Abstract
Many of the mutations reported as potentially causing Lynch syndrome are missense mutations in human mismatch repair (MMR) genes. Here, we used a Saccharomyces cerevisiae-based system to study polymorphisms and suspected missense mutations in human MMR genes by modeling them at the appropriate S. cerevisiae chromosomal locus and determining their effect on mutation rates. We identified a number of weak alleles of MMR genes and MMR gene polymorphisms that are capable of interacting with other weak alleles of MMR genes to produce strong polygenic MMR defects. We also identified a number of alleles of MSH2 that act as if they inactivate the Msh2-Msh3 mispair recognition complex thus causing weak MMR defects that interact with an msh6Δ mutation to result in complete MMR defects. These results indicate that weak MMR gene alleles capable of polygenic interactions with other MMR gene alleles may be relatively common.
Keywords: hereditary nonpolyposis colorectal cancer, lynch syndrome, Saccharomyces cerevisiae, mutator phenotype, genome instability
Lynch syndrome, also called hereditary nonpolyposis colorectal cancer (HNPCC), is an inherited cancer susceptibility syndrome characterized by predisposition to develop colorectal cancer and other cancers (1–3). Inherited DNA mismatch repair (MMR) gene defects underlie many cases of Lynch syndrome (4–7). The two major Lynch syndrome genes are MSH2 and MLH1, encoding MutS and MutL homologs, respectively (4–7). Defects in other MMR genes are found in Lynch syndrome much less frequently but are found in other types of suspected inherited cancer susceptibility such as Turcots syndrome and familial cancers associated with weaker family histories, later age of onset, and potentially a different cancer spectrum (1, 8–14). Large numbers of Lynch syndrome cases have been screened for mutations in MMR genes and consequently large numbers of mutations and genetic variants have been published and reported in public databases.
Many reported mutations in MSH2 and MLH1 are missense mutations (1, 4–6); unfortunately, it is difficult to assess the functional and clinical significance of such mutations. Some studies report data demonstrating cosegregation of a missense variant with disease and lack of the variant in normal controls; however, such data do not definitively distinguish a pathogenic mutation from a rare, nonpathogenic variant. Functional studies have been performed to determine if missense variants affect protein function as loss-of-function would provide evidence for a pathogenic mutation. The approaches used in these studies include bioinformatic and structure-based predictions; evaluation of mutations in human cells, mutant mice, and human cell extract-based MMR reactions; biochemical evaluation of mutant proteins; and a number of Saccharomyces cerevisiae-based assays (15–27). Although most assays used have limitations, useful functional data for a number of potential mutations have emerged.
Results
Identification of Human MMR Gene Mutations and Polymorphisms that Are Weak Alleles.
Here, we evaluated 11 reported polymorphisms in MSH2, MLH1, MSH6, and PMS2 (PMS1 in S. cerevisiae) and 14 reported missense mutations in these genes by making equivalent mutations at the analogous position in the corresponding chromosomal gene in S. cerevisiae and evaluating their effect on MMR using mutator assays (Tables 1 and 2; S. cerevisiae allele designations are used in the text of this report). Some of these variants have been characterized in other studies using S. cerevisiae-based assays (16–19, 23, 25) but mostly by testing them as plasmid-borne mutations (16–19, 23)(Table 2). The mutator assays used include CAN1 gene inactivation that detects mutations inactivating the CAN1 gene and hom3-10 and lys2-10A frameshift reversions that detect mutations that revert +1T and +1A insertions, respectively, present in mononucleotide repeats (28–30). The latter two assays are extremely sensitive. Of the 11 polymorphisms tested, six resulted in a mutation rate indistinguishable from the wild-type rate. Five of the polymorphisms caused weak but significant mutator phenotypes in one of the mutator assays, almost always the sensitive lys2-10A assay. Of the 14 suspected pathogenic mutations tested, four caused effectively complete MMR defects, three caused small but significant mutator phenotypes in multiple mutator assays, three caused a small but significant mutator phenotype in a single mutator assay, and four caused no apparent defect. Mutations and polymorphisms causing little or no MMR defects have been reported in other functional studies (17–19, 23). That we detected a large proportion of weak or silent alleles may reflect the fact that the mutations we selected for study were mostly from cases where there was little if any clinical data supporting potential pathogenicity.
Table 1.
