Abstract
The epidermal growth factor receptor (EGFR), a tyrosine kinase, is central to human tumorigenesis. Typically, three classes of drugs inhibit tyrosine kinase pathways: blocking antibodies, small kinase inhibitors, and soluble ligand receptor traps/decoys. Only the first two types of EGFR-binding inhibitory drugs are clinically available; notably, no EGFR decoy has yet been developed. Here we identify small molecules mimicking EGFR and that functionally behave as soluble decoys for EGF and TGFα, ligands that would otherwise activate downstream signaling. After combinatorial library selection on EGFR ligands, a panel of binding peptides was narrowed by structure–function analysis. The most active motif was CVRAC (EGFR 283–287), which is necessary and sufficient for specific EGFR ligand binding. Finally, a synthetic retro-inverted derivative, D(CARVC), became our preclinical prototype of choice. This study reveals an EGFR-decoy drug candidate with translational potential.
Keywords: peptide, cancer, EGFR, cetuximab, phage display
The epidermal growth factor receptor (EGFR) is a member of the ErbB family of tyrosine kinase receptors (1, 2). Several lines of evidence indicate that the EGFR is abnormally activated in many types of epithelial tumors. The first therapeutic agent targeted to the EGFR is a monoclonal antibody, cetuximab, which blocks ligand binding and thus inhibits tyrosine kinase activity (3). In the past few years, it has become clear that specific somatic EGFR mutations present in non-small-cell lung cancer potentiate responses to certain low molecular weight tyrosine kinase inhibitors and monoclonal antibodies (1, 4–8); mutation of the K-ras gene also has been associated with survival in patients with advanced colon cancer treated with cetuximab (9). These agents, both antibodies and tyrosine kinase inhibitors, prevent ligand-induced receptor activation and downstream signaling and result in cell cycle arrest, promotion of apoptosis, and inhibition of angiogenesis (10, 11).
There are three general classes of agents that inhibit tyrosine kinase receptors: blocking antibodies, small kinase inhibitors, and soluble ligand traps or receptor decoys. However, only agents belonging to the first two classes are currently available for therapeutic intervention: monoclonal antibodies directed at the ligand-binding extracellular domain of the receptor (e.g., cetuximab, panitumumab, zalutumumab, nimotuzumab, and matuzumab) and low-molecular-weight inhibitors of intracellular tyrosine kinase activity (e.g., gefitinib, erlotinib, and lapatinib). Extensive research has recently been done to find EGFR molecular decoys such as Argos, an antagonist of EGFR signaling that was identified in Drosophila (12, 13), or a recombinant form of the extracellular domain of ErbB4 that antagonizes ligand-induced receptor tyrosine phosphorylation (14). Because the EGFR is a central target in oncology, and given the success of this approach with other important ligand-receptor tyrosine kinases such as the vascular endothelial growth factor (VEGF) receptors (15, 16), we reasoned that the combinatorial discovery and translational development of a human EGFR-targeted soluble decoy might result in a unique class of drugs.
We have designed an in tandem approach that comprises mapping of interactive sites on EGFR ligands, followed by the chemical generation and evaluation of derivative consensus motif analogs. We first performed a combinatorial library screening in representative EGFR ligands in vitro to select and identify a panel of consensus motifs. We subsequently used solid-phase synthesis to produce pertinent peptides and peptidomimetic drug candidates. Finally, we evaluated one such EGFR drug decoy candidate—a synthetic, low-molecular-weight, retro-inverted, water-soluble peptidomimetic—by in vitro, in cellulo, and in vivo assays and demonstrated that it has anti-tumor activity. Aside from the retro-inversion approach, which generates degradation-resistant D-peptidomimetics (17), we have also used cyclization in an attempt to improve the bioavailability of our prototype; our small lead molecule, derivatized from a native EGFR cyclic motif, is a structural and functional drug decoy of this tyrosine kinase receptor with tumor targeting attributes and potential for translational applications.
Results and Discussion
Combinatorial Screening on a Panel of Ligands that Bind to the EGFR.
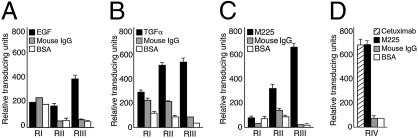
We established a combinatorial approach in a search for consensus protein-interacting sites within the EGFR. First, we screened a random library displaying the general peptide arrangement CX7C on three representative EGFR ligands [namely EGF, tumor growth factor alpha (TGFα), and cetuximab] and selected for phage binding in consecutive rounds. We observed serial enrichment in all selections (Fig. 1 A–C). BSA, VEGF, and irrelevant IgG served as negative controls. As predicted, cetuximab (formerly C225 or IMC225; marketed as Erbitux) showed an overlapping binding profile with its parental murine 225 (M225) version (Fig. 1 C and D). After the third round of selection we observed marked phage binding to each immobilized EGFR ligand, relative to negative controls, as follows: EGF, 8-fold relative to BSA (t test, P < 0.001) and 8-fold relative to VEGF (P < 0.001); TGFα, 22-fold relative to BSA (P < 0.001) and 15-fold relative to VEGF (P < 0.001); M225, 10-fold relative to BSA (P < 0.001) and 8-fold relative to irrelevant IgG (P < 0.001); and cetuximab, 10-fold relative to BSA (P < 0.001) and 8-fold relative to irrelevant IgG (P < 0.001).
Fig. 1.
