Abstract
Mice that accurately model the genetic diversity found in human cancer are valuable tools for interrogating disease mechanisms and investigating novel therapeutic strategies. We performed insertional mutagenesis with the MOL4070LTR retrovirus in Mx1-Cre, KrasG12D mice and generated a large cohort of T lineage acute lymphoblastic leukemias (T-ALLs). Molecular analysis infers that retroviral integration within Ikzf1 is an early event in leukemogenesis that precedes KrasG12D expression and later acquisition of somatic Notch1 mutations. Importantly, biochemical analysis uncovered unexpected heterogeneity, which suggests that Ras signaling networks are remodeled during multistep tumorigenesis. We tested tumor-derived cell lines to identify biomarkers of therapeutic response to targeted inhibitors. Whereas all T-ALLs tested were sensitive to a dual-specificity phosphoinosityl 3-kinase/mammalian target of rapamycin inhibitor, biochemical evidence of Notch1 activation correlated with sensitivity to γ-secretase inhibition. In addition, KrasG12D T-ALLs were more responsive to a MAP/ERK kinase inhibitor in vitro and in vivo. Together, these studies identify a genetic pathway involving Ikzf1, KrasG12D, and Notch1 in T lineage leukemogenesis, reveal unexpected diversity in Ras-regulated signaling networks, and define biomarkers of drug responses that may inform treatment strategies.
Keywords: K-ras, retroviral insertional mutagenesis, T cell leukemia, targeted therapeutics, Ikaros
T lineage acute lymphoblastic leukemia (T-ALL) is characterized by aberrant clonal proliferation and tissue invasion of lymphoblasts (reviewed in ref. 1). Although intensive treatment protocols have markedly improved the outcomes of children and adolescents with T-ALL, cure rates for adults remain below 50%, and the prognosis is poor for patients who relapse at any age (1, 2). Modern treatment regimens also carry a substantial risk of adverse late effects (3). Thus, the development of more effective and less toxic therapies that are based on the underlying molecular lesions is a high priority.
Recent studies have advanced our understanding of T-ALL pathogenesis (reviewed in refs. 4–6). Genes encoding transcription factors, such as TAL1, LYL1, LMO1, and LMO2, are frequently deregulated by chromosomal translocations in T-ALL (7). NOTCH1 was first implicated in leukemogenesis through a t(7;9) chromosomal translocation that truncates and constitutively activates the Notch1 protein (8), and gain-of-function NOTCH1 mutations within the heterodimerization (HD) and/or proline-, glutamic acid-, serine-, and threonine-rich (PEST) domains are found in ≈55% of primary human T-ALL specimens (9). Emerging data also support an important role for aberrant Ras signaling in T-ALL. NRAS and KRAS2 mutations are found in 10–15% of cases (10, 11), whereas the NF1 tumor suppressor gene is inactivated in ≈3% (12). Chromosomal translocations that result in fusions of ABL and JAK2, kinases that are known to activate Ras, are also found in T-ALL (11). More recently, somatic gain-of-function JAK1 mutations were discovered in 18–27% of adult and in 2% of pediatric T-ALL cases, respectively (13, 14). These leukemias demonstrated elevated levels of phosphorylated ERK and Akt, which are important effectors of activated Ras. The PTEN tumor suppressor, which encodes a lipid phosphatase that negatively regulates the phosphoinosityl 3-kinase (PI3K)/Akt signaling pathway, is mutated in 5–8% of T-ALLs, and reduced expression was observed in an additional 17% of cases (15, 16). Recent studies that uncovered PI3K pathway mutations in ≈50% of pediatric T-ALLs underscore the central role of this Ras effector cascade in leukemic growth (17–19).
Observations in mice further implicate hyperactive Ras in T-ALL pathogenesis. Transgenic mice overexpressing Nras or Rasgrp1 develop T lineage lymphomas (20, 21). In addition, thymic lymphomas are observed in ≈30% of mice harboring a latent oncogenic KrasG12D allele that is activated by spontaneous recombination (22). Furthermore, the observation that most of these mice do not develop T-ALL infers that additional mutations are required. Using the IFN-regulated Mx1-Cre transgene to activate a conditional mutant KrasLSL-G12D allele in hematopoietic cells causes an aggressive myeloproliferative disease (MPD) (23, 24). Interestingly, transferring bone marrow from these mice into irradiated recipients results in T-ALL (25, 26), and limit dilution experiments showed that one to three Kras mutant hematopoietic stem cells were sufficient to initiate T-ALL in vivo (26, 27). Consistent with data from other murine T-ALL models, these leukemias acquired somatic Notch1 mutations (4, 25, 26, 28).
Retroviral insertional mutagenesis (RIM) in mice is a robust strategy that has been used to identify genes that are involved in human leukemia, including Ikzf1 (Ikaros), Notch1, and Lmo2 in T-ALL (29–32). We used the MOL4070LTR retrovirus (33) to perform RIM in Mx1-Cre, KrasG12D mice. Here we show that aberrant Ikaros expression due to viral integrations is a frequent early event and that somatic Notch1 mutations arise later and cooperate with oncogenic Kras in leukemogenesis. We generated a large panel of tumor-derived cell lines for biochemical and preclinical studies. Analysis of Ras and Notch1 signaling uncovered unexpected heterogeneity in T-ALL cell lines and in primary leukemias. Exposing T-ALL cells to targeted agents in vitro and in vivo uncovered markers of drug response and revealed synergistic effects of γ-secretase with MAP/ERK kinase (MEK) or PI3K/mammalian target of rapamycin (mTOR) inhibitors. These data demonstrate the value of using diverse panels of related cancers for identifying and ordering mutations, interrogating cancer signaling networks, and discovering molecular markers of drug sensitivity.
