Abstract
The gene SCN9A is responsible for three human pain disorders. Nonsense mutations cause a complete absence of pain, whereas activating mutations cause severe episodic pain in paroxysmal extreme pain disorder and primary erythermalgia. This led us to investigate whether single nucleotide polymorphisms (SNPs) in SCN9A were associated with differing pain perception in the general population. We first genotyped 27 SCN9A SNPs in 578 individuals with a radiographic diagnosis of osteoarthritis and a pain score assessment. A significant association was found between pain score and SNP rs6746030; the rarer A allele was associated with increased pain scores compared to the commoner G allele (P = 0.016). This SNP was then further genotyped in 195 pain-assessed people with sciatica, 100 amputees with phantom pain, 179 individuals after lumbar discectomy, and 205 individuals with pancreatitis. The combined P value for increased A allele pain was 0.0001 in the five cohorts tested (1277 people in total). The two alleles of the SNP rs6746030 alter the coding sequence of the sodium channel Nav1.7. Each was separately transfected into HEK293 cells and electrophysiologically assessed by patch-clamping. The two alleles showed a difference in the voltage-dependent slow inactivation (P = 0.042) where the A allele would be predicted to increase Nav1.7 activity. Finally, we genotyped 186 healthy females characterized by their responses to a diverse set of noxious stimuli. The A allele of rs6746030 was associated with an altered pain threshold and the effect mediated through C-fiber activation. We conclude that individuals experience differing amounts of pain, per nociceptive stimulus, on the basis of their SCN9A rs6746030 genotype.
Keywords: Nav1.7, nociception, pain, SCN9A, single nucleotide polymorphisms
The ability to sense noxious events (nociception) is of profound importance to complex organisms, as shown by its presence in animals as diverse as snails, reptiles, amphibians, and mammals (1–4). As a sense, pain serves as an adaptive mechanism that protects us from tissue damage by alerting us to events that are capable of producing injury and evokes behaviors that promote tissue healing. However, the development of maladaptive persistent pain states in response to tissue injury is common, with one in six adults suffering from a chronic pain condition (5). The search for effective analgesics with acceptable side effects has long been the goal of doctors and biomedical researchers. Because current therapies have limited efficacy, with up to 50% of treated subjects receiving inadequate pain relief (6), there exists a significant need to develop better therapies.
Our understanding of the neurobiology of pain pathways has accelerated substantially since the mid-20th century (7). Within the last decade the molecular biology of membrane receptors and channels that respond to pain-provoking stimuli have been identified and characterized, e.g., the TRPV1 heat receptor (which is also activated by capsaicin, the active ingredient in hot chilli peppers) (8). More recent findings have demonstrated the unexpected importance of other molecules in pain appreciation in humans, including catechol-O-methyltransferase (COMT) and tetrahydrobiopterin (9, 10), which modulate nociceptive and inflammatory pain, and SCN9A (11–17), mutations of which can result in syndromes of either excessive pain or insensitivity to pain.
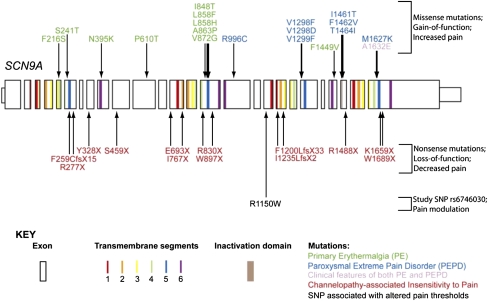
SCN9A encodes the α-subunit of the voltage-gated sodium channel Nav1.7, which is expressed at high density in nociceptive neurons (18). A number of clinical effects of Nav1.7 mutations have been reported in recent years (Fig. 1) (11–13, 15). Primary erythermalgia (OMIM 133020) and paroxysmal extreme pain disorder (OMIM 167400) are distinct syndromes associated with abnormal attacks of severe pain (19, 20), and are caused by SCN9A mutations that increase Nav1.7 channel activity (20). Nonsense mutations in SCN9A lead to a lifelong inability to sense pain in the autosomal recessive condition channelopathy-associated insensitivity to pain (OMIM 243000) (11, 21). With the exception of having never felt pain and being anosmic, affected individuals are completely normal, suggesting that Nav1.7 is a vital component in the perception of pain in humans and that functions of the protein associated with its expression outside the nociceptive system are redundant. The data raise the possibility that almost complete analgesia may be possible by targeting this ion channel in humans.
Fig. 1.
Schematic of the SCN9A gene showing the position of known mutations (arrows) that result in human disorders of pain appreciation. SCN9A encodes the voltage-gated sodium channel, Nav1.7, which is composed of four repeating domains, each comprising six transmembrane segments. The regions of the gene that encode the transmembrane segments and the inactivation domain are indicated (see colored key).