Screening of a combinatorial random peptide library on EGFR ligands EGF, TGFα, and cetuximab. (A) EGF panning. VEGF and BSA were used as negative control proteins in A and B. (B) TGFα panning. (C) M225 monoclonal antibody (the original murine version of cetuximab) was immobilized onto microtiter wells at a concentration of 2 μg. The CX7C phage library was incubated with each of the target proteins. Shown are the relative transducing units (TU) obtained from each well coated with M225, mIgG, or BSA after three rounds of selection (RI–RIII). (D) Specificity of the peptides recovered from RIII targeting. M225 was recapitulated upon binding to cetuximab on a fourth round of selection (RIV). Results are expressed as mean ± SEM of triplicate wells.
Molecular Interaction Between Selected Peptides and the EGFR.
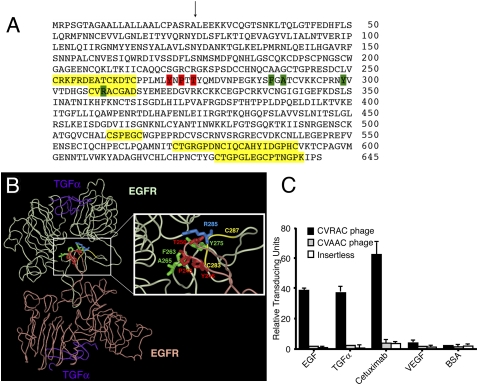
We performed a comprehensive protein similarity analysis of selected peptides (n = 384) to identify sequences resembling the extracellular domain of the EGFR. Overlapping consensus motifs selected in all three EGFR ligands were identified, mapped, and consolidated within the five dominant candidate regions (Cys227-Cys240, Cys283-Asp290, Cys486-Cys491, Cys547-Cys567, and Cys604-Lys618; not accounting for the signal peptide, as indicated) within the primary structure of the receptor (Fig. 2A, yellow highlights). Of note, all such candidate regions contained at least two or more cysteine residues, suggestive of structural motifs.
Fig. 2.
Mapping candidate epitopes within the EGFR. (A) Amino acid sequence corresponding to the extracellular domain of the EGFR (accession no. NP_005219). Leu1 is the first residue after the signal peptide. The arrow designates the signal peptide cleavage site. Yellow highlights indicate five consensus regions to which peptides derived from library screenings (on the ligands EGF, TGFα, and cetuximab) were clustered. Green and red boxes pinpoint the reciprocal residues in the two EGFR molecules involved in dimerization. (B) Location of a cetuximab-binding region within the EGFR structure. Light green and light red ribbons indicate the backbone of each EGFR homodimer. Purple designates the TGFα ligand bound to the EGFR. (Inset) Red and green indicate residues involved in EGFR dimerization (see A). Yellow ribbon shows the location of CVRAC within the EGFR homodimer (residues 283–287). (C) CVRAC-displaying phage binds specifically to cetuximab, EGF, and TGF-α. VEGF and BSA served as negative controls for binding. Recombinant proteins were coated onto microtiter wells at 10 μg/mL, and wells were incubated with either CVRAC-phage or CVAAC-phage (alanine scanning control). An insertless phage was an additional negative control. Phage input was 109 TU per well. Results are expressed as mean ± SEM of triplicate wells.
To understand these findings at a protein interactive level, we generated a consensus motif panel (n = 15) of synthetic linear and cyclic peptides (Table S1) and used binding to the anti-EGFR monoclonal antibody cetuximab as an initial functional screen (Fig. S1) to minimize the number of candidate ligands. We previously expanded this epitope mapping approach to show that selection of random peptide libraries on the repertoire of circulating immunoglobulins from cancer patients (18, 19) can identify immunogenic tumor antigens as molecular targets (20). Moreover, we and other investigators applied similar methodology to therapeutic (21–23) or diagnostic (24) antibodies in a strategy that could reveal mechanisms of action (22), identify biological reagents for immunization (23), or discover as yet unrecognized antigens (24).
The best concentration-dependent ligand peptide in this binding assay (Fig. S1) was CVRACGAD (residues 283–290), one of the candidate regions encompassing a residue involved in receptor dimerization (25–27) within the EGFR (Fig. 2 A and B). Furthermore, binding of the minimized motif CVRAC (residues 283–287) was not significantly different from that of the larger peptide CVRACGAD. Indeed, even the smaller cyclic tripeptide Val-Arg-Ala, containing the residue corresponding to Arg285, was sufficient for binding. In Fig. 2B, light green and light red ribbons indicate the backbone of each EGFR homodimer, and purple designates the TGFα ligand bound to the EGFR (28); the Inset details EGFR residues involved in the dimerization site (corresponding to the green and red color coding from Fig. 2A), and the yellow ribbon shows the location and structure of CVRAC within a single chain.
To evaluate whether this motif had selective EGFR-decoy attributes, we designed and generated phage constructs displaying the cyclic peptide CVRAC or the corresponding negative control CVAAC, in which Arg has been changed to Ala (through site-directed mutagenesis), and measured binding to EGFR ligands. The CVRAC-phage preferentially bound to the receptor ligands EGF (17-fold relative to CVAAC-phage, 38-fold relative to insertless phage; Student’s t test, P < 0.001), TGFα (13-fold relative CVAAC-phage, 23-fold relative to insertless phage; P < 0.001), and cetuximab (23-fold relative CVAAC-phage, 51-fold relative to insertless phage; P < 0.001), but not to the negative control proteins VEGF or BSA. A negative control insertless phage (P < 0.001) or CVAAC-phage (P < 0.001) showed no binding preference (Fig. 2C). These data indicate that the region Cys283–Cys287 of EGFR is implicated in its binding to native ligands and targeted antibodies.