List of mutations tested in this study
| Chromosomal alleles | ||||
| Strain no. | Gene | Human allele | S. cerevisiae allele | Source |
| Mutations | ||||
| RDKY7071 | MLH1 | R217C | R214C | a |
| RDKY7091 | MLH1 | R265H | R265H | a |
| RDKY7092 | MLH1 | V326A | I326A | a |
| RDKY7093 | MLH1 | K84E | K81E | a |
| RDKY7094 | MLH1 | V716M | L731M | a |
| RDKY7095 | MLH1 | T117M | T114M | a |
| RDKY7100 | MSH2 | C333R | C345R | a |
| RDKY7101 | MSH2 | D506Y | D524Y | a |
| RDKY7102 | MSH2 | E198G | E194G | a; 4, 33 |
| RDKY7103 | MSH2 | I194T | L190T | b |
| RDKY7104 | MSH2 | E701K | E720K | b |
| RDKY7107 | MSH6 | S144I | S107I | a |
| RDKY4493 | MSH6 | V509A | V410A | a |
| RDKY7110 | PMS1 | E705K | E738K | a |
| Polymorphisms | ||||
| RDKY7096 | MLH1 | D132H | E129H | a |
| RDKY7097 | MLH1 | H718Y | H733Y | a |
| RDKY7098 | MLH1 | I219L | V216L | a |
| RDKY7099 | MLH1 | S406N | S415N | a |
| RDKY7105 | MSH2 | G322D | G317D | a |
| RDKY7106 | MSH2 | N127S | N123S | a |
| RDKY7108 | MSH6 | S503C | S406C | a |
| RDKY4488 | MSH6 | L396V | L301V | a |
| RDKY7109 | MSH6 | K1358DfsX1 | Asp1239X | a |
| RDKY7111 | PMS1 | T485K | T505K | a |
| RDKY7112 | PMS1 | N775S | Q808S | a |
| Plasmid alleles | ||||
| Plasmid no. | Gene | Human allele | S. cerevisiae allele | Source |
| Mutations | ||||
| pAG207 | MSH2 | C199R | C195R | a; 17 |
| pAG80 | MSH2 | D167H | D163H | a; 17 |
| pAG25 | MSH2 | G122S | c; 17 | |
| pAG204 | MSH2 | I145M | I141M | a; 17 |
| pAG203 | MSH2 | N127S | N123S | a; 17 |
| pAG413 | MSH2 | Q61P | Q61P | a; 17 |
| pAG401 | MSH2 | T44M | T44M | a; 17 |
| pAG414 | MSH2 | Y98C | Y104C | a; 17 |
| pRDK1523 | MSH2 | T33P | T33P | a |
| pRDK1524 | MSH2 | V34E | I34E | a |
| pRDK1525 | MSH2 | D49V | D49V | a |
| pRDK1526 | MSH2 | L93P | L99P | a |
| pRDK1527 | MSH2 | R96H | L102H | a |
| pRDK1528 | MSH2 | Y103C | Y109C | a |
| pRDK1529 | MSH2 | V163D | V159D | a |
| pRDK1530 | MSH2 | L187P | L183P | a |
a, inSIGHT database (6) (insight-group.org); b, Available on request; c, E. coli mutation (17).
Table 2.
Mismatch repair defects caused by mutations and polymorphisms tested
| Mismatch repair defect* |
|||||
| Strain no. | Genotype | Thr+ | Lys+ | Canr | Refs† |
| Controls | |||||
| RDKY3590 | Wild-type | 1 | 1 | 1 | |
| RDKY3591‡ | msh2Δ | 1105 | 7480 | 55 | |
| RDKY3684§ | msh6Δ | 19 | 182 | 18 | |
| Mutations | |||||
| RDKY7071 | mlh1-R214C | NS | NS | NS | 18, 23 |
| RDKY7091 | mlh1-R265H | 71 | 163 | 10.4 | 18, 23 |
| RDKY7092 | mlh1-I326A | 4.4 | 3.9 | 3.8 | 18, 19, 23 |
| RDKY7093 | mlh1-K81E | 2389 | 4835 | 26 | 18 |
| RDKY7094 | mlh1-L731M | NS | NS | NS | 18 |
| RDKY7095 | mlh1-T114M | 2952 | 8730 | 40 | 18, 19 |
| RDKY7100 | msh2-C345R | 2623 | 6625 | 35 | |
| RDKY7101 | msh2-D524Y | NS | NS | NS | 16 |
| RDKY7102 | msh2-E194G | 20 | 13.6 | NS | 17 |
| RDKY7103 | msh2-L190T | NS | NS | NS | |
| RDKY7104 | msh2-E720K | NS | 2.4 | NS | |
| RDKY7107 | msh6-S107I | NS | 2.0 | NS | |
| RDKY4493 | msh6-V410A | NS | 2.3 | NS | |
| RDKY7110 | pms1-E738K | 2092 | 5183 | 59 | 25 |
| Polymorphisms | |||||
| RDKY7096 | mlh1-E129H | NS | 4.0 | NS | 18 |
| RDKY7097 | mlh1-H773Y | NS | NS | NS | 18 |
| RDKY7098 | mlh1-V216L | NS | NS | NS | 18 |
| RDKY7099 | mlh1-S415N | NS | 2.3 | NS | 18 |
| RDKY7105 | msh2-G317D | NS | NS | NS | 16, 23 |
| RDKY7106 | msh2-N123S | NS | 2.6 | NS | |
| RDKY7108 | msh6-S406C | NS | 1.9 | NS | |
| RDKY4488 | msh6-L301V | NS | NS | 4.2 | |
| RDKY7109 | msh6-Asp1239 | NS | NS | NS | |
| RDKY7111 | pms1-T505K | NS | NS | NS | |
| RDKY7112 | pms1-Q808S | NS | NS | NS | |
*Individual experiments were performed in which the mutation rates for one or more mutants and at least one wild-type strain were determined in the same experiment. Mutant and wild-type rates were compared within each experiment and in only those cases where the P value for the significance was P < 0.05 in a Mann Whitney test the fold-increase in mutation rate relative to wild-type was calculated; in two of the cases where a fold-increase is reported, the P value was 0.025, whereas in the remaining cases the P values were < 0.005. Each mutant rate was independently determined from two to eight times and the average fold-increase in mutation rate is reported. In addition, we required that the increase in mutation rate be greater than one standard deviations of the average for 24 independent determinations of the wild-type mutation rate (Thr+ = 1.85 +/− 1.9 x 10−9, Lys+ = 1.38 +/− 0.92 x 10−8, Canr = 1.08 +/− 0.83 x 10−7). Otherwise, NS (no significant difference) is indicated.
