Abstract
Cisplatin is one of the most commonly used anticancer drugs. It kills cancer cells by damaging their DNA, and hence cellular DNA repair capacity is an important determinant of its efficacy. Here, we investigated the repair of cisplatin-induced DNA damage in mouse liver and testis tissue extracts prepared at regular intervals over the course of a day. We find that the XPA protein, which plays an essential role in repair of cisplatin damage by nucleotide excision repair, exhibits circadian oscillation in the liver but not in testis. Consequently, removal of cisplatin adducts in liver extracts, but not in testis extracts, exhibits a circadian pattern with zenith at ∼5 pm and nadir at ∼5 am. Furthermore, we find that the circadian oscillation of XPA is achieved both by regulation of transcription by the core circadian clock proteins including cryptochrome and by regulation at the posttranslational level by the HERC2 ubiquitin ligase. These findings may be used as a guide for timing of cisplatin chemotherapy.
Keywords: chemotherapy, circadian clock, DNA repair
Chronochemotherapy is the administration of chemotherapeutic drugs at specific times of the day so as to optimize efficacy and minimize side effects of the drug (1, 2). Cisplatin is one of the three most commonly used chemotherapeutic drugs (3) for which chronotherapy is thought to have some beneficial effects. Factors that modulate the efficacy of cisplatin therapy include drug uptake and efflux, DNA adduct formation, DNA repair, and cellular proliferation (4, 5). Cisplatin produces DNA intra- and interstrand diadducts and DNA–protein crosslinks (4), and it is well established that the intrastrand diadducts Pt-(GpG), Pt-(ApG), and Pt-(GpXpG), that constitute up to 90% of the total DNA lesions, are the main cause of its cytotoxicity and hence its therapeutic effects. These lesions are removed exclusively by nucleotide excision repair (excision repair) in mammalian cells and hence the status of excision repair is an important factor in the success of chemotheraphy with cisplatin (4, 6).
In humans and mice, excision repair is carried out by the coordinated action of six core repair factors, RPA, XPA, XPC, TFIIH, XPG, and XPF-ERCC1, which remove the damage in the form of 24–32 nt-long oligomers; the resulting gap is filled by DNA polymerases and ligated (7–9).
Recently, we found that the rate of excision repair of a UV photoproduct in the mouse brain exhibits a daily rhythm (10). Furthermore, it appears that this rhythmic pattern is due to the circadian (circa = about, dies = day) oscillation of the XPA (xeroderma pigmentosum A) protein that is one of the rate-limiting factors in excision repair. Even though in that study the damaged-DNA substrate was a UV photoproduct, we suggested that the findings were relevant to the repair of cisplatin because nucleotide excision repair is the only repair system capable of removing bulky DNA lesions produced by UV or by UV-mimetic agents such as cisplatin (11). Although cisplatin is used for treating certain brain cancers, the blood–brain barrier is a serious impediment for its general use in brain tumors (12). In contrast, cisplatin is the drug of choice in testicular and ovarian cancers and a main component of combination therapy regimens for many other cancers, including head and neck, lung, gastric, and colorectal cancers (3). Therefore, in this study we have tested the effect of the circadian clock on the repair capacity of cisplatin–DNA adducts in peripheral organs with the aim of providing a guide for cisplatin chronochemotheraphy. We chose mouse liver and testis for our experiments for practical reasons, because extracts from these organs are of relatively good quality for biochemical assays. We find that excision repair of cisplatin–DNA adducts in mouse liver exhibits a robust circadian rhythm with the zenith in the late afternoon hours (∼5 pm) and the nadir in the early morning hours (∼5 am). We further show that this oscillation is caused by the circadian rhythmicity of Xpa transcription and translation, coupled with a short half-life of the protein. In contrast, in the testis, which is known to lack a circadian rhythm (13–15), Xpa transcription, XPA protein levels, and excision activity remained constant and at a moderately high level over the course of the day.
The circadian clock in mice and humans is generated by a transcriptional-translational feedback loop: The Clock and BMal1 transcriptional activators bind to the promoters of Cryptochrome 1 and 2 (Cry1/2) and Period 1 and 2 (Per1/2) repressor genes and activate their transcription. The Cry and Per proteins, in turn, enter the nucleus after a time lag, bind to Clock and BMal1, and inhibit their own transcription as well as the transcription of clock-controlled output genes (16). This core clock circuitry is consolidated by secondary feedback loops at the transcriptional level as well as by posttranslational modifications and proteolytic degradation of the core clock proteins. In particular, the ubiquitination of Cry proteins by Fbxl3 (17–19) and of Per proteins by βTRCP (20, 21) and subsequent proteolysis of these transcriptional repressors by the proteasome are essential for a robust core clock as well as robust oscillations of the clock-controlled genes.