Potential Attributes of a Short Cyclic Motif as an EGFR-Like Interacting Site.
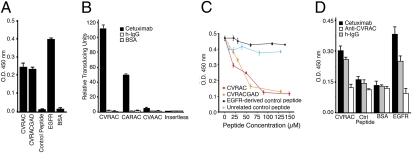
To evaluate CVRAC as a potential drug lead in the development of an EGFR molecular decoy, we first demonstrated that two synthetic cyclic peptides containing the minimal three-residue cyclic loop CVRAC (i.e., outside a phage display context) bind to cetuximab (Fig. 3A); EGFR served as a positive control and an unrelated peptide as a negative control. Having confirmed that these soluble peptides could recapitulate the EGFR-like binding attributes to a certain extent, we next developed an assay to evaluate the capacity of such peptide ligands to inhibit the binding of cetuximab to the EGFR. By ELISA, the two synthetic peptides, but not two negative control peptides (one with an unrelated sequence and another with an EGFR-derived sequence from region II), blocked the binding of cetuximab to the EGFR in a specific and concentration-dependent manner (Fig. 3B). In both assays (Fig. 3 A and B), the binding activities of the synthetic shorter peptide (CVRAC) and the longer peptide (CVRACGAD) were indistinguishable from each other; therefore, we selected the smaller one as a better candidate drug lead for derivatization. These data suggest that the interaction of cetuximab with the EGFR is at least partially mediated by CVRAC, a functional, cyclic interacting site embedded within the sequence of the EGFR.
Fig. 3.
Molecular interaction of CVRAC, cetuximab, and EGFR. (A) Synthetic peptides (CVRACGAD or CVRAC), compared to an unrelated control peptide (SDNRYIGSW), specifically bind to cetuximab. BSA served as an additional negative control, and the EGFR as a positive control. (B) Concentration-dependent inhibition of binding of cetuximab to the EGFR by the synthetic peptides CVRACGAD and CVRAC, in comparison with negative controls: an EGFR sequence-derived peptide (CQKCDPSC) and an unrelated negative control peptide. (C) Phage displaying alanine scanning versions of the CVRAC peptide (CARAC and CVAAC) were used to identify critical residues on the basis of their capacity to bind to cetuximab. Insertless phage served as a negative control. (D) Polyclonal antibody against CVRAC recognized the EGFR. Bars represent mean ± SEM.
To confirm that the interaction of CVRAC with cetuximab was specific and to identify the residue(s) responsible for peptide binding, we constructed phage displaying alanine scanning versions of the peptide and performed binding assays to cetuximab or to a negative isotype control (Fig. 3C). CVRAC-displaying phage exhibited marked binding to immobilized cetuximab, in comparison with the negative controls BSA (131-fold; Student’s t test, P < 0.001) and isotype antibody (81-fold; P < 0.001); moreover, CVRAC-displaying phage bound to a greater extent, relative to negative controls that included insertless phage (96-fold; P < 0.001) and CVAAC-displaying phage (48-fold; t test, P < 0.001). Consistently, CVAAC-displaying phage lacked binding entirely, but CARAC-displaying phage retained partial activity (∼45% of the CVRAC-displaying phage binding activity), data indicating again that the arginine residue (corresponding to Arg285 within the full-length EGFR) is critical for the interaction of the displayed peptide with cetuximab. Specificity was indicated by the lack of binding to BSA or to the isotype control (Fig. 3C).
We hypothesized that, if the interacting site Cys283–Cys287 within the EGFR exhibits receptor-like properties or biological activity, a synthetic motif might also elicit a cross-reactive humoral immune response. To test this hypothesis, we immunized rabbits with keyhole limpet hemocyanin (KLH) conjugated to the synthetic CVRAC peptide and evaluated the reactivity of purified antibodies by ELISA. Polyclonal antibodies against the soluble motif CVRAC specifically recognized the EGFR (Fig. 3D). These data demonstrate the generation of antibodies against the native receptor and indicate that the interacting site CVRAC within the EGFR does behave as a hapten.
The Motif CVRAC is Biologically Active.
Having established the potential of the EGFR interacting site CVRAC in vitro, we evaluated the cognate synthetic motif in tumor cell lines. The representative colon cancer cell line GEO and the head-and-neck cancer cell line HN5 were chosen because (i) they express the EGFR and represent common human cancers in which EGFR-targeted therapy is used clinically (1, 29, 30) and (ii) their respective pattern of tumor response to cetuximab has been well established (31, 32).
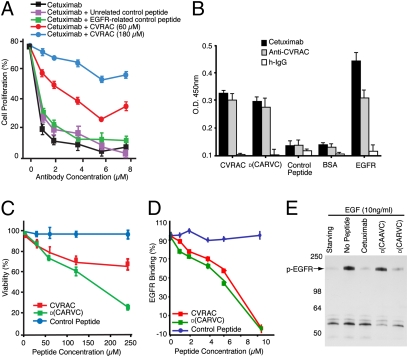
As an experimental baseline, we first confirmed that treatment of these tumor cells in vitro with cetuximab consistently and reproducibly inhibited cell proliferation. To evaluate the biological activity of the synthetic motif CVRAC, we incubated HN5 tumor cells (Fig. 4A) with increasing equimolar concentrations of CVRAC or negative control peptides. In both cell lines, we observed a concentration-dependent inhibition by CVRAC of the antibody-dependent inhibition of proliferation, presumably due to the binding to cetuximab as a soluble EGFR decoy. These results demonstrate that CVRAC is biologically active in the context of EGFR-expressing human tumor cells in vitro and is a drug candidate meriting further development.