Results
MOL4070LTR Induces T-ALL in Mx1-Cre, KrasLSL-G12D Mice.
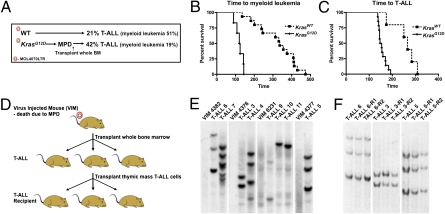
We injected neonatal Mx1-Cre, KrasLSL-G12D mice (KrasG12D) and wild-type littermates (KrasWT) with MOL4070LTR to induce diverse retroviral integrations within the expanding hematopoietic compartment. KrasG12D expression was then activated at 3 weeks of age by administering a single dose of polyinosinic-polycytidilic acid (pIpC). All KrasG12D mice that were infected with MOL4070LTR developed MPD without overt evidence of acute leukemia. We reasoned that the rapid progression of the MPD might provide insufficient time for retrovirally induced hematologic malignancies to emerge. To test this idea, we exploited the fact that the KrasG12D-induced MPD is not transplantable into sublethally irradiated mice (23, 24) and transferred bone marrow cells from 48 moribund KrasG12D mice into 3–5 recipients that received 450 cGy of radiation (Fig. 1A). We refer to bone marrow donors with MPD as “virus-injected mice” throughout this article. Twenty bone marrows from virus-injected mice (42%) induced T-ALL, and nine others (19%) resulted in transplantable myeloid malignancies (Fig. 1A and Fig. S1). By contrast, the frequencies of T-ALL and myeloid malignancies in KrasWT littermates that received MOL4070LTR and were observed for ≈15 months were 21% and 51%, respectively (Fig. 1A). Latency was determined by adding the time to death from MPD in the virus-injected mice and the time to the development of a myeloid malignancy (Fig. 1B) or T-ALL (Fig. 1C) in transplant recipients. KrasG12D expression reduced acute myeloid leukemia latency from 336 to 122 days (P < 0.0001; Fig. 1B) and T-ALL latency from 226 to 151 days (P < 0.001; Fig. 1C).
Fig. 1.
Incidence, latency, and clonality of MOL4070LTR-induced T-ALL. (A) Incidence of T-ALL vs. myeloid leukemia. (B) Time to myeloid leukemia. (C) Time to T-ALL. (D) Overview of T-ALLs generated by transplanting bone marrow from virus-injected mice (VIM) with MPD into primary and secondary recipients. (E) Southern blotting with a MOL4070LTR probes does not reveal a dominant clone in VIM but shows a unique pattern of insertions in each T-ALL. Paired VIM and recipient T-ALLs are separated by space between the blots. (F) Southern blotting demonstrates the same integration pattern in secondary recipients (R1 and R2).
Primary recipients with T-ALL showed a large thymic mass, modest leukocytosis with lymphoblasts visible on blood smears, and extensive infiltration of multiple tissues (Fig. S2). T-ALL cells of thymic origin readily induced T-ALL in secondary recipients (Fig. 1D). KrasG12D and KrasWT T-ALLs from primary and secondary recipient mice are arrested at an immature stage of development, and most express CD4 and CD8. Southern blot analysis of primary T-ALLs revealed a clonal integration pattern that was not detected in the marrows of donor virus-injected mice (Fig. 1 D and E). Interestingly, a single donor bone marrow could initiate independent T-ALLs in different primary recipients (Fig. 1 D and E), which invariably demonstrated a stable pattern of retroviral insertions in secondary transplants (Fig. 1 D and F). Secondary recipients developed aggressive leukemia with reduced latency, bone marrow effacement, and variable infiltration of the thymus.
MOL4070LTR Integrations Disrupt the Ikzf1 Locus in KrasG12D T-ALLs.
We cloned retroviral integration sites from 24 KrasG12D T-ALLs and 6 KrasWT T-ALLs (Table S1). The criteria used to define common insertion sites (CIS) were (i) genes must be identified in at least two independent tumors; (ii) an individual integration must be recovered more than five times (of 192 independent sequenced clones per T-ALL); and (iii) one integration must be within 30 kb upstream/downstream of the identified gene. Ikzf1 (Ikaros) was the most frequent CIS (n = 9) and was restricted to KrasG12D tumors. All nine integrations were within the Ikzf1 gene, and most were predicted to disrupt Ikaros function (Table S1). Consistent with this, RT-PCR analysis of leukemias with Ikzf1 integrations revealed aberrant cDNA expression. Western blotting demonstrated absence of the high-molecular-weight Ikaros protein, which is associated with full biologic activity, and expression of truncated dominant negative Ikaros isoforms (Fig. S3A). Interestingly, whereas KrasG12D T-ALLs without Ikzf1 insertions showed robust expression of the high-molecular-weight form of Ikaros (n = 3), thymus lysates from KrasG12D mice without T-ALL unexpectedly showed reduced expression of this isoform (Fig. S3A). We performed a retroviral transduction/transplantation experiment to assess the effect of expressing a dominant negative Ikaros isoform in Kras mutant bone marrow cells (32, 34). Recipient mice that were transplanted with these cells developed T-ALL (Fig. S3B).