†References to other studies concerning the indicated mutations.
‡The mutation rates for RDKY3591 msh2Δ and paired wild-type RDKY3590 were [Thr+ = 4.66 x 10−6, Lys+ = 1.21 x 10−4, Canr = 5.62 x 10−6] and [Thr+ = 4.21 x 10−9, Lys+ = 1.61 x 10−8, Canr = 1.02 x 10−7], respectively. The mutation rates for msh2Δ, mlh1Δ, and pms1Δ strains were not significantly different from each other.
Of the 14 mutations and polymorphisms tested here that have also been tested in other studies (Table 2), our results differed from those of at least one published study in seven cases (mlh1-R214C, mlh1-R265H, msh2-D524Y, msh2-E194G, mlh1-E129H, mlh1-S415N, and msh2-G317D). It seems likely that these differences are because our chromosomal mutation-based assay system in which cells can be grown in rich, nonselective media is more sensitive and accurate for characterizing small differences relative to low wild-type mutation rates than plasmid complementation-based assay systems used in other studies.
Identification of Weak MMR Gene Alleles that Interact with Other Weak MMR Gene Alleles.
We previously described a method for screening for enhancer mutations that cause little or no MMR defect alone but significantly enhance the MMR defect caused by a second weakly MMR defective allele (28). To further evaluate the potential significance of polymorphisms and weakly defective mutations in MMR genes, we selected two weak alleles (msh2-E194G, mlh1-I326A) and three polymorphisms (msh2-G317D, msh6 -L301V, mlh1-E129H) and screened 6,000–12,000 mutagenized survivors for enhancer mutations. In the case of the msh2-E194G mutation, 12 enhancer mutations (msh6-T312I, msh6-E624K, msh6-G477D, pms1-P343L, msh6-G1064E, msh6-T474I, msh6-E395K, msh6-E442K, pms1-R610K, mlh1-G144D, mlh3-D556N, msh6-G1066R) were found. Reconstruction experiments demonstrated that although each single mutant had little if any MMR defect, the double mutants could be almost completely MMR defective (Table 3). Enhancer screens with the mlh1-I326A mutation and the mlh1-E129H polymorphism had a lower yield of three and two enhancer mutations each, respectively (mlh1-I326A - msh2-T743I, msh2-P640S, pms1-G173E; mlh1-E129H - msh6-G1064E, pms1-E191K), whereas enhancer screens with the msh2-G317D and msh6-L301V polymorphisms did not yield enhancer mutations (of note, one dominant msh6 mutation was identified in the msh2-G317D screen; Table S1) (31). These screens are not saturated given they would be expected to yield allele-specific, likely rare mutations; however, it seems likely that the differences in yields obtained reflect differences in the diversity of enhancer mutations each allele might be capable of interacting with, given the similar number of mutations tested in each screen.
Table 3.
Genetic interactions with the msh2-E194G mutation
| Mutation Rate |
||||
| Strain no. | Genotype | Thr+ | Lys+ | Canr |
| RDKY3590 | Wild-type | 2.0 × 10−9 (1) | 1.1 × 10−8 (1) | 9.0 × 10-8 (1) |
| RDKY7102 | msh2-E194G | 2.3 × 10−8 (11.5) | 1.54 × 10−7 (14) | 1.35 × 10−7 (1.5) |
| RDKY7114 | msh6-E624K | 6.32 × 10−9 (3.2) | 1.22 × 10−7 (11.1) | 2.7 × 10−7 (3) |
| RDKY7115 | msh6-E624K msh2-E194G | 1.44 × 10−6 (720) | 2.19 × 10−5 (1991) | 1.53 × 10−6 (17) |
| RDKY7116 | msh6-G477D | 2.07 × 10−9 (1) | 2.08 × 10−8 (1.9) | 1.32 × 10−7 (1.5) |
| RDKY7117 | msh6-G477D msh2-E194G | 6.55 × 10−7 (327.5) | 3.11 × 10−6 (283) | 6.81 × 10−7 (7.6) |
| RDKY7118 | pms1-P343L | 1.37 × 10−7 (68.5) | 1.12 × 10−5 (1018) | 2.84 × 10−7 (3.2) |
| RDKY7119 | pms1-P343L msh2-E194G | 7.6 × 10−7 (380) | 1.26 × 10−5 (1145) | 5.14 × 10−7 (5.7) |
| RDKY7120 | msh6-T312I | 3.73 × 10−9 (1.9) | 5.87 × 10−8 (5.3) | 3.41 × 10−7 (3.8) |
| RDKY7121 | msh6-T312I msh2-E194G | 1.67 × 10−6 (835) | 2.9 × 10−5 (2636) | 1.82 × 10−6 (20.2) |
| RDKY3684 | msh6Δ | 3.8 × 10−8 (19) | 2.0 × 10−6 (182) | 1.62 × 10−6 (18) |
| RDKY7122 | msh2-E194G msh6Δ | 2.4 × 10−6 (1200) | 6.49 × 10−5 (5900) | 3.20 × 10−6 (35.5) |
The number in parentheses is the fold-increase in mutation rate relative to the wild-type strain RDKY3590. For comparison, mutation rates for the RDKY3591 msh2Δ strain were [Thr+ = 4.66 × 10−6, Lys+ = 1.21 × 10−4, Canr = 5.62 × 10−6], which are representative of a complete loss of MMR.