Fig. 4.
The retro-inverso peptidomimetic of the CVRAC motif is recognized by cetuximab and inhibits binding of cetuximab to the EGFR. (A) Human HN5 tumor cells were treated with increasing concentrations of cetuximab (black line). Cells were also exposed to either 60 μM (red line) or 180 μM (blue line) CVRAC. Unrelated control peptide (purple line) or EGFR-related control peptide (green line) had no effect on cetuximab activity. A representative experiment is depicted. Experiments were repeated four times with similar results. Bars represent mean ± SEM. (B) Binding of retro-inverso D-form peptides (plated at 10 μg/mL) to cetuximab in an ELISA-based assay. Equivalent amounts of IgGs (cetuximab, anti-CVRAC, or h-IgG) were analyzed for binding to CVRAC or to its retro-inverso peptidomimetic D(CARVC). (C) Effect of the synthetic peptides on HN5 tumor cells. Cells were incubated with increasing concentrations (up to 250 mM) of the peptide CVRAC, the retro-inverso peptidomimetic D(CARVC), or a negative control peptide. Viability in the absence of peptide was set to 100%. (D) Inhibition of EGFR:cetuximab association, monitored by SPR in the presence of synthetic peptides or peptidomimetic D(CARVC). Bars represent mean ± SEM. (E) Analysis of receptor autophosphorylation in cells stimulated with EGF or control media for 15 min, after which cetuximab or synthetic peptides were added with the growth factor to evaluate inhibition. Receptors were immunoprecipitated with antibodies against phosphorylated (p)EGFR and were immunoblotted with anti-phosphotyrosine IgG. This representative experiment shows that D(CARVC) specifically inhibits the phosphorylation of the EGFR in human HN5 tumor cells.
Design, Synthesis, and Development of a Small Drug Decoy.
Peptide-based drugs are often susceptible to degradation by proteolytic enzymes; consequently, the biological activity of a peptide depends directly on its stability in serum. Retro-inverted peptide modification (i.e., reversal of the direction of the primary peptide sequence plus inversion of the chirality of each individual residue to the D-enantiomer) of biologically active motifs has been shown to increase the stability of peptidomimetic drug candidates (33), because most natural mammalian proteases do not cleave D-residue nonpeptide bonds. In general, this retro-inversion approach can result in peptidomimetics with strong topological correlation to the parent peptide because the resulting side-chain disposition is similar (i.e., the positions of side-chains are preserved) but carbonyl and amide groups are interconverted (i.e., the positions of carbonyl and amino groups in the backbone of the peptide are exchanged).
Through solid-phase synthesis, we designed and produced a compound based on the EGFR-interacting site CVRAC (Fig. S2). Because the function of any peptidomimetic-based drug candidate generated through retro-inversion methodology must be empirically validated, we used several assays (Fig. 4 B–D) to determine the activity and biological properties of the retro-inverted drug prototype D(CARVC). We first asked whether antibodies produced against the peptide CVRAC would also recognize D(CARVC) by ELISA (Fig. 4B). D(CARVC) retained binding activity to the antibody cetuximab; in addition, polyclonal anti-CVRAC antibodies recognized both the peptide CVRAC and the drug D(CARVC). The EGFR served as an immobilized positive control, and BSA or control peptides (CVAAC) served as immobilized negative controls. Negative control IgG showed only minimal background binding relative to the specific binding mediated by either anti-CVRAC antibodies or cetuximab (Fig. 4B). This result indicates that antibody recognition of the peptide CVRAC and the drug D(CARVC) is similar in this assay. In summary, the hapten-carrier adduct KLH-CVRAC induces a humoral immune response that recognizes either the peptide CVRAC or the drug D(CARVC) as a hapten.
Next, we determined whether the peptide CVRAC or the drug D(CARVC) would affect the proliferation of HN5 cells. Tumor cells exposed to either CVRAC or D(CARVC) proliferated much less in vitro than those exposed to the control peptide (Fig. 4C); this marked effect was specific and concentration dependent. In addition to HN5 cells, similar results were also observed with GEO cells and with EF43.fgf-4 cells (Fig. S3). Finally, D(CARVC) activity on an equimolar basis appeared to be more potent, possibly due to the expected proteolytic degradation of the peptide CVRAC in this prolonged (120-h) functional assay.
The capacity of CVRAC and D(CARVC) to block EGFR binding to cetuximab was further assayed by surface plasmon resonance (SPR). An immobilized anti-human Fc monoclonal antibody was used to capture cetuximab; subsequently, the EGFR, either alone or in the presence of the synthetic peptides, was introduced. Both CVRAC and D(CARVC) markedly inhibited the binding of the EGFR to cetuximab (Fig. 4D), relative to the control peptide. The inhibition was concentration dependent, with an IC50 value of ∼5.4 mM for CVRAC and ∼4.8 mM for D(CARVC); the observed similar activity of the two agents reflects the lack of enzymatic activity in this serum-free assay. No binding inhibition was observed with control peptides at equimolar concentrations (Fig. 4D). We also observed low-affinity interactions (micromolar range) with both agents by NMR spectroscopy.