We identified three other CIS including Rasgrp1 integrations, which occurred in three of six of the KrasWT tumors and are likely to be functionally redundant with KrasG12D expression (Table S1).
Somatic Notch1 Mutations in MOL4070LTR-Induced T-ALLs.
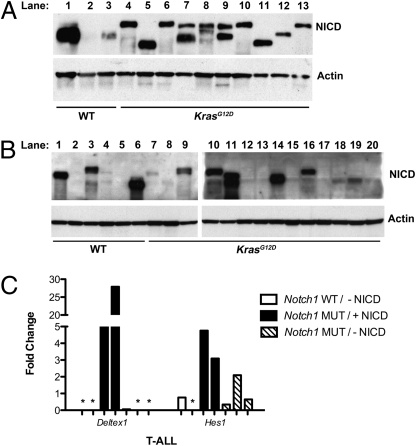
Western blotting with an antibody that detects the Notch1 intracellular domain (NICD) revealed evidence of pathway activation in 90% of primary KrasG12D T-ALLs (Fig. 2A), and sequence analysis identified truncating PEST domain mutations that are similar to those observed in human T-ALL (Table S2). We detected Notch1 mutations in all 30 KrasG12D T-ALLs examined. By contrast, only 3 of 9 KrasWT T-ALLs (33%) contained Notch1 mutations. Similarly, all 11 KrasG12D T-ALL cell lines shown in Table 1 carried Notch1 mutations, compared with 1 of 5 KrasWT lines. As expected, each cell line harbored the same mutation as the parental T-ALL. Interestingly, however, we observed variable levels of NICD in T-ALL cell lines with Notch1 mutations, including a number in which NICD was not visualized by Western blotting (Fig. 2B, lanes 12, 13, 15, and 17, and Table 1).
Fig. 2.
Notch1 is deregulated in MOL4070LTR-induced T-ALL. Western blot analysis of NICD expression in (A) primary T-ALLs and (B) T-ALL cell lines. (C) Expression of Deltex1 and Hes1 were assessed by quantitative RT-PCR in cell lines without a Notch1 mutation or detectable NICD by Western blotting (Notch1 WT/-NICD; n = 2); with a Notch1 mutation and detectable NICD (Notch1 MUT/+NICD; n = 2); and with a Notch1 mutation but not detectable NICD (Notch1 MUT/-NICD; n = 3). Asterisks (*) indicate undetectable levels.
Table 1.
Characteristics of T-ALL cell lines
| Primary tumor | Cell line ID | KRasG12D genotype | PD0325901 sensitivity | Notch1 mutation | NICD detected | GSI sensitivity | Ikaros status |
| 7 | 7 | + | +++ | + | ++ | + | WT |
| 9 | 9A | + | +++ | + | ++ | +++ | DomNeg |
| 12 | 12 | + | +++ | + | ++ | + | Null |
| 6 | 6 | + | +++ | + | + | +++ | DomNeg |
| 9 | 9B | + | +++ | + | ++ | +++ | DomNeg |
| 17 | 17 | + | ++ | + | + | + | Null |
| 15 | 15 | + | +++ | + | + | ++ | DomNeg |
| 3 | 3B | + | +++ | + | — | — | WT |
| 29 | 29 | + | +++ | + | — | — | WT |
| 3 | 3A | + | +++ | + | — | — | DomNeg |
| 22 | 22 | + | ++ | + | — | — | DomNeg |
| C1 | C1 | — | + | + | +++ | ++ | Null |
| C3 | C3 | — | — | — | + | ++ | DomNeg |
| C7 | C7 | — | + | — | — | — | WT |
| C2 | C2 | — | — | — | ++ | +++ | Null |
| 11C | 11C | — | — | — | — | — | WT |
To further assess the relationship of Notch1 mutations and NICD protein expression to Notch pathway activation, we used quantitative real-time PCR to assay the Notch target genes Deltex1 and Hes1 in seven T-ALL cell lines (Fig. 2C). Elevated Deltex1 expression was observed in two cell lines in which NICD was visualized by Western blotting but not in three lines with Notch1 mutations that lacked NICD (Fig. 2C). As expected, Deltex1 expression was not detected in two T-ALL cell lines without Notch1 mutations (Fig. 2C). Although less dramatic, Hes1 expression demonstrated a similar overall pattern, with the highest levels observed in the two cell lines with detectable NICD (Fig. 2C). Together, these data support the idea that NICD is a biochemical marker of Notch pathway activation in T-ALL cell lines.
The coexistence of retroviral integrations and Notch1 mutations in individual T-ALLs allowed us to ascertain the likely order in which these genetic changes were acquired. The outcome shown in Fig. 1 D and E, in which a single virus-injected KrasG12D bone marrow gave rise to genetically distinct T-ALLs, was especially informative. In these cases, each T-ALL carried a different Notch1 mutation. Further analysis using allele-specific PCR did not detect Notch1 mutations in the bone marrows of virus-injected mice (Fig. S4). On the basis of the sensitivity of these assays, it is formally possible that ≈200 or fewer cells of the 2 × 106 that were transplanted could have harbored a Notch1 mutation. Together, these studies provide strong evidence for sequential acquisition of Ikzf1, Kras, and Notch1 mutations during leukemogenesis and support the idea that Notch1 mutations are acquired after transplantation.