The msh2-E194G mutation screen yielded large numbers of enhancer mutations. This mutation alters an amino acid located at the boundary of Msh2 domains I and II and would be predicted to interfere with the correct positioning of domain I. Domain I is required for Msh2 to function in the Msh2-Msh3 complex but not the Msh2-Msh6 complex (32) and consistent with this, the msh2-E194G msh6Δ double mutant had a synergistic increase in mutation rate in the frameshift reversion assays compared to the respective single mutants (Table 3). This result and the observation that it is possible to isolate many enhancer mutations that cause much weaker MMR defects than a msh6Δ mutation indicates that the msh2-E194G mutation sensitizes the Msh6-dependent pathway to inactivation in addition to causing a defect in the Msh3 pathway. The human mutation, msh2-E198G, was initially found in a Lynch syndrome family with microsatellite stable tumors consistent with a weak MMR defect (4, 33). Four of the affected mutation carriers also inherited a missense variant in MLH3 (34). However, our studies suggest that the mlh3 missense mutation might not be an enhancing mutation as the human msh2-E198G mutation (Sc msh2-E194G) would be predicted to already cause a defect in the Msh3 pathway where Mlh3 functions (35).
Identification of Weak Lynch Syndrome-Associated MSH2 Alleles that Inactivate the Msh2-Msh3 Complex.
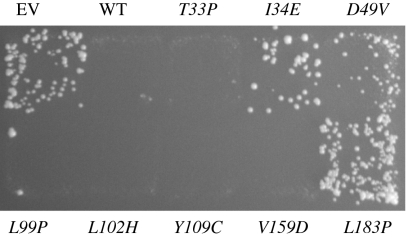
Interestingly, numerous Lynch syndrome-associated msh2 missense mutations affecting Msh2 domains I and II have been reported, and many of those tested in functional assays cause no MMR defect (6, 17). These mutations potentially inactivate the function of the Msh2-Msh3 complex but not the Msh2-Msh6 complex, and such mutations would be expected to cause synergistic increases in the rate of accumulating frameshift mutations when combined with an msh6Δ mutation. To investigate this possibility, we first tested 16 such mutations for MMR defects by determining their ability to complement an msh2Δ mutation as plasmid borne alleles using patch tests. Of these, eight had been previously tested in functional assays (17) and eight had not been previously studied. We confirmed that five of the previously studied mutant alleles (T44M, Q63P, Y104C, N123S, I141M) were fully complementing and had no effect on MMR, and one was a loss of function mutation (C195R). Two of these were not fully complementing (G122S, D164H) and caused weak MMR defects (Table 4). Of the newly studied mutations, we found that three mutations (L102H, Y109C, V159D) had no effect on MMR, three mutations (I34E, D49V, L183P) were loss of function mutations, and two mutations (T33P, L99P) were not fully complementing and caused weak MMR defects (Fig. 1). Each of the alleles that caused either no or weak MMR defects were then tested as plasmid borne alleles in an msh2Δ msh6Δ double mutant in patch tests to determine if they caused synergistic increases in the rate of accumulating frameshift mutations when combined with an msh6Δ mutation, and the interacting mutations were then characterized by performing quantitative mutation rate assays. Six mutations did not cause an MMR defect and did not cause synergistic increases in the hom3-10 frameshift reversion assay in combination with an msh6Δ mutation in patch tests (T44M, Q63P, Y104C, N123S, I141M, V159D) and were not studied further. Of the mutations studied further, five caused synergistic increases in mutation rates in combination with an msh6Δ mutation in the hom3-10 frameshift reversion assay (T33P, domain I; L99P, domain I; L102H, domain I; Y109C, domain I; G122S, domain I) and hence behaved as if they inactivated the Msh2-Msh3 complex, whereas one mutation (D164H, domain II) caused at most a very small synergistic increase in mutation rate in combination with an msh6Δ mutation and was likely a weakly MMR defective mutation (Table 4).
Table 4.