Finally, to determine whether EGFR activation was inhibited after treatment with the candidate drug, we incubated tumor cells with synthetic peptides, D(CARVC), or cetuximab in the presence or absence of EGF. Treatment of tumor cells with EGF led to the tyrosine phosphorylation of a specific 170-kDa protein (Fig. 4E); as expected, no phosphorylation was observed in non-EGFR-expressing control cells. Both cetuximab and D(CARVC)—but not the negative control drug [D(CAAVC), a synthetic peptidomimetic with a mutation corresponding to EGFR Arg285Ala]—effectively inhibited the EGF-induced tyrosine phosphorylation in this functional assay (Fig. 4E), thus suggesting a new EGFR inhibitory mechanism. To dissect the downstream signal transduction cascade in this setting, we examined proliferative, survival, and migratory pathways. In preliminary experiments with human HN5 head and neck cells, D(CARVC) appears to differentially inhibit EGF-induced phosphorylation of ERK and Akt but not p38. Future studies will be required to fully understand the signal transduction modulation mediated by D(CARVC).
Tumor Targeting and Preclinical Validation of D(CARVC).
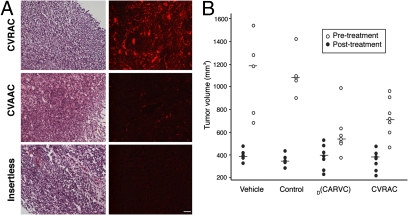
Having demonstrated the biological activity of our EGFR-derived agents in vitro, we next attempted to evaluate their potential for use in vivo (Fig. 5). We reasoned that, to behave as EGFR decoys, these agents should home to an EGFR “ligand-rich” tumor microenvironment (i.e., with high local concentrations of the native ligands EGF and/or TGFα). Therefore, the capacity of CVRAC-displaying phage to target tumors in vivo was determined by administration of CVRAC-displaying phage or controls (either CVAAC-displaying phage or insertless control phage) i.v. into immunocompetent (BALB/c) female mice bearing mammary tumors (Fig. 5A) (34). We chose to test a standard tumor model in which EF43.fgf-4 cells are administered s.c. to induce very rapid growth of highly vascularized tumors in immunocompetent mice (34). In histological sections of fixed tissue, there was marked staining of the tumors in mice receiving CVRAC-displaying phage but only background staining in control organs. In contrast, negative control phage (either CVAAC-displaying phage or insertless phage) were not detected in tumors (Fig. 5A) or in control organs (Fig. S4 A and B). We used the same isogenic tumor model (34) to evaluate whether the peptide CVRAC or the drug prototype D(CARVC) could suppress tumor growth in vivo. Tumor-bearing mice received vehicle alone, the peptide CVRAC, the drug D(CARVC), or control peptides (Fig. 5B). We observed differences in tumor growth as early as 5 days after treatment. At the end of 2 weeks, mice treated with D(CARVC) exhibited significantly smaller tumor volumes (550 ± 50 mm3, P = 0.02), relative to tumor-bearing mice that received negative control peptide (1,120 ± 120 mm3; t test). Tumors in mice treated with control peptide behaved similarly to tumors in mice receiving vehicle alone (1,200 ± 135 mm3), data indicating that a control peptide had no measurable effect. The CVRAC peptide also showed therapeutic efficacy in vivo but, because of enzymatic degradation, the tumor responses observed were somewhat inferior to those of D(CARVC). These results confirm that ligand-directed viral particles and EGFR-derived peptidomimetics target tumors. Consistent with an EGFR-decoy activity, these results likely represent homing due to high concentrations of EGFR ligands in the tumor microenvironment in vivo. It has long been established that D-amino acid oxidase is the only mammalian enzyme that catabolizes D-peptidomimetics in the kidney (17) and thus it is likely that the drug excretion mechanism will be renal; however, follow-up studies will clarify the drug stability in vivo and in metabolism.
Fig. 5.
CVRAC-targeted phage homes to tumors. (A) Phage displaying the peptide CVRAC or CVAAC, or insertless negative control phage, were administered i.v. into mice bearing EF43.fgf-4-derived tumors. An anti-phage antibody was used for staining. H&E staining, with the corresponding fluorescence-based immunostaining, is shown in tumors. Tumor-bearing mice received CVRAC phage, CVAAC phage, or insertless control phage as indicated. Cohorts of tumor-bearing mice (n = 5 mice/group) were used. A representative experiment is shown. (Scale bar, 100 μm.) (B) Treatment of tumor-bearing mice with peptides and peptidomimetics. BALB/c mice bearing EF43.fgf-4-derived tumors were divided into size-matched cohorts (n = 7 mice/group); individual tumor volumes are represented before (black circles) and after (white circles) treatment. Peptides and peptidomimetics were administered at 750 μg/mouse/dose for 5 days. Shown are mean tumor volumes ± SEM.
Mechanism of Action as a Candidate Drug Decoy.