Signaling in T-ALL Cell Lines.
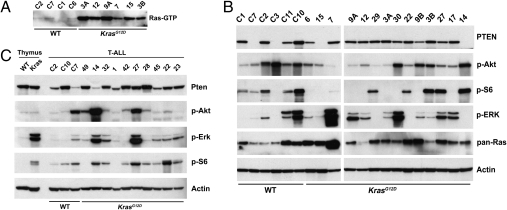
We interrogated Ras signaling in cell lines generated from KrasWT and KrasG12D T-ALLs (Table 1). K-RasG12D accumulates in the active, GTP-bound conformation and exerts its oncogenic effects, in part, by deregulating the Raf1/MEK/ERK and PI3K/Akt/mTOR/S6 effector pathways. KrasG12D T-ALL cell lines uniformly showed markedly elevated levels of Ras-GTP compared with lines generated from KrasWT leukemias (Fig. 3A). However, the levels of phosphatase and tensin homolog (PTEN) and of phosphorylated ERK, Akt, and S6 (p-ERK, p-Akt, and p-S6) were highly variable in cell lines of both Kras genotypes, and changes in “upstream” pathway components were not highly correlated with downstream proteins. For example, absent or reduced PTEN expression was not uniformly associated with increased p-Akt and p-S6 levels (Fig. 3B). Western blot analysis also revealed variable levels of p-ERK, p-Akt, p-S6, and PTEN in primary T-ALLs (Fig. 3C). By contrast, KrasG12D expression in untransformed primary thymocytes consistently induced ERK and S6 activation (Fig. 3C). The striking biochemical heterogeneity in KrasG12D T-ALLs indicates that oncogenic K-RasG12D signaling is modulated in the context of multistep tumorigenesis.
Fig. 3.
Ras signaling in T-ALL. (A) Ras-GTP levels are elevated in KrasG12D cell lines deprived of serum for 16 h. (B and C) PTEN p-Akt, p-S6, and p-ERK levels in cell lines under normal growth conditions (B) and in primary T-ALLs (C).
Sensitivity of KrasG12D and KrasWT T-ALL Cell Lines to Chemical Inhibitors.
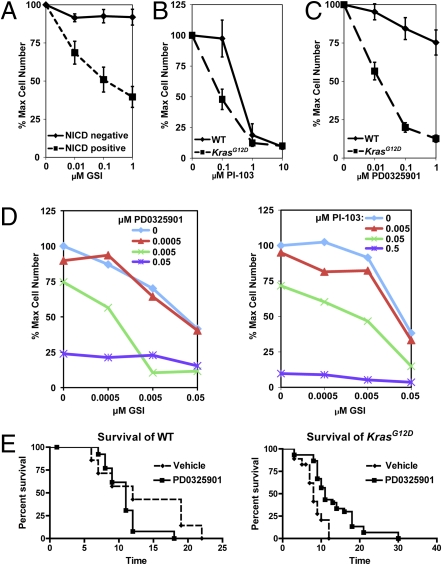
Notch1 activation requires cleavage by the γ-secretase protease. Importantly, because many T-ALL–associated Notch1 mutant proteins remain dependent on γ-secretase for biologic activity, treatment with γ-secretase inhibitors (GSIs) is a rational therapeutic strategy for T-ALL (4). We investigated the effects of Compound E, a potent GSI, on the proliferation of T-ALL cell lines. These studies revealed variable sensitivity, which correlated poorly with the presence of a mutation in either Notch1 or Kras (Table 1). However, 10 cell lines in which NICD was detected by Western blotting were sensitive to Compound E, whereas none of 6 lines without visible NICD were sensitive (Table 1, Fig. 4A, and Fig. S5).
Fig. 4.
Effects of small molecule inhibitors on T-ALL cell lines and primary tumors. Cell lines were grown for 5–7 days in the presence of each compound, and the number of viable cells was determined. (A) Effects of Compound E, a GSI. The solid line displays the growth of NICD negative cell lines (n = 6), and dashed line shows NICD positive lines (n = 10; P = 0.04–<0.001). (B) Effects of PI-103, a dual-specificity PI3K/mTOR inhibitor. KrasWT cell lines are summated in the solid line (n = 2), and KrasG12D cell lines are shown as a dashed line (n = 8; P = 0.03–0.8). (C) Effects of PD0325901, a MEK inhibitor. The KrasWT cell lines are shown in the solid line (n = 5), and the KrasG12D cells are presented in the dashed line (n = 11; P = 0.0009–<0.0001). Error bars represent SEM. (D) Combined inhibitor treatment. A cell line was grown in increasing concentrations of both (Left) GSI and PD0325901 or (Right) GSI and PI-103. The blue line (GSI alone) shifts to the left (green and purple lines) when combined with PD0325901 or PI-103, thereby reducing the IC50 of GSI. (E) PD0325901 prolongs survival in mice transplanted with KrasG12D T-ALLs. Mice were transplanted with three independent primary KrasWT (Left) or six independent KrasG12D (Right) T-ALLs and were treated with vehicle (dashed line) or PD0325901 (solid line). There was no significant difference in the survival (in days) of mice engrafted with KrasWT leukemias (P = 0.13). In contrast, PD0325901 treatment increased survival in recipients of KrasG12D T-ALLs (P < 0.0005). KrasWT vehicle n = 7, KrasWT PD0325901 n = 13, KrasG12D vehicle n = 15, and KrasG12D PD0325901 n = 30.