Although subjects with major pain phenotypes are very rare, a role for more common polymorphisms in determining interindividual pain perception is beginning to emerge (22). Within this context there are multiple single nucleotide polymorphisms (SNPs) in the human SCN9A gene, raising the possibility that allelic differences in this gene might influence nociception in the general population. In this study we have investigated the association between SCN9A SNPs and pain perception in five cohorts of subjects with a range of pathological conditions for whom we had individual measures of reported pain and corresponding DNA samples for genotyping. The study was performed to establish whether common polymorphisms in SCN9A have an effect on nociception, and if so, what might be the pathophysiological basis as assessed by measurements both of in vitro Nav1.7 channel function and of pain perception in normal subjects.
Results
Association Analysis of Nav1.7 Variants with Pain Cohorts.
In an initial cohort of 578 subjects with osteoarthritis (OA) (see also SI Subjects and Methods and Fig. S1), we screened 27 SNPs in SCN9A and found that 5 showed a significant association with the pain score in a linear regression model that included adjustments for age, gender, body mass index (BMI), and age–gender interaction (Table 1): rs6432896, P = 0.048; rs7604448, P = 0.036; rs10930214, P = 0.027; rs6746030, P = 0.016; and rs7595255, P = 0.02. The corresponding allele frequencies and the unadjusted mean pain scores for all SNPs divided according to genotype are shown in Tables S1 and S2. Examination of the pair-wise linkage disequilibrium data suggests that of the five SNPs that exhibited a significant association with pain, the last four were in linkage disequilibrium. The magnitude of the effect on the pain score ranged from 0.44 to 0.76/rare allele [on the Western Ontario and McMaster Osteoarthritis Index (WOMAC) scale of 0–20] with the largest effect and lowest P value being observed with the amino acid changing SNP rs6746030.
Table 1.
Linear regression analysis for pain score (trend effect per rare allele) in osteoarthritis cohort
| SNP | Mean effect size | Lower 95% confidence interval | Upper 95% confidence interval | P value |
| rs4447616 | −0.32 | −0.77 | 0.13 | 0.1598 |
| rs10171225 | 0.28 | −0.26 | 0.81 | 0.3094 |
| rs4286289 | −0.08 | −0.56 | 0.41 | 0.7595 |
| rs4605385 | 0.09 | −0.48 | 0.67 | 0.7464 |
| rs6432896 | 0.44 | 0.00 | 0.88 | 0.0476 |
| rs12994338 | 0.08 | −0.42 | 0.58 | 0.7546 |
| rs13017637 | −0.18 | −0.63 | 0.27 | 0.4375 |
| rs12620053 | 0.10 | −0.35 | 0.55 | 0.6623 |
| rs12619987 | −0.02 | −0.69 | 0.64 | 0.9422 |
| rs11688164 | −0.05 | −0.67 | 0.58 | 0.8845 |
| rs13402540 | 0.18 | −0.27 | 0.62 | 0.4323 |
| rs6747673 | −0.03 | −0.46 | 0.40 | 0.8822 |
| rs6432894 | −0.05 | −0.51 | 0.41 | 0.8335 |
| rs4453709 | −0.03 | −0.47 | 0.41 | 0.8918 |
| rs4443014 | 0.34 | −0.18 | 0.85 | 0.2019 |
| rs4561679 | 0.60 | −0.06 | 1.27 | 0.0754 |
| rs4371369 | 0.33 | −0.11 | 0.77 | 0.1431 |
| rs7604448 | 0.63 | 0.04 | 1.21 | 0.0361 |
| rs10930214 | 0.52 | 0.06 | 0.99 | 0.0266 |
| rs6746030 | 0.76 | 0.14 | 1.38 | 0.0158 |
| rs12621853 | 0.08 | −0.51 | 0.68 | 0.7853 |
| rs17748381 | −0.36 | −1.08 | 0.36 | 0.3226 |
| rs10170041 | 0.30 | −0.14 | 0.74 | 0.1791 |
| rs16851799 | −0.02 | −0.46 | 0.43 | 0.9457 |
| rs7595255 | 0.73 | 0.11 | 1.34 | 0.0210 |
| rs6432885 | −0.25 | −0.68 | 0.18 | 0.2529 |
| rs3750904 | 1.44 | −1.84 | 4.72 | 0.3888 |
Significant results are shown in boldface. “Mean effect size” is the mean difference in pain score per additional rare allele, e.g., AA versus Aa and Aa versus aa calculated from the regression analysis. “P value” is based on a linear regression analysis for pain score trend effect per rare allele adjusting for age, gender, body mass index, and age/gender interaction.
