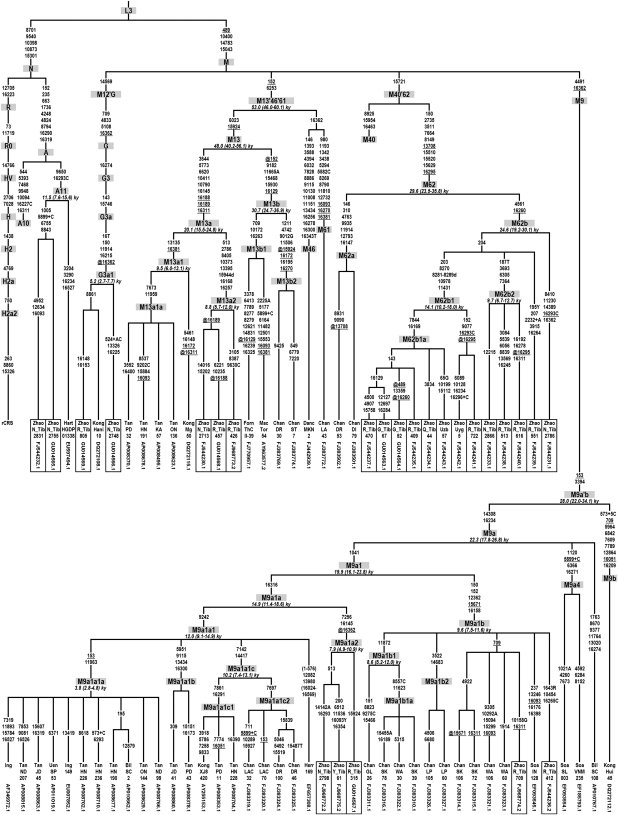
Detailed knowledge about the human mitochondrial (mt)DNA phylogeny (1) is accumulating rapidly because of the growing number of available complete mtDNA sequences from diverse populations across the world. The work by Zhao et al. (2) makes a valuable contribution to this knowledge by describing 25 complete mtDNA sequences from Tibetan and neighboring populations. However, the phylogenetic tree shown in their figure 3 presents several conflicts regarding haplogroup nomenclature, as detailed below. As a solution, I hereby provide an updated mtDNA phylogeny (Fig. 1) with revised haplogroup nomenclature containing the 25 sequences by Zhao et al. plus an additional 53 sequences that are relevant for the tree topology. Starting from the left of Fig. 1, four changes are proposed.
Fig. 1.
Revised mtDNA phylogeny with newly proposed haplogroup nomenclature based on 78 complete mtDNA sequences. The 25 sequences in boxes are from Zhao et al. (2). Mutations are transitions unless an exact base change is specified; R or Y indicate heteroplasmic states. Insertions and deletions are indicated with + and d, respectively. C insertions at positions 309 and 315, deletions of an AC dinucleotide at positions 523–524, and transitions at position 16519 were omitted. Homoplasic mutations are underlined only when they are recurrent within one of the clades A, M12’G, M13’46’61, M40’62, or M9. In the case that they are back mutations of an ancestral mutation, they are preceded by @. Age estimates for haplogroups (± SE intervals) were calculated using entire genome substitutions according to ref. 4. Sample names refer to the sources Bilal et al. (6), Chandrasekar et al. (5), Dancause et al. (7), Fornarino et al. (8), Hartmann et al. (3), Herrnstadt et al. (9), Ingman et al. (10), Kong et al. (11), Macaulay et al. (12), Soares et al. (13), Tanaka et al. (14), Ueno et al. (15), and Zhao et al. (2), and they can be looked up in GenBank through their respective accession numbers.
First, in Build 7 (Nov. 10, 2009) of our mtDNA classification tree (1), the label A10 was used for a different haplogroup than the one that is named A10 by Zhao et al. (2). Therefore, I propose to relabel Zhao et al.’s A10 to A11. Moreover, the defining mutations of haplogroup A11 can be refined to 9650–16293C by considering the sequence with accession number EU597494 (3), which was probably overlooked by Zhao et al.
Second, there is an additional bifurcation within haplogroup M13 as noted previously (4), so that Zhao et al.’s M13, M13a, and M13b must be relabeled M13a, M13a1, and M13a2, respectively.