Interaction between plasmid MSH2 alleles and an msh6Δ mutation
| MSH2 | RDKY2706 msh2Δ |
RDKY4136 msh2Δmsh6Δ |
|||
| Mutation rate |
Mutation rate |
||||
| Plasmid no. | Allele | Thr+ | Canr | Thr+ | Canr |
| pMSH2 | MSH2 | 21.27 × 10−8 (1) | 2.81 × 10−7 (1) | 4.54 × 10−8 (1) | 4.29 × 10−6 (1) |
| pRS413 | Vector | 1.45 × 10−5 (1142) | 6.49 × 10−6 (23) | 1.19 × 10−5 (262) | 9.37 × 10−6 (2.2) |
| pAG80 | msh2-D164H | 1.24 × 10−7 (9.8) | 7.92 × 10−7 (2.8) | 7.87 × 10−7 (13) | 3.48 × 10−6 (0.8) |
| pAG25 | msh2-G122S | 1.11 × 10−7 (8.7) | 4.65 × 10−7 (1.7) | 2.49 × 10−6 (55) | 7.37 × 10−6 (1.7) |
| pRDK1523 | msh2-T33P | 4.25 × 10−7 (33) | 4.01 × 10−6 (14.3) | 2.86 × 10−5 (630) | 2.21 × 10−5 (5.2) |
| pRDK1526 | msh2-L99P | 6.43 × 10−7 (51) | 9.97 × 10−7 (3.5) | 1.70 × 10−5 (374) | 1.13 × 10−5 (2.6) |
| pRDK1527 | msh2-L102H | 1.44 × 10−8 (1.1) | 3.04 × 10−7 (1.1) | 1.15 × 10−5 (253) | 1.09 × 10−5 (2.5) |
| pRDK1528 | msh2-Y109C | 5.70 × 10−8 (4.5) | 4.04 × 10−7 (1.44) | 2.75 × 10−5 (606) | 2.02 × 10−5 (4.7) |
All of the indicated MSH2 alleles were made by site-directed mutagenesis in the pRS413 HIS3 MSH2 vector pMSH2; two of the plasmid mutants (pAG80 and pAG25) were obtained from Dr. Alison Gammie (Princeton University, Princeton, NJ). The number in () is the fold-increase in mutation rate relative to the value obtained with the pMSH2 wild-type plasmid.
Fig. 1.
Complementation of a chromosomal msh2Δ mutation by plasmid MSH2 alleles assessed by patch tests. An msh2Δ mutant strain RDKY2706 containing the indicated plasmid MSH2 alleles was patched onto His drop out media and then replicated onto Thr− His− CSM media to assess the frequency of accumulation of hom3-10 (Thr+) revertants. EV indicates Empty Vector.
Discussion
Our studies have used a S. cerevisiae-based system to explore the genetics of polymorphisms and suspected mutations in human MMR genes. The limitations of the system are that only mutations affecting conserved amino acids can be analyzed, and it is tedious to make mutations at the chromosomal MMR gene of interest. The strength of the system is that sophisticated genetic analysis can be used. We found that most polymorphisms are likely silent but that at least five polymorphisms, including one previously associated with increased cancer susceptibility (mlh1-E129H) (36, 37), cause very weak MMR defects. Our results also indicate that although many reported missense mutations in MMR genes are likely to be loss-of-function mutations, numerous reported mutations have either no affect on MMR or cause very weak MMR defects underscoring the need for functional testing before concluding missense mutations are pathogenic. Finally, we identified a number of weak alleles that are capable of interacting with other weak alleles in a number of different MMR genes or that interact with an msh6Δ mutation to produce strong polygenic MMR defects, indicating such alleles may be relatively common. These latter results reveal a level of genetic complexity that is relevant to the genetics of cancer susceptibility not previously demonstrated for human MMR genes.
How might weak alleles result in increased development of cancer? It is possible that such defects simply lead to low rates of accumulating mutations resulting in the need for increased numbers of cell generations before sufficient numbers of mutations occur in critical tumor suppressor genes and proto-oncogenes, compared to complete loss of MMR. Alternatively, such mutations might be separation-of-function mutations that primarily cause DNA damage response defects rather than MMR defects (27, 38). These types of defects would have lower penetrance than complete MMR defective mutations. Our results demonstrate two other potential mechanisms. One is that inherited weak alleles might participate in polygenic interactions with other inherited weak alleles. However, unless the interacting alleles were common, the cancers due to inheritance of two such alleles would likely be seen as isolated cancer cases rather than showing the dominant, high penetrance segregation patterns characteristic of Lynch syndrome families. Furthermore, becasue two “second hits” would be required to inactivate the wild-type alleles in the resulting double heterozygotes, it is likely that cancer in these cases would show a later age of onset than seen in Lynch syndrome. The other mechanism is that in patients with an inherited weak allele, somatic second hits of the type seen here could occur in a different MMR gene resulting in strong MMR defects; this type of genetic interaction could be identified by sequencing appropriate target MMR genes in tumors from Lynch syndrome cases where an inherited weak allele has been implicated. In the case of mutations affecting Msh2 domains I and II, the MSH6 gene would be the most likely target for enhancing mutations.