We also evaluated the effects of the dual-specificity PI3K/mTOR inhibitor PI-103 (35) on the proliferation of T-ALL cell lines and found that all were sensitive to 1 μM of this drug, irrespective of Kras genotype or pathway activation as determined by Western blotting (Figs. 3B and 4B, Table 1, and Fig. S6 A and B). By contrast, there was substantial variability in the responses of T-ALL cell lines to the MEK inhibitor PD0325901. Interestingly, KrasG12D T-ALL cell lines showed enhanced sensitivity to PD0325901, which was independent of p-ERK levels (Figs. 3B and 4C, Table 1, and Fig. S6 C and D).
Most cell lines that were sensitive to Compound E were also sensitive to PD0325901 (Table 1). We therefore examined whether either inhibitor influences both targets. However, PD325901 had no effect on cleaved Notch1 levels, and Compound E did not reduce p-ERK levels (Fig. S7). The finding that many of the same cell lines were sensitive to PD0325901 and Compound E suggested that blocking both pathways might lead to an enhanced therapeutic effect. Indeed, there is an additive effect on proliferation when these inhibitors are combined at low doses (Fig. 4D and Fig. S8A). Similarly, exposing cells lines to both PI-103 and Compound E increased the inhibitory effects of either drug alone (Fig. 4D and Fig. S8B).
PD0325901 Treatment of Primary T-ALLs.
To test the prediction that Kras genotype might influence MEK dependence in vivo, we established cohorts of eight recipient mice that were transplanted with each of six primary KrasG12D or three KrasWT T-ALLs (n = 72 total recipients). Five mice in each group were randomly assigned to treatment with PD0325901, and three received control vehicle. The mice were treated for 4 weeks or until death. Consistent with data from the T-ALL cell lines, therapeutic response to MEK inhibition was associated with the KrasG12D genotype (Fig. 4E). Whereas treatment with PD0325901 was associated with a significant increase in median survival in recipients that were injected with KrasG12D T-ALLs (P > 0.0005), this inhibitor had no beneficial effect on the KrasWT T-ALLs (Fig. 4E). As in human cancer, there was considerable variability among individual T-ALLs with respect to aggressiveness and degree and duration of response.
Discussion
We dissected the molecular events involved in T-ALL pathogenesis and tested potential therapeutic strategies in a diverse panel of T-ALLs generated by RIM. Ikaros is essential for lymphoid development and is mutated or aberrantly expressed in human T and B lineage ALL (36, 37). Our data implicating aberrant Ikaros expression as an initiating event in T lineage leukemia are consistent with previous studies in genetically engineered mice (38). Beverly and Copabianco (32) performed a RIM screen in transgenic mice that express a potent NICD allele (NIC mice) and recovered Ikzf1 as a CIS. They also showed that expressing mutant Ikaros proteins in NIC bone marrow rapidly induced T-ALL in irradiated recipients (32). Subsequent studies also revealed somatic Notch1 PEST domain mutations in Ikaros transgenic mice (39, 40).
In contrast to previous RIM studies (30, 41), we found that Notch1 activation invariably resulted from PEST domain mutations rather than from retroviral integrations within the Notch1 locus. A study in which Chiang et al. (28) used a transduction/transplantation system to compare a series of Notch1 mutant alleles suggests an explanation for this finding. These authors showed that truncated Notch1 proteins generated as a result of chromosomal translocations or retroviral integrations within the Notch1 gene are much more potent than PEST domain mutations. These data raise the possibility that truncated Notch1 alleles may be initiating or early events in T-ALL, whereas biologically “weaker” HD and PEST domain mutations may be obligate cooperating events (4, 28). Our data support this hypothesis. We identified Ikzf1 disruption as an early event that cooperates strongly with subsequent oncogenic Kras expression in T lineage leukemogenesis (Fig. 5A). Furthermore, RT-PCR analysis of independent T-ALLs that emerged from the same virus-injected mouse provided strong evidence that Notch1 mutations are acquired after transplantation (Fig. 5A). On the basis of these results, we hypothesize that antecedent Ikzf1 and Kras mutations create a field of thymocytes with aberrant proliferation, survival, and/or differentiation that only require a Notch1 PEST domain mutation to induce full leukemic transformation.
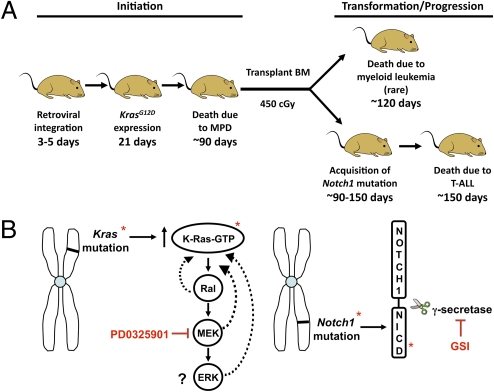
Fig. 5.