Four of the SNPs that were significantly associated with the pain score are located in introns and are not changes that commonly affect mRNA stability or splicing, nor do they form part of conserved motifs. SNP rs6746030, however, is located in exon 18 and affects the amino acid at position 1150 of Nav1.7, with the more frequent allele encoding an arginine (NaV1.7–1150R) and the less frequent encoding a tryptophan (NaV1.7–1150W). Because of its strongest effect in the regression analysis and the predicted amino acid change, SNP rs6746030 was selected for investigation in additional pain cohorts.
Analysis of a Finnish cohort, comprising 195 individuals with sciatica, showed a significant association of rs6746030 with a Visual Analog Pain Score (P = 0.039) under a recessive genetic model. In a group of 100 Danish amputees, using the same recessive model, rs6746030 was found to be significantly associated with phantom pain experience (P = 0.011). In both cohorts, as in the individuals with osteoarthritis, the minor A allele was associated with a greater experience of pain. Furthermore, in a cohort of 179 individuals with lumbar root pain the pain score also tended to increase with the number of minor A alleles at SNP rs6746030 (Table S3 and SI Results). The mean adjusted pain scores for the different genotypes were 0.47 (G/G), 0.65 (G/A), and 0.97 (A/A) on a scale with values ranging between −1.3 and 2.7, giving a p value for an additive model analysis of 0.088.
We also investigated a cohort of patients with pancreatitis pain, but found no difference in the distribution of rs6746030 alleles either between patients and controls or between patients who had or had not needed surgery to control pain (SI Results and Table S4). For those patients experiencing ongoing pain, however, both the mean pain score and the composite pain score were higher in subjects with the minor A allele than in those without the A allele, although these differences were not statistically significant.
We noted that in each individual study the trend was for the minor A allele to be associated with a greater experience of pain. We therefore generated a combined P value of all five cohorts, using a modification of the inverse normal method (23). The combined P value for the five cohorts was 0.0001 (Table 2), indicating that across a variety of study conditions, the degree of pain experienced was strongly dependent on an individual's rs6746030 genotype.
Table 2.
Results for the five clinical cohorts showing the effect of the minor A allele on pain sensitivity
| Cohort | No. of affected individuals | rs6746030 allele for which most pain was felt | P value for A allele association |
| 1. Osteoarthritis | 578 | A | 0.016* |
| 2. Sciatica | 195 | A | 0.039* |
| 3. Postamputation | 100 | A | 0.011* |
| 4. Postdiscectomy | 179 | A | 0.088 |
| 5. Chronic pancreatitis | 205 | A = G | 0.732 |
| 6. Meta-analysis | 1257 | A | 0.0001* |
*Cohort results reached statistical significance.
Functional Analysis of Nav1.7 Variants.
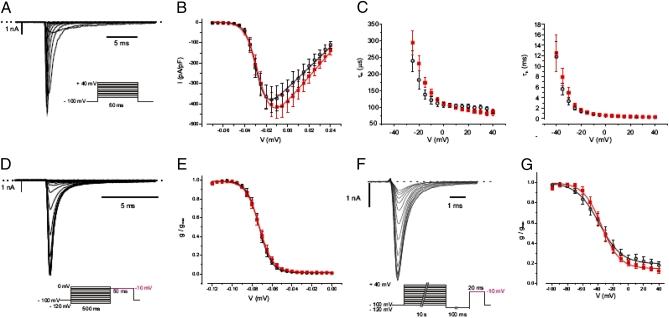
To investigate the functional effects of the rs6746030 SNPs, we compared the electrophysiological properties of Nav1.7 channels containing the two different amino acid variants at position 1150. NaV1.7–1150R and NaV1.7–1150W were coexpressed with the β1 and β2 subunits of voltage-gated sodium channels in HEK293 cells, and the properties of the expressed channels were investigated in whole-cell patch-clamp recordings by using a range of protocols to evaluate different parameters of channel activation and inactivation (Fig. 2 and SI Subjects and Methods). No differences between the variants were observed in the peak current amplitude or the voltage dependence and time constants of activation and fast inactivation. However, inactivation of voltage-gated sodium channels involves at least two inactivated states, the slower of which is evident only after prolonged depolarizations. To analyze slow inactivation, we measured the current amplitude after a 10-s depolarization to different voltages and a subsequent short hyperpolarization step to recover only the fast inactivating states. Slow inactivation occurred at more positive potentials than fast inactivation and reached only∼80%. Although the extent of slow inactivation was similar for both channel variants, we observed a significantly steeper voltage dependence of NaV1.7–1150W compared with NaV1.7–1150R currents (k = 10.7 ± 0.4 mV and 13.7 ± 1.2 mV, respectively; n = 7 each; P = 0.042; Fig. 2G).