Materials and Methods
S. cerevisiae were grown on standard media, either yeast extract/peptone/dextrose (YPD), or complete supplement mixture (CSM) medium (US Biological) lacking specific amino acids to select for plasmid markers and/or Lys+ and Thr+ revertants; 60 mg/L canavanine was added to CSM–arg medium to select for canavanine resistance. All S. cerevisiae strains are isogenic derivatives of S288C. The wild-type strain RDKY 3590 (28) has the genotype MATa ura3-52 leu2Δ1 trp1Δ63 hom3-10 lys2::InsE-A10 and was the parent for all strains containing the chromosomal mutations studied. The wild-type strain RDKY 3023 (39) has the genotype MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 ade2Δ1 ade8 hom3-10 lys2-Bgl and was derived from the same parental strain background as RDKY 3590. The strains RDKY 2706 and RDKY 4136, which were used for all plasmid complementation tests, were derived from RDKY 3023 have an msh2Δ::hisG mutation or both msh2Δ::hisG and msh6Δ::hisG mutations, respectively. The plasmid the ARS CEN HIS3 MSH2 plasmid pMSH2 and derivatives containing the S. cerevisiae MSH2 alleles T44M, Q63P, Y104C, N123S, G122S, I141M, D164H, and C195R were kindly provided by Dr. Alison Gammie (Princeton University, Princeton, NJ); the matched vector, pRS413, was from our laboratory collection. pMSH2 derivatives containing the MSH2 alleles T33P, I34E, D49V, L99P, L102H, Y109C, V159D, and L183P were made by site directed mutagenesis. The plasmids are listed in Table 1 and the strains are described in detail in Table 5. All of the methods used in the described studies including those for site directed mutagenesis, verification by DNA sequencing, transferring mutations to the relevant chromosomal locus, construction of strains by gene disruption or intercrossing, and evaluation of mutator phenotypes by patch tests or fluctuation rate analysis were standard methods or were exactly as previously described (28, 29, 40, 41).
Table 5.
S. cerevisiae strains used in this study
| Strain no. | Relevant genotype | Source |
| RDKY3590 | MATa, ura3-52, leu2∆1, trp1∆63, hom3-10, lys2-10A | 28 |
| RDKY3023 | MATa, ura3-52, leu2∆1, trp1∆63, hom3-10, lys2∆Bgl, his3∆200, ade2∆1::hisG, ade8 | 39 |
| RDKY2706 | MATa, ura3-52, leu2∆1, trp1∆63, hom3-10, lys2∆Bgl, his3∆200, ade2∆1::hisG, ade8, msh2∆::hisG | This study |
| RDKY4136 | MATa, ura3-52, leu2∆1, trp1∆63, hom3-10, lys2∆Bgl, his3∆200, ade2∆1::hisG, ade8, msh∆2::hisG, msh6∆::hisG | This study |
| RDKY3591 | MATa, ura3-52, leu2∆1, trp1∆63, hom3-10, lys2-10A, msh2∆::hisGURA3hisG | This study |
| RDKY3684 | MATa, ura3-52, leu2∆1, trp1∆63, hom3-10, lys2-10A msh6∆::hisG | This study |
| RDKY7071 | RDKY3590 mlh1-R214C | This study |
| RDKY7091 | RDKY3590 mlh1-R265H | This study |
| RDKY7092 | RDKY3590 mlh1-I326A | This study |
| RDKY7093 | RDKY3590 mlh1-K81E | This study |
| RDKY7094 | RDKY3590 mlh1-L731M | This study |
| RDKY7095 | RDKY3590 mlh1-T114M | This study |
| RDKY7096 | RDKY3590 mlh1-E129H | This study |
| RDKY7097 | RDKY3590 mlh1-H733Y | This study |
| RDKY7098 | RDKY3590 mlh1-V216L | This study |
| RDKY7099 | RDKY3590 mlh1-S415N | This study |
| RDKY7100 | RDKY3590 msh2-C345R | This study |
| RDKY7101 | RDKY3590 msh2-D524Y | This study |
| RDKY7102 | RDKY3590 msh2-E194G | This study |
| RDKY7103 | RDKY3590 msh2-L190T | This study |
| RDKY7104 | RDKY3590 msh2-E720K | This study |
| RDKY7105 | RDKY3590 msh2-G317D | This study |
| RDKY7106 | RDKY3590 msh2-N123S | This study |
| RDKY7107 | RDKY3590 msh6-S107I | This study |
| RDKY7108 | RDKY3590 msh6-S406C | This study |
| RDKY7109 | RDKY3590 msh6-Asp1239X | This study |
| RDKY7110 | RDKY3590 pms1-E738K | This study |
| RDKY7111 | RDKY3590 pms1-T505K | This study |
| RDKY7112 | RDKY3590 pms1-Q808S | This study |
| RDKY4488 | RDKY3590 msh6-L301V | This study |
| RDKY4493 | RDKY3590 msh6-V410A | This study |
| RDKY7113 | RDKY3590 msh6-R1024C | This study |
| RDKY7114 | RDKY3590 msh6-E624K | This study |
| RDKY7115 | RDKY3590 msh2-E194G, msh6-E624K | This study |
| RDKY7116 | RDKY3590 msh6-G477D | This study |
| RDKY7117 | RDKY3590 msh2-E194G, msh6-G477D | This study |
| RDKY7118 | RDKY3590 pms1-P343L | This study |
| RDKY7119 | RDKY3590 msh2-E194G, pms1-P343L | This study |
| RDKY7120 | RDKY3590 msh6-T312I | This study |
| RDKY7121 | RDKY3590 msh2-E194G, msh6-T312I | This study |
| RDKY7122 | RDKY3590 msh2-E194G, msh6∆::hisG | This study |