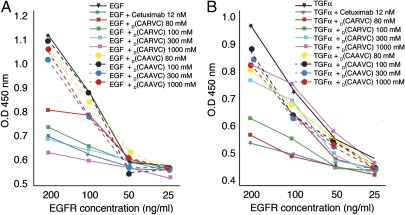
Given the promising results observed in vivo, we further investigated the mechanism of action of D(CARVC) as an EGFR-targeted soluble decoy. Decreasing molar concentrations of EGFR were immobilized in vitro, after which the native ligand (EGF or TGFα) was used to establish the baseline for the binding of EGF to the EGFR or of TGFα to the EGFR. As predicted, cetuximab displaced EGF at low nanomolar concentrations and thus served as a positive control. The drug D(CARVC) displaced EGF and yielded a concentration-dependent effect (from 30 to 1,000 mM). Moreover, the magnitude of ligand displacement elicited by D(CARVC) at 300 mM was similar to that of cetuximab in this assay (Fig. 6A). Finally, we observed concentration-dependent displacement of TGFα by D(CARVC) (Fig. 6B). These data support the EGFR-decoy effect as a mechanism of action of our prototype.
Fig. 6.
The prototype peptidomimetic drug D(CARVC) functions through an EGFR-decoy mechanism. (A) D(CARVC) displaces EGF from the EGFR. The EGFR was coated onto 96-well plates at decreasing concentrations. Increasing molar concentrations of the synthetic peptidomimetic D(CARVC) were used to evaluate competitive inhibition of EGF binding (squares). D(CAAVC) was used as a negative peptidomimetic control at the same concentrations (circles). Cetuximab (12 nM) served as a positive control for the displacement of EGF from the EGFR. (B) D(CARVC) displaces the binding of TGFα from the EGFR. Evaluation is shown of the competitive inhibition of the binding of TGFα to the EGFR by increasing molar concentrations (as indicated) of the synthetic peptidomimetic D(CARVC). Bars represent mean ± SEM.
Li et al. (35) reported that cetuximab interacts with domain III of the soluble extracellular region of the EGFR. Notably, the EGFR ligand we selected, peptide CVRAC, is on domain II; moreover, other phage display assays on cetuximab (23) selected yet another set of ligand peptides without a clear relation to the reported EGFR–cetuximab x-ray crystal structure. In another study, the anti-ErbB2 monoclonal antibody pertuzumab was found to bind to ErbB2 near the center of domain II, noteworthy because the central portions of domain II in ErbB2 and EGFR are highly homologous and represent the major part of the dimerization interface observed in the EGFR (25, 28). In support of our results, one of the alanine scan mutants made in ErbB2, involving the corresponding residue (Val286) in CVRAC on the EFGR, is essential for the binding and dimerization of pertuzumab to its receptor (36). Those data notwithstanding, whether or not CVRAC and/or other cetuximab ligand peptides are involved in a form of “induced fit” (37–39) remains an open question to be addressed in future studies. The experimental data presented here may lead to development of other molecular decoys for EGF- and TGFα-related pathways.
Conclusion
In summary, these results show that the prototype D(CARVC) functions as a specific inhibitor of tumor cell proliferation, with experimental evidence in vitro, in cells, and in vivo to support our initial hypothesis that this unique class of small drug candidates functions through an EGFR-decoy mechanism. In contrast to other EGFR-targeting agents such as cetuximab, this ligand-sequestering drug still may be active and perhaps a suitable candidate for translation in the setting of downstream K-ras gene mutations. Indeed, it has long been demonstrated that human tumors containing KRAS mutations express high levels of ErbB ligands (40–42). Moreover, Fujimoto et al. (43) have shown that KRAS mutations are not sufficient to confer resistance to EGFR inhibition. One can speculate that EGFR ligands can still stimulate tumor growth, even with genetic alterations in the downstream pathway. Although the reasons for these observations are not entirely understood, it is conceivable and it remains to be determined whether or not an EGFR ligand decoy such as D(CARVC) or its derivatives might be effective in such settings. On a larger context, this work illustrates an example of combinatorial peptide library selection and discovery on key receptor–ligand tumor pathways, followed by rapid generation and preclinical evaluation of a targeted anticancer drug lead.
Materials and Methods
Reagents.
Cetuximab is a human (h)–mouse (m) chimeric anti-EGFR IgG1 class monoclonal antibody (44). Monoclonal antibody 528 (isotype IgG2a) and M225 (isotype IgG1) are directed against EGFR. m-IgG and h-IgG were purchased from Sigma. Primary antibodies were anti-EGFR1 (Tyr1068) and anti-phospho-tyrosine (Cell Signaling) and HRP-conjugated goat anti-mouse (Jackson). Synthetic peptides CVRACGAD and CVRAC and peptidomimetics D(CARVC) and D(CAAVC) were purchased from PolyPeptide Laboratory. EGFR-derived and unrelated sequences (such as SDNRYIGSW and CEFESC) also served as controls in assays.
Cell Culture and Cell Viability Assays.
Tumor cell lines HN5, UMSCC 10A, GEO, and EF43.fgf-4 were cultured in standard conditions. Viability was assessed by MTT assays (Sigma) as described (45). For details, see SI Text.
Phage Display Screening and Binding Assays.
Phage peptide screening and binding assays were performed as described (45); see SI Text.
Antibodies Against CVRAC Peptide and ELISA.
Rabbits were immunized with KLH-conjugated CVRAC peptide (46). For details, see SI Text.
Surface Plasmon Resonance.
SPR was used to determine the inhibitory effect of CVRAC or D(CARVC) on the binding of EGFR to cetuximab in a BIAcore 3000. For details, see SI Text.