Fig. 2.
Biophysical properties of NaV1.7–1150R and NaV1.7–1150W currents. (A) Current responses to 50-ms voltage steps of 5-mV increments between −70 and +40 mV from a holding potential of −100 mV in a whole-cell voltage clamp recording applied at 0.5 Hz for a HEK293 cell expressing Nav1.7–1150W. (Inset) The voltage pulse protocol. (B) Current–voltage relationship of the peak currents normalized for cell size [picoamperes per picofarad (pA/pF)] obtained using the experimental setup shown in A. Lines represent fits of the data with a Boltzmann equation: ■, NaV1.7–1150W [V0.5 = −26.0 ± 2 mV, k = 4.8 ± 0.4 mV, Vrev = 64 ± 2 mV (n = 17)]; ○, Nav1.7–1150R [V0.5 = −29.0 ± 1 mV, k = 4.4 ± 0.3 mV, Vrev = 62 ± 2 mV (n = 10)]. (C) Voltage dependence of the activation time constant τm (left) and inactivation time constant τh (right), obtained by fitting current traces elicited by depolarizing pulses from −40 to +40 mV of 50 ms duration from a holding potential of −100 mV (as shown in A) with a Hodgkin–Huxley m3h model (for voltage gated sodium channels). ■, NaV1.7–1150W, O, Nav1.7–1150R. (D) Voltage dependence of the steady-state inactivation of NaV1.7–1150W currents was measured by holding the membrane potential for 500 ms at conditioning voltages from −120–0 mV (at 5-mV increments) before stepping to a test pulse at −10 mV for 50 ms. (Inset) The voltage pulse protocol, which was applied at 0.5 Hz. Only the current responses to the test pulse are shown. (E) Peak currents obtained as in D were normalized to the maximum peak current and plotted against the holding potential applied during the conditioning pulse. Lines represent a fit of the data with a Boltzmann equation: ■, NaV1.7–1150W [V0.5 = −73.0 ± 2 mV, k = 5.5 ± 0.2 mV (n = 17)]; O, Nav1.7–1150R [V0.5 = −73.0 ± 2 mV, k = 5.4 ± 0.2 mV (n = 10)]. (F) Voltage dependence of the slow steady-state inactivation of NaV1.7–1150W currents was measured by holding the membrane potential for 10 s at conditioning voltages from −120 mV to 0 mV (at 5-mV increments) before stepping up for 100 ms to −120 mV to recover channels in the fast inactivated state followed by a test pulse at −10 mV for 20 ms. (Inset) The voltage pulse protocol. Only the current responses to the test pulse are shown. (G) Peak currents obtained as in f were normalized to the maximum peak current and plotted against the holding potential applied during the conditioning pulse. Lines represent a fit of the data with a Boltzmann equation: ■, NaV1.7–1150W [V0.5 = −35 ± 3 mV, k = 11 ± 1 mV, A2 = 15 ± 2% (n = 7)]; O, Nav1.7–1150R [V0.5 = -37 ± 5 mV, k = 14 ± 1 mV; A2 = 18 ± 3% (n = 7)].
Is SNP rs6746030 Associated with Pain Thresholds in Normal Individuals?
To investigate the contribution of specific nociceptive pathways that might mediate the effect of the minor A allele of SCN9A, we analyzed data from a cohort of 186 healthy European–American females who were free of pain at the time of examination and who were phenotyped with a diverse set of experimental noxious stimuli (9). Minor A allele carriers showed a trend to be more sensitive to all types of experimental stimuli, although only those procedures that evoked predominately C-fiber–mediated heat pain reached statistical significance (Figs. S2 and S3 and Table S5).
Discussion
Our data show that a common polymorphism in the SCN9A gene is strongly associated (P = 0.0001) with pain perception in a range of pathological conditions. Of five SCN9A SNPs found initially to be significantly associated with the level of pain experienced from osteoarthritis, only rs6746030 results in a coding change whereas the other four are not predicted to be detrimental. The minor allele of rs6746030 occurs at a frequency of ∼10%, is associated with increased pain perception, and results in an amino acid change from arginine to tryptophan at position 1150 of Nav1.7. This residue lies in the intracellular loop between transmembrane domains II and III of Nav1.7, a region of the channel with unknown function. However, 1150R is an evolutionarily conserved amino acid (Fig. S4), suggesting that it could contribute to the normal function of Nav1.7.