Models. (A) RIM-induced T-ALL in KrasG12D mice. Pups are injected with MOL4070LTR at 3–5 days of age, followed by induction of KrasG12D expression at 21 days. Virus-injected mice (VIM) die of KrasG12D-induced MPD at ≈90 days. Bone marrow is then transplanted into sublethally irradiated recipients. Myeloid malignancies develop with short latency. By contrast, an individual VIM can give rise to multiple independent T-ALLs with different retroviral insertion sites and Notch1 mutations. (B) Relationship of MEK and γ-secretase inhibitor responses to mutations found in T-ALL. (Left) The pharmacologic target of PD0325901 (MEK) is downstream of oncogenic K-Ras-GTP, which allows for negative feedback and remodeling (dotted lines). As a result, ERK phosphorylation may not accurately reflect pathway activation and/or dependence. (Right) Notch1 PEST domain mutations result in the production of mutant Notch1 proteins that remain dependent on γ-secretase for proteolytic cleavage and biologic activity. NICD levels therefore provide a direct biochemical readout of the deleterious effects of a Notch1 mutation, and the ability of GSI treatment to reduce these levels is a robust marker of drug response.
The combinatorial effects of Ikzf1, KrasG12D, and Notch1 mutations on cellular signaling networks are unknown, and molecular markers of responses to targeted agents have not been defined. Whereas K-RasG12D protein expression resulted in markedly elevated Ras-GTP levels, we unexpectedly observed great variability in the activation of downstream effectors. It is possible that T lymphoid cells adapt to the stress of oncogenic K-Ras expression by modulating effector pathways and that cooperating mutations either antagonize or potentiate this adaptive response. Alternatively, mutations that initiate T-ALL could partially dictate the biochemical consequences of a subsequent Ras gene mutation. These potential mechanisms are not mutually exclusive, and our data demonstrate the profound biochemical complexity of primary cancers that arise because of multiple interacting mutations.
Our analysis of a large panel of murine T-ALL cell lines uncovered a complex pattern of GSI response that is consistent with the heterogeneous responses of human T-ALL lines (9). We unexpectedly found a poor correlation between the presence of a Notch1 mutation and GSI sensitivity and instead identified NICD expression as a highly predictive biomarker of drug response. T-ALL clones that acquire Notch1 mutations in vivo uniformly express NICD (Fig. 2A); however, NICD expression is no longer detected in many tumor-derived cell lines (Fig. 2B, lanes 12, 13, 15, and 17). This apparent paradox is not due to widespread clonal infidelity, given that Southern blot analysis of paired primary T-ALLs and tumor-derived cell lines revealed the same pattern of MOL4070LTR integrations. A plausible explanation for the loss of NICD expression in many T-ALL cell lines is that aberrant Notch1 signaling is required for leukemic growth in vivo but is dispensable in vitro. If this is true, data from preclinical trials of agents targeting Notch1 signaling that use human and murine cell lines should be interpreted with caution. Our observation that T-ALL cell lines with Notch1 mutations can survive and proliferate in the absence of NICD expression also suggests outgrowth of a Notch1-indpendent clone as a potential mechanism of acquired GSI resistance.
Despite variable activation of downstream effector pathways in individual T-ALLs, we found that a Kras mutant genotype correlated with sensitivity to the MEK inhibitor PD0325901. This is in contrast to our experience with GSI treatment, whereby a biochemical marker (NICD expression) predicted drug sensitivity but the presence of a Notch1 mutation did not. The relationship between the tumor-associated mutation and the protein targets of PD0325901 and Compound E may explain this difference (Fig. 5B). In cancers with oncogenic Kras mutations that are treated with a MEK inhibitor, the biochemical target is a downstream component of a signaling cascade, which is subject to extensive feedback regulation that may dramatically alter p-ERK levels. Despite this modulation, our data indicate that the Kras mutant T-ALL cells remain dependent on MEK. By contrast, GSIs interfere with the production of a protein that is encoded by mutant Notch1. As such, NICD levels provide a biomarker that directly reflects the functional output of the mutant allele. It follows that growth of T-ALL cells that no longer contain NICD would be insensitive to GSI treatment.
Our data have implications for understanding the biology of T-ALL and improving the treatment of this aggressive cancer. Mutations or rearrangements in KRAS, NRAS, NF1, PTEN, PIK3R1, PIK3CA, AKT, ABL, and JAK1 occur in ≈60% of T-ALL (10–13, 15–19), and identifying mutations in other signaling proteins may uncover new therapeutic targets. The uniform sensitivity of our diverse panel of cell lines to a PI3K inhibitor is consistent with emerging genetic data implicating deregulated PI3K/PTEN/Akt signaling as playing a pivotal role in T lineage leukemogenesis (15–19). Together, these studies and the observed synergy between Compound E and PI-103 support initiating preclinical and clinical trials of PI3K pathway inhibitors—both alone and in combination with GSIs—in T-ALL.