Third, the M16 haplogroup that is referred by Zhao et al. was recently also found by others who labeled it M62 (5), and it shares a basal mutation with haplogroup M40. Acknowledging first publication date, I propose to follow the M62 label. Consequently, Zhao et al.’s M16, M16a, and M16b are now relabeled M62b, M62b1, and M62b2, respectively.
Fourth, the same haplogroup that is called M9c by Zhao et al. was recently also found by others (5), who called it M9d. In addition, the same study (5) used the label M9a2 to designate a different branch than the one labeled M9a2 in Zhao et al. To end the ongoing confusion regarding the nomenclature of haplogroup M9 sublineages, I propose a structurally more logical nomenclature for this haplogroup avoiding the labels M9c, M9d, and M9a2. As a consequence, Zhao et al.’s M9a, M9a1, M9a2, and M9c are now relabeled M9a1a, M9a1a1, M9a1a2, and M9a1b, respectively.
I believe that these changes are necessary to reach universally accepted mtDNA haplogroup nomenclature, which is essential to the comparison of different mtDNA studies. The revisions will also be shown in the next build of the mtDNA tree available at http://www.phylotree.org (1).
Footnotes
The author declares no conflict of interest.
References
- 1.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30:E386–E394. doi: 10.1002/humu.20921. [DOI] [PubMed] [Google Scholar]
- 2.Zhao M, et al. Mitochondrial genome evidence reveals successful Late Paleolithic settlement on the Tibetan Plateau. Proc Natl Acad Sci USA. 2009;106:21230–21235. doi: 10.1073/pnas.0907844106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmann A, et al. Validation of microarray-based resequencing of 93 worldwide mitochondrial genomes. Hum Mutat. 2009;30:115–122. doi: 10.1002/humu.20816. [DOI] [PubMed] [Google Scholar]
- 4.Soares P, et al. Correcting for purifying selection: An improved human mitochondrial molecular clock. Am J Hum Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chandrasekar A, et al. Updating phylogeny of mitochondrial DNA macrohaplogroup m in India: Dispersal of modern human in South Asian corridor. PLoS One. 2009;4:e7447. doi: 10.1371/journal.pone.0007447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilal E, et al. Mitochondrial DNA haplogroup D4a is a marker for extreme longevity in Japan. PLoS One. 2008;3:e2421. doi: 10.1371/journal.pone.0002421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dancause KN, Chan CW, Arunotai NH, Lum JK. Origins of the Moken Sea Gypsies inferred from mitochondrial hypervariable region and whole genome sequences. J Hum Genet. 2009;54:86–93. doi: 10.1038/jhg.2008.12. [DOI] [PubMed] [Google Scholar]
- 8.Fornarino S, et al. Mitochondrial and Y-chromosome diversity of the Tharus (Nepal): a reservoir of genetic variation. BMC Evol Biol. 2009;9:154. doi: 10.1186/1471-2148-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrnstadt C, et al. Reduced-median-network analysis of complete mitochondrial DNA coding-region sequences for the major African, Asian, and European haplogroups. Am J Hum Genet. 2002;70:1152–1171. doi: 10.1086/339933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 11.Ingman M, Gyllensten U. Rate variation between mitochondrial domains and adaptive evolution in humans. Hum Mol Genet. 2007;16:2281–2287. doi: 10.1093/hmg/ddm180. [DOI] [PubMed] [Google Scholar]
- 12.Kong QP, et al. Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences. Am J Hum Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kong QP, et al. Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum Mol Genet. 2006;15:2076–2086. doi: 10.1093/hmg/ddl130. [DOI] [PubMed] [Google Scholar]
- 14.Macaulay V, et al. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- 15.Soares P, et al. Climate change and postglacial human dispersals in Southeast Asia. Mol Biol Evol. 2008;25:1209–1218. doi: 10.1093/molbev/msn068. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka M, et al. Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genome Res. 2004;14:1832–1850. doi: 10.1101/gr.2286304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ueno H, et al. Analysis of mitochondrial DNA variants in Japanese patients with schizophrenia. Mitochondrion. 2009;9:385–393. doi: 10.1016/j.mito.2009.06.003. [DOI] [PubMed] [Google Scholar]