Isolation of enhancer mutations was performed exactly as previously described (28). Briefly, a derivative of RDKY 3590 containing one of the indicated mutations or polymorphisms (msh2-E194G, mlh1-I326A, msh2-G317D, msh6 -L301V, mlh1-E129H) at the relevant chromosomal locus was mutagenized with Ethyl Ethane Sulphonate and 6,000–12,000 survivors were plated onto YPD plates. The resulting colonies were then replica plated onto CSM plates that either lacked lysine or threonine, or lacked arginine and also contained canavanine to identify mutator mutants. The initial mutants were subjected to two cycles of retesting and then each mutator mutant identified was transformed with an ARS CEN plasmid containing the wild-type gene that would complement the initial mutation present in the starting strain to identify those mutants whose mutator phenotype was fully complemented by this plasmid. For all complementable mutants, the starting mutant gene was sequenced to identify those mutants where the starting mutant gene only contained the starting mutation and had not acquired a loss of function mutation. Then, as relevant, the MSH2, MSH3, MSH6, MLH1, PMS1, MLH3, EXO1 and POL30 genes from each mutant were sequenced; this resulted in the identification of a second mutation in one of these genes in each starting mutant. Finally, each mutant was transformed with an ARS CEN plasmid containing the gene predicted to complement the second mutation to verify that the mutator phenotype of each mutant required both the starting mutation and the second mutation.
Supplementary Material
Acknowledgments
The authors thank Drs. Jill M. Dowen, Richard Fishel, and Christopher D. Putnam for helpful discussions. Supported by National Institutes of Health Grant GM50006.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000798107/DCSupplemental.
References
- 1.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 2.Vasen HF, et al. Cancer risk in families with hereditary nonpolyposis colorectal cancer diagnosed by mutation analysis. Gastroenterology. 1996;110:1020–1027. doi: 10.1053/gast.1996.v110.pm8612988. [DOI] [PubMed] [Google Scholar]
- 3.Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: The syndrome, the genes, and historical perspectives. J Natl Cancer Inst. 1995;87:1114–1125. doi: 10.1093/jnci/87.15.1114. [DOI] [PubMed] [Google Scholar]
- 4.Lagerstedt Robinson K, et al. Lynch syndrome (hereditary nonpolyposis colorectal cancer) diagnostics. J Natl Cancer Inst. 2007;99:291–299. doi: 10.1093/jnci/djk051. [DOI] [PubMed] [Google Scholar]
- 5.Casey G, et al. Colon Cancer Family Registry Conversion analysis for mutation detection in MLH1 and MSH2 in patients with colorectal cancer. JAMA. 2005;293:799–809. doi: 10.1001/jama.293.7.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peltomäki P, Vasen H. Mutations associated with HNPCC predisposition—Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–276. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodríguez-Moranta F, et al. Gastrointestinal Oncology Group of the Spanish Gastroenterological Association Clinical performance of original and revised Bethesda guidelines for the identification of MSH2/MLH1 gene carriers in patients with newly diagnosed colorectal cancer: Proposal of a new and simpler set of recommendations. Am J Gastroenterol. 2006;101:1104–1111. doi: 10.1111/j.1572-0241.2006.00522.x. [DOI] [PubMed] [Google Scholar]
- 8.Senter L, et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wijnen J, et al. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet. 1999;23:142–144. doi: 10.1038/13773. [DOI] [PubMed] [Google Scholar]
- 10.Kolodner RD, et al. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–5074. [PubMed] [Google Scholar]
- 11.Hendriks YM, et al. Cancer risk in hereditary nonpolyposis colorectal cancer due to MSH6 mutations: Impact on counseling and surveillance. Gastroenterology. 2004;127:17–25. doi: 10.1053/j.gastro.2004.03.068. [DOI] [PubMed] [Google Scholar]
- 12.Hampel H, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer) N Engl J Med. 2005;352:1851–1860. doi: 10.1056/NEJMoa043146. [DOI] [PubMed] [Google Scholar]
- 13.Miyaki M, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–272. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- 14.Akiyama Y, et al. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920–3923. [PubMed] [Google Scholar]
- 15.Jeyaprakash A, Das Gupta R, Kolodner R. Saccharomyces cerevisiae pms2 mutations are alleles of MLH1, and pms2-2 corresponds to a hereditary nonpolyposis colorectal carcinoma-causing missense mutation. Mol Cell Biol. 1996;16:3008–3011. doi: 10.1128/mcb.16.6.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drotschmann K, Clark AB, Kunkel TA. Mutator phenotypes of common polymorphisms and missense mutations in MSH2. Curr Biol. 1999;9:907–910. doi: 10.1016/s0960-9822(99)80396-0. [DOI] [PubMed] [Google Scholar]