We have identified NICD expression as a potential biomarker for assessing clinical responsiveness to GSIs. This may be particularly relevant in two contexts. First, human T-ALLs that do not express elevated levels of NICD at diagnosis may not respond to GSI treatment irrespective of NOTCH1 mutation status. Second, our data support the idea that human T-ALLs with elevated levels of NICD are likely to respond to GSI treatment, and that this will be associated with reduced NICD levels. An intriguing question is whether tumors that subsequently relapse will reactivate NICD expression even in the face of GSI exposure or will acquire the capacity to proliferate without NICD. Studies of this nature would shed light on potential mechanisms of acquired drug resistance. On the other hand, our observation that tumor genotype (i.e., KrasG12D) was a more important determinant of response to the MEK inhibitor PD0325901 than the biochemical readout of p-ERK levels suggests that cancer cells can remain dependent on a specific upstream mutation despite heterogeneous effects on pathway components. These data, in turn, infer that it will be important to compare potential biochemical and genetic biomarkers to predict how cancers will respond to drugs that do not directly target mutant oncoproteins. Insertional mutagenesis is a powerful strategy for approaching the genetic and biochemical complexity of human cancer, for harnessing “chemical genetic” strategies to correlate tumor biology with therapeutic responses, and for generating diverse reagents for preclinical trials of specific agents that can inform human treatment protocols.
Materials and Methods
Mice.
All animal experiments conformed to national regulatory standards and were approved by the University of California, San Francisco Committee on Animal Research. C57Bl6/129Sv F1 pups were injected with MOL4070LTR intraperioneally at 3–5 days of age and with pIpC at 21 days of age. For adoptive transfer of acute leukemias, 2 × 106 cells were injected retroorbitally into 8–12 week old wild-type C57Bl6/129Sv F1 recipient mice that had been irradiated with 450 cGy.
Southern Analysis.
Genomic DNA was digested with HindIII, followed by electrophoresis and capillary transfer to Hybond-N filters (Amersham). Filters were hybridized with a MOL4070 LTR probe that was labeled with radioactive α-dCTP using Rediprime II (Amersham).
T-ALL Cell Lines.
Single-cell suspensions were plated in lymphocyte media (DME-H21, 20% FBS, 10 mM Hepes, penicillin/streptomycin, L-glutamine, 50 μM β-mercaptoethanol, nonessential amino acids, sodium pyruvate, 10 ng/mL IL-2, and 10 ng/mL IL-7). For proliferation assays, 3–5 × 105 cells were plated per well in a 24-well plate in lymphocyte media plus drug or DMSO (vehicle). After a minimum of four cell doublings in vehicle wells, cells were counted on a Vi-Cell XR (Beckman-Coulter). Cell counts were then normalized to vehicle treated cells.
Preclinical Studies.
T-ALL transplant recipients with evidence of leukemia were randomly assigned to receive PD0325901 at a dose of 5 mg/kg per day by oral gavage or a control vehicle. Mice were weighed weekly to adjust the dose and observed daily during treatment (28 days).
Other Methods.
Procedures for Shotgun cloning, retroviral transduction and adoptive transfer, Western blotting, Notch1 mutation analysis, and quantitative RT-PCR can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Judith Leopold (Pfizer) for providing PD0325901 and David Tuveson and Tyler Jacks for providing KrasG12D mice; Charles Mullighan for the Murine Stem Cell Virus Ik6 construct and for helpful comments and suggestions; Jon Aster and Michelle Hermison for insightful comments; and Michelle Kang for technical guidance. This work was supported by National Institutes of Health Grants U01 CA84221, R37 CA72614, and K08 CA119105, by a Specialized Center of Research award from the Leukemia and Lymphoma Society of America (LLSA 7019-04), by the William Lawrence and Blanche Hughes Foundation, the Jeffrey and Karen Peterson Family Foundation, and by the Frank A. Campini Foundation. S.C.K. is an LLSA Scholar, and M.D. is a supported by an LLSA Fellowship. The Intramural Research Program of the National Cancer Institute Center for Cancer Research supports research in the laboratory of L.W. at the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001064107/DCSupplemental.