The WOMAC self-administered questionnaire specifically documents pain relevant to osteoarthritis and is validated for determining pain levels, but relies on a subjective assessment by the patient. All such measures of pain severity are intrinsically imprecise because they necessarily rely on subjective reporting using pain scales such as the WOMAC questionnaire or the Visual Analog Pain Scale. The consequent variability in quantifying pain is a major contributor to reducing the power of this type of study. Nevertheless, the observed difference in pain intensity felt by homozygous wild-type subjects and carriers of the genetic variants is ≈0.8 WOMAC points per rare allele (on the WOMAC pain scale of 0–20). This compares with a mean improvement of opiates over placebo in OA pain studies of ≈1 point (on an 11-point scale) (24). The results suggested that carriers of the rs6746030 A allele in the general population would be expected to experience more pain for a given stimulus than their fellows.
Further evidence that the minor A allele of SCN9A SNP rs6746030 influences the degree of pain experienced comes from the studies in people with sciatica, amputees with phantom limb pain, and patients with postdiscectomy lumbar root pain (same trend but not reaching significance). We found no association between pain experienced in pancreatitis and SNP rs6746030. However, when the combined P value was calculated across all studies, a p value of 0.0001 was produced, confirming a significant contribution of the rs6746030 A allele to human pain experience. Furthermore, when pain responses to a diverse set of experimental procedures were examined, the association of the rs6746030 a allele was found to be most strongly associated with C-fiber activation. This result suggests that C-fiber activation, which underlies the sensation of diffuse, dull, aching pain represents the nociceptive pathway mediating A-allele–driven clinical pain (22). Both clinical and experimental pain studies support an additive model for the effect of rs6746030 alleles, with most pain being felt by carriers of the AA genotype and the least pain with the GG genotype.
During detailed electrophysiological assessment of Nav1.7–1150R and 1150W currents, we observed a statistically significant difference in the steepness of the voltage dependence of slow inactivation. The steeper slope of Nav1.7–1150W currents (encoded by the minor A allele) would tend to make these channels less susceptible to slow inactivation at potentials negative to −35 mV. Because slow inactivation of Nav1.7 is believed to play a role in setting the threshold and firing frequency of nociceptive neurons, the lesser slow inactivation of the rare A allelic variant at voltages close to the resting potential would tend to make the neurons more sensitive to painful stimuli (25). However, the observed biophysical effect is rather small and might be insufficient to explain the associated phenotype, because alteration of the slow inactivation of the Nav1.7 conductance alone in the absence of altered voltage-dependent activation failed to change the firing properties in a computer model of nociceptive neurons (26). It is important to note that another recent study failed to observe a change in voltage dependence of slow inactivation, but instead reported a small depolarizing shift in the voltage dependence of activation for Nav1.7–1150W compared to Nav1.7–1150R currents (27). By contrast, most Nav1.7 mutations associated with hyperexcitability and erythromelalgia have been reported to shift the voltage dependence of activation in the opposite, hyperpolarizing direction (26, 28). Nevertheless, Nav1.7–1150W produced hyperexcitability when expressed in small dorsal root ganglion neurons (27), a finding that is consistent with the clinical results presented here. The exact mechanism by which the minor A allele enhances pain sensitivity therefore remains unclear and might involve factors not present in the heterologous expression system, factors that affect expression, processing, or interaction of the Nav1.7 protein in nociceptive neurons.
The work presented here provides additional evidence that human pain experience is under genetic influence. Although small association studies have limitations, and each of these studies will need replication in independent cohorts, the trend for carriers of the minor A allele to experience more pain in a variety of nociceptive situations is striking and supported by a strong combined P value of 0.0001. Pain experienced in common conditions such as osteoarthritis and back pain is influenced by disease severity, pain sensitivity, and responses to analgesics. The contribution of each common polymorphic genetic variant with common pain conditions is expected to be relatively modest, unlike the major effects observed in Mendelian familial diseases such as channelopathy-associated insensitivity to pain (22).
Our results suggest that, like COMT and GCH1, SCN9A is a substantial contributor to human pain sensitivity and clinical pain conditions (9). Activating mutations in SCN9A lead to severe episodic pain, inactivating mutations cause a complete absence of pain, and we now can show that SNP rs6746030 quantitatively alters pain sensitivity and pain experienced in common conditions such as osteoarthritis and spinal nerve root pain. The findings support the idea that pharmacological substances that modify the function of SCN9A (Nav1.7) would be potentially useful in treating these conditions. As individuals appear to have differing genetic susceptibilities to pain, future studies should be directed toward understanding whether responsiveness to different classes of analgesics is also genetically determined.
Subjects and Methods
Study Subjects.
For further descriptions of the study subjects, see SI Subjects and Methods. Relevant ethical approval was obtained for the inclusion of all individuals in each study cohort.
Cohort 1: Osteoarthritis.