Immunoprecipitation and Western Blot Analysis.
Cells were lysed and lysates were used for immunoprecipitation assays as described (45). An anti-phosphotyrosine antibody (PY20) was used for EGFR activation assays. For details, see SI Text.
Tumor Targeting.
Selective phage homing to tumors was evaluated as described (34, 47). See SI Text for details.
Immunohistochemistry.
Immunostaining was performed as described (48); see SI Text.
Sequence Alignment.
We used peptide-matching software (49) to identify motifs resembling targeted ligands. For details, see SI Text.
Statistics.
The appropriate statistic test was used for the analysis of assays as indicated. Results were considered statistically significant if P < 0.05. For details, see SI Text.
Supplementary Material
Acknowledgments
We thank Drs. K. Ang and J. M. Kurie for discussions and Dr. E. H. Sage for reading the manuscript. This work was supported in part by grants from the Department of Defense and National Institutes of Health and by awards from the Gillson–Longenbaugh Foundation, the Marcus Foundation, and AngelWorks (to W.A. and R.P.). M.C.-V. is a fellow of the Susan G. Komen Breast Cancer Foundation.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915146107/DCSupplemental.
References
- 1.Gusterson BA, Hunter KD. Should we be surprised at the paucity of response to EGFR inhibitors? Lancet Oncol. 2009;10:522–527. doi: 10.1016/S1470-2045(09)70034-8. [DOI] [PubMed] [Google Scholar]
- 2.Baselga J. Targeting tyrosine kinases in cancer: The second wave. Science. 2006;312:1175–1178. doi: 10.1126/science.1125951. [DOI] [PubMed] [Google Scholar]
- 3.Kawamoto T, et al. Growth stimulation of A431 cells by epidermal growth factor: Identification of high-affinity receptors for epidermal growth factor by an anti-receptor monoclonal antibody. Proc Natl Acad Sci USA. 1983;80:1337–1341. doi: 10.1073/pnas.80.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 5.Mellinghoff IK, et al. Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N Engl J Med. 2005;353:2012–2024. doi: 10.1056/NEJMoa051918. [DOI] [PubMed] [Google Scholar]
- 6.Paez JG, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 7.Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- 8.Scott AM, et al. A phase I clinical trial with monoclonal antibody ch806 targeting transitional state and mutant epidermal growth factor receptors. Proc Natl Acad Sci USA. 2007;104:4071–4076. doi: 10.1073/pnas.0611693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karapetis CS, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 10.Mendelsohn J, Baselga J. Epidermal growth factor receptor targeting in cancer. Semin Oncol. 2006;33:369–385. doi: 10.1053/j.seminoncol.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Dassonville O, Bozec A, Fischel JL, Milano G. EGFR targeting therapies: Monoclonal antibodies versus tyrosine kinase inhibitors. Similarities and differences. Crit Rev Oncol Hematol. 2007;62:53–61. doi: 10.1016/j.critrevonc.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Klein DE, Nappi VM, Reeves GT, Shvartsman SY, Lemmon MA. Argos inhibits epidermal growth factor receptor signalling by ligand sequestration. Nature. 2004;430:1040–1044. doi: 10.1038/nature02840. [DOI] [PubMed] [Google Scholar]
- 13.Klein DE, Stayrook SE, Shi F, Narayan K, Lemmon MA. Structural basis for EGFR ligand sequestration by Argos. Nature. 2008;453:1271–1275. doi: 10.1038/nature06978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmore JL, Riese DJ,, 2nd secErbB4-26/549 antagonizes ligand-induced ErbB4 tyrosine phosphorylation. Oncol Res. 2004;14:589–602. doi: 10.3727/0965040042707907. [DOI] [PubMed] [Google Scholar]
- 15.Holash J, et al. VEGF-Trap: A VEGF blocker with potent antitumor effects. Proc Natl Acad Sci USA. 2002;99:11393–11398. doi: 10.1073/pnas.172398299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim ES, et al. Potent VEGF blockade causes regression of coopted vessels in a model of neuroblastoma. Proc Natl Acad Sci USA. 2002;99:11399–11404. doi: 10.1073/pnas.172398399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meister A. Biochemistry of the Amino Acids. 2nd Ed. Vol. 1. New York: Academic Press; 1965. p. 364. [Google Scholar]
- 18.Mintz PJ, et al. Fingerprinting the circulating repertoire of antibodies from cancer patients. Nat Biotechnol. 2003;21:57–63. doi: 10.1038/nbt774. [DOI] [PubMed] [Google Scholar]
- 19.Vidal CI, et al. An HSP90-mimic peptide revealed by fingerprinting the pool of antibodies from ovarian cancer patients. Oncogene. 2004;23:8859–8867. doi: 10.1038/sj.onc.1208082. [DOI] [PubMed] [Google Scholar]
- 20.Arap MA, et al. Cell surface expression of the stress response chaperone GRP78 enables tumor targeting by circulating ligands. Cancer Cell. 2004;6:275–284. doi: 10.1016/j.ccr.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 21.Binder M, Otto F, Mertelsmann R, Veelken H, Trepel M. The epitope recognized by rituximab. Blood. 2006;108:1975–1978. doi: 10.1182/blood-2006-04-014639. [DOI] [PubMed] [Google Scholar]