We and others previously reported that the XPA protein and transcript (and the excision repair activity for UV photoproducts) were controlled by the circadian clock in the mouse brain (10, 22). Here, we have directly measured the Xpa transcript levels in the liver and testis and found that Xpa mRNA exhibits circadian rhythm in liver, and this oscillation is dependent on the circadian clock. In addition, since solely altering the transcription of a gene may not be sufficient to confer an oscillatory mode for its activity profile unless the gene product has a relatively short lifetime, we determined the half-life of XPA protein in cultured cells and found it to be similar to those of Cry1/2 that are known to be processed specifically by Fbxl3 ubiquitin ligase. We reasoned that XPA may also be a target of a specific E3 ligase. We found that the HERC2 protein that, based on sequence data, is classified as a putative E3 ligase of the HECT class (23), binds specifically to XPA and ubiquitinates it. Importantly, downregulation of HERC2 leads to stabilization of XPA and increased repair activity of cisplatin damage in cultured cells. Similarly, in Cry1/2 double knockout mice (CryDKO), the Xpa transcript and protein levels cease to oscillate, and the liver extracts exhibit high levels of cisplatin excision activity at all tested time points. In contrast, Cry mutations have no effect on Xpa transcript and protein levels in the testis and hence no effect on cisplatin repair activity over the course of the day. Taken together, our study provides some mechanistic foundation for cisplatin chronochemotherapy for the vast majority of organs that contain a strong circadian regulatory component.
Results
Effect of Circadian Time on Excision Repair of Cisplatin–DNA Adducts by Mouse Liver Extracts.
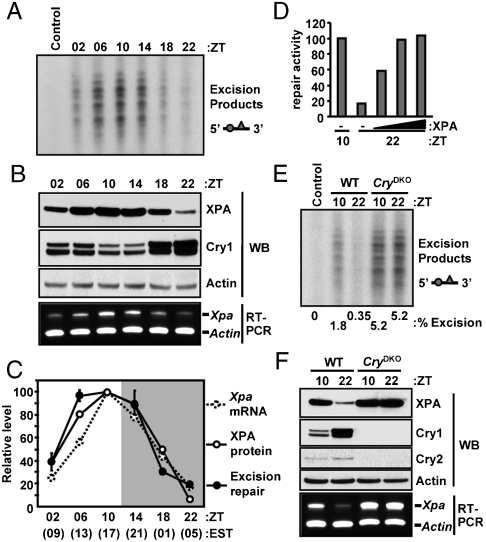
Cisplatin, and its second and third generation derivatives, make two major DNA adducts, Pt-(GpG) and Pt-(GpTpG), in addition to the much less frequent G-Pt-G interstrand crosslink (3). Extensive data indicate that the intrastrand diadducts are the major cause of cytotoxicity and that nucleotide excision repair is the sole repair system for these adducts (4). Further, even though the Pt-(GpG) adduct is more abundant than the Pt-(GpTpG) adduct, the latter is repaired at about fivefold faster rate than Pt-(GpG) (7, 24). Therefore, we used either linear or circular DNA molecules containing a single Pt-(GpTpG) to assess the repair capacity of mouse tissue extracts. Fig. 1 shows the excision assays conducted with mouse liver extracts harvested at 4 h intervals over a period of 24 h (ZT0 light on and ZT12 light off for mice maintained under 12 h light: 12 h dark schedule). As apparent from the primary data in Fig. 1A and B and the quantitative analysis in Fig. 1C, the repair of cisplatin-(GpTpG) and the Xpa transcript and protein levels exhibited robust and in-phase daily oscillation in the liver. Moreover, the phases of these oscillations were antiphase to the clock transcriptional repressor Cry1 and in synchrony with those of excision activity and XPA oscillation previously reported in mouse brain (10). Also, as previously demonstrated with the mouse brain extracts, supplementing the liver extracts from the nadir of excision activity (ZT22 or 5 am) with recombinant XPA, restored the level of excision repair to that observed at the zenith (ZT10 or 5 pm) (Fig. 1D).