- 17.Gammie AE, et al. Functional characterization of pathogenic human MSH2 missense mutations in Saccharomyces cerevisiae. Genetics. 2007;177:707–721. doi: 10.1534/genetics.107.071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi M, et al. Functional analysis of human MLH1 variants using yeast and in vitro mismatch repair assays. Cancer Res. 2007;67:4595–4604. doi: 10.1158/0008-5472.CAN-06-3509. [DOI] [PubMed] [Google Scholar]
- 19.Shimodaira H, et al. Functional analysis of human MLH1 mutations in Saccharomyces cerevisiae. Nat Genet. 1998;19:384–389. doi: 10.1038/1277. [DOI] [PubMed] [Google Scholar]
- 20.Kondo E, Suzuki H, Horii A, Fukushige S. A yeast two-hybrid assay provides a simple way to evaluate the vast majority of hMLH1 germ-line mutations. Cancer Res. 2003;63:3302–3308. [PubMed] [Google Scholar]
- 21.Nyström-Lahti M, et al. Functional analysis of MLH1 mutations linked to hereditary nonpolyposis colon cancer. Genes Chromosomes Cancer. 2002;33:160–167. [PubMed] [Google Scholar]
- 22.Trojan J, et al. Functional analysis of hMLH1 variants and HNPCC-related mutations using a human expression system. Gastroenterology. 2002;122:211–219. doi: 10.1053/gast.2002.30296. [DOI] [PubMed] [Google Scholar]
- 23.Ellison AR, Lofing J, Bitter GA. Functional analysis of human MLH1 and MSH2 missense variants and hybrid human-yeast MLH1 proteins in Saccharomyces cerevisiae. Hum Mol Genet. 2001;10:1889–1900. doi: 10.1093/hmg/10.18.1889. [DOI] [PubMed] [Google Scholar]
- 24.Guerrette S, Acharya S, Fishel R. The interaction of the human MutL homologues in hereditary nonpolyposis colon cancer. J Biol Chem. 1999;274:6336–6341. doi: 10.1074/jbc.274.10.6336. [DOI] [PubMed] [Google Scholar]
- 25.Deschênes SM, et al. The E705K mutation in hPMS2 exerts recessive, not dominant, effects on mismatch repair. Cancer Lett. 2007;249:148–156. doi: 10.1016/j.canlet.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heinen CD, et al. HNPCC mutations in hMSH2 result in reduced hMSH2-hMSH6 molecular switch functions. Cancer Cell. 2002;1:469–478. doi: 10.1016/s1535-6108(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 27.Yang G, et al. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–150. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- 28.Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: Model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 30.Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–2865. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Das Gupta R, Kolodner RD. Novel dominant mutations in Saccharomyces cerevisiae MSH6. Nat Genet. 2000;24:53–56. doi: 10.1038/71684. [DOI] [PubMed] [Google Scholar]
- 32.Lee SD, Surtees JA, Alani E. Saccharomyces cerevisiae MSH2-MSH3 and MSH2-MSH6 complexes display distinct requirements for DNA binding domain I in mismatch recognition. J Mol Biol. 2007;366:53–66. doi: 10.1016/j.jmb.2006.10.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahlberg S, Liu T, Lindblom P, Lindblom A. Various mutation screening techniques in the DNA mismatch repair genes hMSH2 and hMLH1. Genet Test. 1999;3:259–264. doi: 10.1089/109065799316563. [DOI] [PubMed] [Google Scholar]
- 34.Liu HX, et al. The role of hMLH3 in familial colorectal cancer. Cancer Res. 2003;63:1894–1899. [PubMed] [Google Scholar]
- 35.Flores-Rozas H, Kolodner RD. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin BY, et al. Low allele frequency of MLH1 D132H in American colorectal and endometrial cancer patients. Dis Colon Rectum. 2005;48:1723–1727. doi: 10.1007/s10350-005-0123-8. [DOI] [PubMed] [Google Scholar]
- 37.Lipkin SM, et al. The MLH1 D132H variant is associated with susceptibility to sporadic colorectal cancer. Nat Genet. 2004;36:694–699. doi: 10.1038/ng1374. [DOI] [PubMed] [Google Scholar]
- 38.Marinovic-Terzic I, et al. Apoptotic function of human PMS2 compromised by the nonsynonymous single-nucleotide polymorphic variant R20Q. Proc Natl Acad Sci USA. 2008;105:13993–13998. doi: 10.1073/pnas.0806435105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau PJ, Flores-Rozas H, Kolodner RD. Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol Cell Biol. 2002;22:6669–6680. doi: 10.1128/MCB.22.19.6669-6680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shell SS, Putnam CD, Kolodner RD. Chimeric Saccharomyces cerevisiae Msh6 protein with an Msh3 mispair-binding domain combines properties of both proteins. Proc Natl Acad Sci USA. 2007;104:10956–10961. doi: 10.1073/pnas.0704148104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang ME, Kolodner RD. A biological network in Saccharomyces cerevisiae prevents the deleterious effects of endogenous oxidative DNA damage. Mol Cell. 2005;17:709–720. doi: 10.1016/j.molcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.