- 22.Binder M, et al. Identification of their epitope reveals the structural basis for the mechanism of action of the immunosuppressive antibodies basiliximab and daclizumab. Cancer Res. 2007;67:3518–3523. doi: 10.1158/0008-5472.CAN-06-3919. [DOI] [PubMed] [Google Scholar]
- 23.Riemer AB, et al. Vaccination with cetuximab mimotopes and biological properties of induced anti-epidermal growth factor receptor antibodies. J Natl Cancer Inst. 2005;97:1663–1670. doi: 10.1093/jnci/dji373. [DOI] [PubMed] [Google Scholar]
- 24.Jaalouk DE, et al. The original Pathologische Anatomie Leiden-Endothelium monoclonal antibody recognizes a vascular endothelial growth factor binding site within neuropilin-1. Cancer Res. 2007;67:9623–9629. doi: 10.1158/0008-5472.CAN-07-2737. [DOI] [PubMed] [Google Scholar]
- 25.Ogiso H, et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell. 2002;110:775–787. doi: 10.1016/s0092-8674(02)00963-7. [DOI] [PubMed] [Google Scholar]
- 26.Dawson JP, et al. Epidermal growth factor receptor dimerization and activation require ligand-induced conformational changes in the dimer interface. Mol Cell Biol. 2005;25:7734–7742. doi: 10.1128/MCB.25.17.7734-7742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dawson JP, Bu Z, Lemmon MA. Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure. 2007;15:942–954. doi: 10.1016/j.str.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Garrett TP, et al. Crystal structure of a truncated epidermal growth factor receptor extracellular domain bound to transforming growth factor alpha. Cell. 2002;110:763–773. doi: 10.1016/s0092-8674(02)00940-6. [DOI] [PubMed] [Google Scholar]
- 29.Jonker DJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 30.Bonner JA, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 31.Posner MR, Wirth LJ. Cetuximab and radiotherapy for head and neck cancer. N Engl J Med. 2006;354:634–636. doi: 10.1056/NEJMe058306. [DOI] [PubMed] [Google Scholar]
- 32.Golfinopoulos V, Salanti G, Pavlidis N, Ioannidis JP. Survival and disease-progression benefits with treatment regimens for advanced colorectal cancer: A meta-analysis. Lancet Oncol. 2007;8:898–911. doi: 10.1016/S1470-2045(07)70281-4. [DOI] [PubMed] [Google Scholar]
- 33.Chorev M, Goodman M. A dozen years of retro-inverso peptidomimetics. Acc Chem Res. 1993;26:266–273. [Google Scholar]
- 34.Hajitou A, et al. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 35.Li S, et al. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 36.Hubbard SR. EGF receptor inhibition: Attacks on multiple fronts. Cancer Cell. 2005;7:287–288. doi: 10.1016/j.ccr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 37.Stevens FJ, Chang CH, Schiffer M. Dual conformations of an immunoglobulin light-chain dimer: Heterogeneity of antigen specificity and idiotope profile may result from multiple variable-domain interaction mechanisms. Proc Natl Acad Sci USA. 1988;85:6895–6899. doi: 10.1073/pnas.85.18.6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rini JM, Schulze-Gahmen U, Wilson IA. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992;255:959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- 39.James LC, Roversi P, Tawfik DS. Antibody multispecificity mediated by conformational diversity. Science. 2003;299:1362–1367. doi: 10.1126/science.1079731. [DOI] [PubMed] [Google Scholar]
- 40.Dlugosz AA, et al. Autocrine transforming growth factor alpha is dispensible for v-rasHa-induced epidermal neoplasia: Potential involvement of alternate epidermal growth factor receptor ligands. Cancer Res. 1995;55:1883–1893. [PubMed] [Google Scholar]
- 41.Baba I, et al. Involvement of deregulated epiregulin expression in tumorigenesis in vivo through activated Ki-Ras signaling pathway in human colon cancer cells. Cancer Res. 2000;60:6886–6889. [PubMed] [Google Scholar]
- 42.Sweet-Cordero A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 43.Fujimoto N, et al. High expression of ErbB family members and their ligands in lung adenocarcinomas that are sensitive to inhibition of epidermal growth factor receptor. Cancer Res. 2005;65:11478–11485. doi: 10.1158/0008-5472.CAN-05-1977. [DOI] [PubMed] [Google Scholar]
- 44.Goldstein NI, Prewett M, Zuklys K, Rockwell P, Mendelsohn J. Biological efficacy of a chimeric antibody to the epidermal growth factor receptor in a human tumor xenograft model. Clin Cancer Res. 1995;1:1311–1318. [PubMed] [Google Scholar]
- 45.Cardó-Vila M, Arap W, Pasqualini R. Alpha v beta 5 integrin-dependent programmed cell death triggered by a peptide mimic of annexin V. Mol Cell. 2003;11:1151–1162. doi: 10.1016/s1097-2765(03)00138-2. [DOI] [PubMed] [Google Scholar]
- 46.Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. pp. 74–82. [Google Scholar]
- 47.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 48.Ozawa MG, et al. Angiogenesis with pericyte abnormalities in a transgenic model of prostate carcinoma. Cancer. 2005;104:2104–2115. doi: 10.1002/cncr.21436. [DOI] [PubMed] [Google Scholar]
- 49.Mandava S, Makowski L, Devarapalli S, Uzubell J, Rodi DJ. RELIC—a bioinformatics server for combinatorial peptide analysis and identification of protein-ligand interaction sites. Proteomics. 2004;4:1439–1460. doi: 10.1002/pmic.200300680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.