Fig. 1.
Circadian oscillation of Xpa transcript, XPA protein, and nucleotide excision repair activity in the mouse liver. Mice under 12 h light: 12 h dark (L12∶D12) cycle were sacrificed at the indicated times and their livers were harvested. Zeitgeber (ZT) = 0 is light on and ZT = 12 is light off. In parentheses we also indicate the time of day in conventional Eastern Standard Time (EST). The liver extracts were tested for repair activity and for the indicated protein and mRNA levels. (A) The control lane contained DNA substrate but no extract. The excision products [indicated as a bar with a circle (isotope labeled) and a triangle (Pt adduct)] in the range of 24–32 nt were detected by autoradiography (48) (Fig. S1). Only the part of the autoradiograph containing the excision product is shown; the entire gel is shown in Fig. S2. (B) Xpa mRNA and protein were detected by RT-PCR and immunoblotting, respectively. (C) The excision repair datapoints are averages of two experiments each with two mice (bars = standard error). The mRNA, protein, and repair levels are plotted relative to the maximum values of each (which happen to be at ZT = 10). The actual amount of excision at ZT10 was 1.8%. (D) Liver extracts from ZT22 (nadir of repair activity) were supplemented with increasing amounts of XPA (0.25, 0.5, 1 pmol of XPA) and the excision activity in the supplemented extracts are plotted relative to that of the activity of ZT10 (zenith) extract. (E and F) Liver extracts prepared at the indicated ZTs from wild-type or Cry1/2 double knockout mice (CryDKO) were analyzed for excision repair activity (E) and XPA and Cry proteins and mRNAs (F). The CryDKO extracts contained 2.4- and 2.3-fold more XPA protein and mRNA relative to the ZT10 of WT, respectively.
To ascertain that the circadian oscillation of XPA level and excision repair were dependent on the circadian clock, we analyzed liver extracts from wild-type mice or mice lacking Cry1 and Cry2 (25, 26), essential components of the circadian clock (16). Analysis of excision repair activity from liver extracts harvested at the zenith (ZT10) or nadir (ZT22) revealed that the excision repair activity was three times higher in CryDKO mice compared to the zenith of wild-type mice and did not oscillate with the time of day (Fig. 1E). Likewise, the XPA protein and mRNA levels were higher in the CryDKO mice and did not oscillate (Fig. 1F). Therefore, the oscillation of excision repair activity and Xpa mRNA and protein levels in the mouse liver are all dependent on the circadian clock.
Effect of Circadian Time on Excision Repair of Cisplatin-DNA Adducts by Mouse Testis.
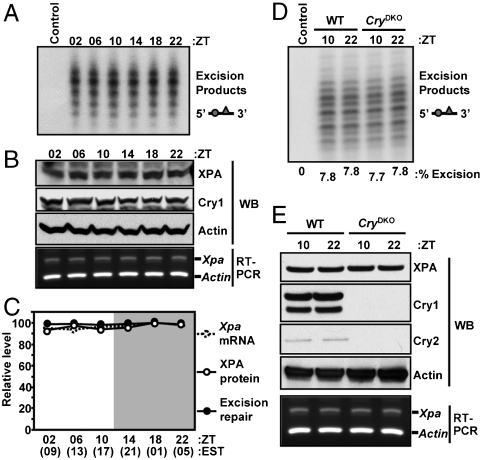
Next, we tested extracts made from mouse testis harvested at 4 h intervals over a 24 h period. In contrast to the results obtained with liver, repair of cisplatin-(GpTpG) did not exhibit daily oscillation in the testis (Fig. 2A and C). To determine the cause of this lack of oscillatory behavior in testis we measured the expression of XPA and the clock protein Cry1, which is known to oscillate in most tissues, over the course of a day. As shown in Fig. 2B and C, neither XPA nor Cry1 oscillate in testis. These findings are consistent with several reports on lack of the circadian clock either at the transcriptional or protein level in the mouse testis (13–15). Interestingly, because of lack of circadian control of Xpa transcription in the testis, excision repair activity is at a constitutively high level, and neither repair (Fig. 2D) nor the level of Xpa mRNA and protein (Fig. 2E) are affected by the presence or absence of the Cry proteins.
Fig. 2.
