmthera.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 120.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458(7239):762–5. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Blais JD, Addison CL, Edge R, et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol. 2006;26(24):9517–32. doi: 10.1128/MCB.01145-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bateman JF, Freddi S, Nattrass G, Savarirayan R. Tissue-specific RNA surveillance? Nonsense-mediated mRNA decay causes collagen X haploinsufficiency in Schmid metaphyseal chondrodysplasia cartilage. Hum Mol Genet. 2003;12(3):217–25. doi: 10.1093/hmg/ddg054. [DOI] [PubMed] [Google Scholar]
- 123.Zetoune AB, Fontaniere S, Magnin D, et al. Comparison of nonsense-mediated mRNA decay efficiency in various murine tissues. BMC Genet. 2008;9:83. doi: 10.1186/1471-2156-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Koditz J, Nesper J, Wottawa M, et al. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood. 2007;110(10):3610–7. doi: 10.1182/blood-2007-06-094441. [DOI] [PubMed] [Google Scholar]
- 125.Shima DT, Adamis AP, Ferrara N, et al. Hypoxic induction of endothelial cell growth factors in retinal cells: identification and characterization of vascular endothelial growth factor (VEGF) as the mitogen. Mol Med. 1995;1(2):182–93. [PMC free article] [PubMed] [Google Scholar]
- 126.Levy AP. Hypoxic regulation of VEGF mRNA stability by RNA-binding proteins. Trends Cardiovasc Med. 1998;8(6):246–50. doi: 10.1016/s1050-1738(98)00020-6. [DOI] [PubMed] [Google Scholar]
- 127.Levy AP, Levy NS, Goldberg MA. Post-transcriptional regulation of vascular endothelial growth factor by hypoxia. J Biol Chem. 1996;271(5):2746–53. doi: 10.1074/jbc.271.5.2746. [DOI] [PubMed] [Google Scholar]
- 128.Galban S, Martindale JL, Mazan-Mamczarz K, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 2003;23(20):7083–95. doi: 10.1128/MCB.23.20.7083-7095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zou T, Mazan-Mamczarz K, Rao JN, et al. Polyamine depletion increases cytoplasmic levels of RNA-binding protein HuR leading to stabilization of nucleophosmin and p53 mRNAs. J Biol Chem. 2006;281(28):19387–94. doi: 10.1074/jbc.M602344200. [DOI] [PubMed] [Google Scholar]
- 130.Das D, Clark TA, Schweitzer A, et al. A correlation with exon expression approach to identify cis-regulatory elements for tissue-specific alternative splicing. Nucleic Acids Res. 2007;35(14):4845–57. doi: 10.1093/nar/gkm485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Barbier J, Dutertre M, Bittencourt D, et al. Regulation of H-ras splice variant expression by cross talk between the p53 and nonsense-mediated mRNA decay pathways. Mol Cell Biol. 2007;27(20):7315–33. doi: 10.1128/MCB.00272-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447(7140):87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 133.Dang Y, Low WK, Xu J, et al. Inhibition of nonsense-mediated mRNA decay by the natural product pateamine A through eukaryotic initiation factor 4AIII. J Biol Chem. 2009 doi: 10.1074/jbc.M109.009985. [DOI] [PMC free article] [PubMed] [Google Scholar]