Clinical information and DNA samples were available on 578 individuals from four OA clinical trials (see SI Subjects and Methods) sponsored by Pfizer. Individuals had radiographically confirmed osteoarthritis of either knee or hip, and all were self-reported as Caucasian. Participants had a WOMAC pain score—a visual analog scale that has been used extensively in patients with osteoarthritis of the hip or knee to judge discomfort and gauge eligibility for surgery.
Cohort 2: Sciatica pain.
A total of 195 Finnish patients with sciatica symptoms were recruited from Oulu University Hospital in an extension of a previous study (29). Inclusion criteria included unilateral pain radiating from the back to below the knee. All patients were confirmed to have a lumbar disk herniation concordant with the symptoms by MRI. A rating on the Visual Analog Pain Scale at baseline was used as the primary outcome for the analysis.
Cohort 3: Phantom pain.
A total of 100 Danish amputees with upper and/or lower limb amputations were studied; all were Caucasian. The primary phenotype was the experience of phantom pain, graded as (1) never, (2) previously, or (3) currently with further subdivision of those who experienced phantom pain within the last month according to the frequency, duration, and intensity of pain episodes.
Cohort 4: Lumbar root pain.
A total of 179 Caucasians from the Maine Lumbar Spine Study were assessed for pain on a seven-point score one year after lumbar discectomy surgery (30). The primary endpoint—persistent leg pain over the first year—was determined by an area-under-the-curve score calculated from four questions about leg pain collected at 3, 6, and 12 months.
Cohort 5: Chronic pancreatitis.
A total of 205 Caucasians with chronic pancreatitis were compared with a control group of 168 healthy adults. Chronic pancreatitis was diagnosed according to established criteria (31). Pain was measured by (1) a modified version of the Gastrointestinal Symptoms Questionnaire and (2) a validated composite pain score that also included the pain pattern, pain perception during the last attack, usage of morphine-like analgesics, and whether surgery for pancreatic pain had ever been necessary.
Cohort 6: Pain-free females.
A total of 186 healthy European–American pain-free females were phenotyped and genotyped. We restricted our analysis to Europeans to avoid the possible effect of population stratification. Each enrolee was quantified for responsiveness to a set of 13 noxious stimuli applied to various anatomical sites (9, 32).
Genotyping.
SNPs were identified in the SCN9A gene from the Hapmap database and chosen for genotyping in the OA cohort on the basis of their allele frequency, potential to have a functional effect, confirmation as a real SNP, and linkage disequilibrium coverage of the gene (Table S1). Genotyping was performed using primer extension methodology or TaqMan assay following the manufacturer's instructions (Applied Biosystems).
Statistical Analysis of Pain Phenotypes.
For OA cohort 1, an initial analysis was undertaken to establish the importance of covariates, including gender, age, and each study. To evaluate SNP associations, separate models were run with each SNP term added to the base model. Genotype effect was initially assessed in a codominant genetic model and compared with models that included a term for deviation from additivity due to either a dominant or a recessive mechanism (SI Subjects and Methods).
Calculation of Combined P Value.
A modification of the inverse normal method (23) was used to combine association P values for the clinical cohorts. The modification takes into account the direction of the phenotypic effect. This is achieved by converting two-sided P values (p) into one-sided (q), noting the direction of the association with the allele A. If the direction of association is positive, then q = p/2; otherwise, q = 1 − p/2. After these values are combined into an overall P value, the result is doubled if the combined value is smaller than 1/2; otherwise, it is subtracted from 1 and then doubled. Overall and Rhoades (34) proposed a similar approach based on the Fisher combination method. Because the effect direction is not chosen beforehand, they recommend evaluating both directional hypotheses in turn, computing two one-sided P values and then doubling the minimum value (33). However, this is not needed with Stouffer's approach because of the symmetry of the inverse normal transformation: the two one-sided P values are the same with this approach, once converted back to the two-sided. Stouffer's method allows P values to be weighted (34). The weights were set to the square roots of the sample sizes.
Electrophysiology.
Both rs6746030 alleles were generated in a cDNA clone encoding the most common SCN9A splice variant found in dorsal root ganglia (NM_002977) (18) using the QuikChange XL site-directed mutagenesis kit (Stratagene). HEK293A cells (QBiogene) were transiently transfected with plasmids expressing either NaV1.7–1150R (common allele) or NaV1.7–1150W (minor allele) + DsRed2 and SCN1B + SCN2B + EGFP using lipofectamine 2000, and experiments were performed 2–3 days after transfection on cells positive for red and green fluorescence, as previously described (11). Briefly, electrodes had resistances of 2.5–3 MΩ when filled with pipette solution, and 70% series resistance compensation was used throughout. Currents were zero and leak subtracted using a p/4 protocol. The bath solution contained (mM): 3 KCl, 140 NaCl, 2 CaCl2, 1 MgCl2, 10 hepes, and 1 glucose (pH 7.4 with NaOH). The patch pipette solution contained (mM): 107 CsF, 10 NaCl, 1 CaCl2, 2 MgCl2, 10 Hepes, 10 TEACl, and 10 EGTA (pH 7.2 with CsOH). See SI Subjects and Methods for additional details.