References
- 1.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 2.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354:166–178. doi: 10.1056/NEJMra052603. [DOI] [PubMed] [Google Scholar]
- 3.Bhatia S, Sklar C. Second cancers in survivors of childhood cancer. Nat Rev Cancer. 2002;2:124–132. doi: 10.1038/nrc722. [DOI] [PubMed] [Google Scholar]
- 4.Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armstrong SA, Look AT. Molecular genetics of acute lymphoblastic leukemia. J Clin Oncol. 2005;23:6306–6315. doi: 10.1200/JCO.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 6.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–390. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 7.O'Neil J, Look AT. Mechanisms of transcription factor deregulation in lymphoid cell transformation. Oncogene. 2007;26:6838–6849. doi: 10.1038/sj.onc.1210766. [DOI] [PubMed] [Google Scholar]
- 8.Ellisen LW, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 9.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 10.Wiemels JL, et al. RAS mutation is associated with hyperdiploidy and parental characteristics in pediatric acute lymphoblastic leukemia. Leukemia. 2005;19:415–419. doi: 10.1038/sj.leu.2403641. [DOI] [PubMed] [Google Scholar]
- 11.Graux C, Cools J, Michaux L, Vandenberghe P, Hagemeijer A. Cytogenetics and molecular genetics of T-cell acute lymphoblastic leukemia: From thymocyte to lymphoblast. Leukemia. 2006;20:1496–1510. doi: 10.1038/sj.leu.2404302. [DOI] [PubMed] [Google Scholar]
- 12.Balgobind BV, et al. Leukemia-associated NF1 inactivation in patients with pediatric T-ALL and AML lacking evidence for neurofibromatosis. Blood. 2008;111:4322–4328. doi: 10.1182/blood-2007-06-095075. [DOI] [PubMed] [Google Scholar]
- 13.Flex E, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205:751–758. doi: 10.1084/jem.20072182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong EG, et al. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin Cancer Res. 2008;14:3716–3721. doi: 10.1158/1078-0432.CCR-07-4839. [DOI] [PubMed] [Google Scholar]
- 15.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13:1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maser RS, et al. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gutierrez A, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Remke M, et al. High-resolution genomic profiling of childhood T-ALL reveals frequent copy-number alterations affecting the TGF-beta and PI3K-AKT pathways and deletions at 6q15-16.1 as a genomic marker for unfavorable early treatment response. Blood. 2009;114:1053–1062. doi: 10.1182/blood-2008-10-186536. [DOI] [PubMed] [Google Scholar]
- 19.Larson Gedman A, et al. The impact of NOTCH1, FBW7 and PTEN mutations on prognosis and downstream signaling in pediatric T-cell acute lymphoblastic leukemia: A report from the Children's Oncology Group. Leukemia. 2009;23:1417–1425. doi: 10.1038/leu.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haupt Y, Harris AW, Adams JM. Retroviral infection accelerates T lymphomagenesis in E mu-N-ras transgenic mice by activating c-myc or N-myc. Oncogene. 1992;7:981–986. [PubMed] [Google Scholar]
- 21.Klinger MB, Guilbault B, Goulding RE, Kay RJ. Deregulated expression of RasGRP1 initiates thymic lymphomagenesis independently of T-cell receptors. Oncogene. 2005;24:2695–2704. doi: 10.1038/sj.onc.1208334. [DOI] [PubMed] [Google Scholar]
- 22.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 23.Braun BS, et al. Somatic activation of oncogenic Kras in hematopoietic cells initiates a rapidly fatal myeloproliferative disorder. Proc Natl Acad Sci USA. 2004;101:597–602. doi: 10.1073/pnas.0307203101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan IT, et al. Conditional expression of oncogenic K-ras from its endogenous promoter induces a myeloproliferative disease. J Clin Invest. 2004;113:528–538. doi: 10.1172/JCI20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kindler T, et al. K-RasG12D-induced T-cell lymphoblastic lymphoma/leukemias harbor Notch1 mutations and are sensitive to gamma-secretase inhibitors. Blood. 2008;112:3373–3382. doi: 10.1182/blood-2008-03-147587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, et al. Oncogenic Kras-induced leukemogeneis: Hematopoietic stem cells as the initial target and lineage-specific progenitors as the potential targets for final leukemic transformation. Blood. 2009;113:1304–1314. doi: 10.1182/blood-2008-01-134262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabnis AJ, et al. Oncogenic Kras initiates leukemia in hematopoietic stem cells. PLoS Biol. 2009;7:e59. doi: 10.1371/journal.pbio.1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chiang MY, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–3194. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girard L, et al. Frequent provirus insertional mutagenesis of Notch1 in thymomas of MMTVD/myc transgenic mice suggests a collaboration of c-myc and Notch1 for oncogenesis. Genes Dev. 1996;10:1930–1944. doi: 10.1101/gad.10.15.1930. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki T, et al. New genes involved in cancer identified by retroviral tagging. Nat Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 31.Yanagawa S, et al. Identification of Notch1 as a frequent target for provirus insertional mutagenesis in T-cell lymphomas induced by leukemogenic mutants of mouse mammary tumor virus. J Virol. 2000;74:9786–9791. doi: 10.1128/jvi.74.20.9786-9791.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beverly LJ, Capobianco AJ. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell. 2003;3:551–564. doi: 10.1016/s1535-6108(03)00137-5. [DOI] [PubMed] [Google Scholar]
- 33.Wolff L, Koller R, Hu X, Anver MR. A Moloney murine leukemia virus-based retrovirus with 4070A long terminal repeat sequences induces a high incidence of myeloid as well as lymphoid neoplasms. J Virol. 2003;77:4965–4971. doi: 10.1128/JVI.77.8.4965-4971.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 35.Knight ZA, et al. A pharmacological map of the PI3-K family defines a role for p110alpha in insulin signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullighan CG, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 37.Rebollo A, Schmitt C. Ikaros, Aiolos and Helios: Transcription regulators and lymphoid malignancies. Immunol Cell Biol. 2003;81:171–175. doi: 10.1046/j.1440-1711.2003.01159.x. [DOI] [PubMed] [Google Scholar]
- 38.Georgopoulos K. Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 39.Dumortier A, et al. Notch activation is an early and critical event during T-Cell leukemogenesis in Ikaros-deficient mice. Mol Cell Biol. 2006;26:209–220. doi: 10.1128/MCB.26.1.209-220.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantha S, et al. Activating Notch1 mutations are an early event in T-cell malignancy of Ikaros point mutant Plastic/+ mice. Leuk Res. 2007;31:321–327. doi: 10.1016/j.leukres.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 41.Uren AG, et al. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.