URLs.
SNPs are detailed in dbSNP at http://www.ncbi.nlm.nih.gov/SNP/. Genes and the human genome can be viewed and accessed at UCSC Bioinformatics Genome Browser (http://genome.ucsc.edu/) and at the Human Mendelian disease database (OMIM) (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM&itool=toolbar).
Supplementary Material
Acknowledgments
We thank Adeline Nicholas, Gemma Thornton, Baljinder Matharu, and John Yates for help in preparing this manuscript. We also thank Steven J. Atlas and Robert B. Keller, the principal clinical investigators of the Maine Lumbar Spine Study, for allowing us to investigate their patients; and Monique H. M. Derikx for clinical data collection. St John's College in Cambridge, United Kingdom, the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences, the Wellcome Trust, and Pfizer contributed to the funding of this project.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.G.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913181107/DCSupplemental.
References
- 1.Erickson HH, Kitchell RL. Pain perception and alleviation in animals. Fed Proc. 1984;43:1307–1312. [PubMed] [Google Scholar]
- 2.Kavaliers M. Evolutionary and comparative aspects of nociception. Brain Res Bull. 1988;21:923–931. doi: 10.1016/0361-9230(88)90030-5. [DOI] [PubMed] [Google Scholar]
- 3.Machin KL. Fish, amphibian, and reptile analgesia. Vet Clin North Am Exot Anim Pract. 2001;4:19–33. doi: 10.1016/s1094-9194(17)30048-8. [DOI] [PubMed] [Google Scholar]
- 4.Shevelkin AV, Kozyrev SA, Nikitin VP, Sherstnev VV. In vivo investigation of genome activity and synaptic plasticity of neurons in snails during learning. Neurosci Behav Physiol. 2005;35:595–603. doi: 10.1007/s11055-005-0099-9. [DOI] [PubMed] [Google Scholar]
- 5.Mäntyselkä PT, Turunen JH, Ahonen RS, Kumpusalo EA. Chronic pain and poor self-rated health. JAMA. 2003;290:2435–2442. doi: 10.1001/jama.290.18.2435. [DOI] [PubMed] [Google Scholar]
- 6.Stewart WF, Ricci JA, Chee E, Morganstein D, Lipton R. Lost productive time and cost due to common pain conditions in the US workforce. JAMA. 2003;290:2443–2454. doi: 10.1001/jama.290.18.2443. [DOI] [PubMed] [Google Scholar]
- 7.Scholz J, Woolf CJ. Can we conquer pain? Nat Neurosci. 2002;5(Suppl):1062–1067. doi: 10.1038/nn942. [DOI] [PubMed] [Google Scholar]
- 8.Pingle SC, Matta JA, Ahern GP. Capsaicin receptor: TRPV1 a promiscuous TRP channel. Handb Exp Pharmacol. 2007;179:155–171. doi: 10.1007/978-3-540-34891-7_9. [DOI] [PubMed] [Google Scholar]
- 9.Diatchenko L, et al. Genetic basis for individual variations in pain perception and the development of a chronic pain condition. Hum Mol Genet. 2005;14:135–143. doi: 10.1093/hmg/ddi013. [DOI] [PubMed] [Google Scholar]
- 10.Tegeder I, et al. GTP cyclohydrolase and tetrahydrobiopterin regulate pain sensitivity and persistence. Nat Med. 2006;12:1269–1277. doi: 10.1038/nm1490. [DOI] [PubMed] [Google Scholar]
- 11.Cox JJ, et al. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fertleman CR, et al. SCN9A mutations in paroxysmal extreme pain disorder: Allelic variants underlie distinct channel defects and phenotypes. Neuron. 2006;52:767–774. doi: 10.1016/j.neuron.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, et al. Mutations in SCN9A, encoding a sodium channel alpha subunit, in patients with primary erythermalgia. J Med Genet. 2004;41:171–174. doi: 10.1136/jmg.2003.012153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drenth JP, et al. SCN9A mutations define primary erythermalgia as a neuropathic disorder of voltage gated sodium channels. J Invest Dermatol. 2005;124:333–338. doi: 10.1111/j.0022-202X.2005.23737.x. [DOI] [PubMed] [Google Scholar]
- 15.Estacion M, et al. Nav1.7 gain-of-function mutations as a continuum: A1632E displays physiological changes associated with erythromelalgia and paroxysmal extreme pain disorder mutations and produces symptoms of both disorders. J Neurosci. 2008;28:1079–11088. doi: 10.1523/JNEUROSCI.3443-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi JS, et al. Mexiletine-responsive erythromelalgia due to a new Na(v)1.7 mutation showing use-dependent current fall-off. Exp Neurol. 2009;216:383–389. doi: 10.1016/j.expneurol.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 17.Drenth JP, Waxman SG. Mutations in sodium-channel gene SCN9A cause a spectrum of human genetic pain disorders. J Clin Invest. 2007;117:3603–3609. doi: 10.1172/JCI33297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raymond CK, et al. Expression of alternatively spliced sodium channel alpha-subunit genes. Unique splicing patterns are observed in dorsal root ganglia. J Biol Chem. 2004;279:46234–46241. doi: 10.1074/jbc.M406387200. [DOI] [PubMed] [Google Scholar]
- 19.Waxman SG, Dib-Hajj S. Erythermalgia: Molecular basis for an inherited pain syndrome. Trends Mol Med. 2005;11:555–562. doi: 10.1016/j.molmed.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 20.Rush AM, et al. A single sodium channel mutation produces hyper- or hypoexcitability in different types of neurons. Proc Natl Acad Sci USA. 2006;103:8245–8250. doi: 10.1073/pnas.0602813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg YP, et al. Loss-of-function mutations in the Nav1.7 gene underlie congenital indifference to pain in multiple human populations. Clin Genet. 2007;71:311–319. doi: 10.1111/j.1399-0004.2007.00790.x. [DOI] [PubMed] [Google Scholar]
- 22.Diatchenko L, Nackley AG, Tchivileva IE, Shabalina SA, Maixner W. Genetic architecture of human pain perception. Trends Genet. 2007;23:605–613. doi: 10.1016/j.tig.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Stouffer SA, Suchman EA, DeVinney LC, Star SA, Jr, Williams RM. Adjustment During Army Life. Vol. 1. Princeton, NJ: Princeton University Press; 1949. The American Soldier. [Google Scholar]
- 24.Cepeda MS, Camargo F, Zea C, Valencia L. Tramadol for osteoarthritis: A systematic review and metaanalysis. J Rheumatol. 2007;34:543–555. [PubMed] [Google Scholar]
- 25.Cummins TR, Howe JR, Waxman SG. Slow closed-state inactivation: A novel mechanism underlying ramp currents in cells expressing the hNE/PN1 sodium channel. J Neurosci. 1998;18:9607–9619. doi: 10.1523/JNEUROSCI.18-23-09607.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheets PL, Jackson JO, II, Waxman SG, Dib-Hajj SD, Cummins TR. A Nav1.7 channel mutation associated with hereditary erythromelalgia contributes to neuronal hyperexcitability and displays reduced lidocaine sensitivity. J Physiol. 2007;581:1019–1031. doi: 10.1113/jphysiol.2006.127027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Estacion M, et al. A sodium channel gene SCN9A polymorphism that increases nociceptor excitability. Ann Neurol. 2009;66:862–866. doi: 10.1002/ana.21895. [DOI] [PubMed] [Google Scholar]
- 28.Cummins TR, Sheets PL, Waxman SG. The roles of sodium channels in nociception: Implications for mechanisms of pain. Pain. 2007;131:243–257. doi: 10.1016/j.pain.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Virtanen IM, et al. Phenotypic and population differences in the association between CILP and lumbar disc disease. J Med Genet. 2007;44:285–288. doi: 10.1136/jmg.2006.047076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atlas SJ, et al. The Maine Lumbar Spine Study, Part II. 1-year outcomes of surgical and nonsurgical management of sciatica. Spine (Phila Pa 1976) 1996;21:1777–1786. doi: 10.1097/00007632-199608010-00011. [DOI] [PubMed] [Google Scholar]
- 31.van Esch AA, Wilder-Smith OH, Jansen JB, van Goor H, Drenth JP. Pharmacological management of pain in chronic pancreatitis. Dig Liver Dis. 2006;38:518–526. doi: 10.1016/j.dld.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Diatchenko L, et al. Catechol-O-methyltransferase gene polymorphisms are associated with multiple pain-evoking stimuli. Pain. 2006;125:216–224. doi: 10.1016/j.pain.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 33.Overall JE, Rhoades HM. Beware of a half-tailed test. Psychol Bull. 1986;100:121–122. [PubMed] [Google Scholar]
- 34.Whitlock MC. Combining probability from independent tests: The weighted Z-method is superior to Fisher's approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.