Circadian clock-independent repair of cisplatin-(GpTpG) adducts in mouse testis. Testicular extracts were prepared from mice under L12∶D12 conditions and were tested for repair activity with the cisplatin-(GpTpG) substrate (A) and for the indicated protein and mRNA levels (B). (C) The excision repair data points are averages of two experiments (bars = standard error). The mRNA, protein, and repair values are expressed in percentages relative to the ZT10 values. The actual amount of excision at ZT10 was 7.8%. (D) Extracts prepared at the indicated times from testes of either wild-type or CryDKO mice were analyzed for excision repair activity, and the percentages of excised products are indicated below the lanes. (E) The proteins were detected by probing immunoblots with the indicated antibodies and the mRNA levels were analyzed by RT-PCR.
XPA Protein Has a Short Lifetime.
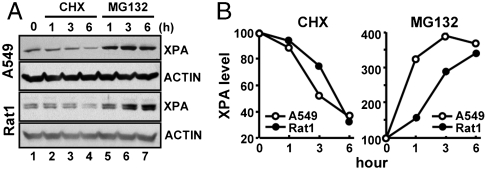
The findings that XPA exhibits high amplitude oscillation in the mouse brain (10) and liver (Fig. 1C), reaching a maximum in the afternoon and dropping precipitously at night with a 5- to 10-fold decrease in protein level within a 6-h period at night, was of special interest. We reasoned that such a drop could not be ascribed to clock-controlled transcriptional oscillation alone, and that XPA might be controlled at a posttranslational level as well. Hence, we tested XPA for proteolytic degradation using human (A549) and rat (Rat1) cell lines that are commonly used in cell biology and circadian clock research as representative of human and rodent cells, respectively. The two cell lines were treated with either cycloheximide (CHX) to inhibit protein synthesis or with MG132 to inhibit proteolysis by the proteosome, and then the XPA levels were measured for a 6 h period following treatment. As is seen in Fig. 3A, inhibition of protein synthesis by CHX (lanes 2–4) leads to a steady drop in XPA levels in both cell lines, whereas there is no change in the control protein Actin over the course of the experiment. In contrast, when MG132 is included in the culture medium to inhibit proteolysis, the levels of XPA steadily increased in both cell lines (lanes 5–7). Quantitative analysis of the data (Fig. 3B) shows that XPA decays with a first-order rate constant of k1 ∼ 6 × 10-5 sec-1 from which a half-life of t1/2 ∼ 200 min is calculated, which is in accord with the rapid drop in XPA protein levels in the brain and liver in the evening hours when the XPA gene transcription is rapidly declining as well, such that the proteolytically degraded XPA cannot be replenished by newly synthesized protein.
Fig. 3.
Degradation of XPA by the proteasome in human and mouse cell lines. (A) Immunoblots of extracts from A549 and Rat1 cells that were treated with either 20 μg/ml of protein synthesis inhibitor cycloheximide (CHX) or 40 μM proteosome inhibitor (MG132) for the indicated times. Whole cell lysates were separated by SDS-PAGE and probed with the appropriate antibodies. (B) The amounts are expressed relative to the control samples (time 0) which were taken immediately before the addition of either CHX or MG132.
Specific Interaction of XPA with a Putative Ubiquitin Ligase.
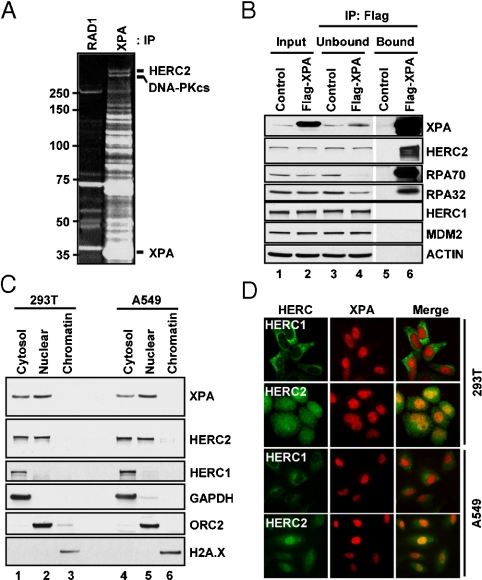
Next, we wished to identify the ubiquitin ligase that ubiquitinates XPA and targets it for proteolysis. We constructed a HEK293 cell line that contains an inducible and Flag epitope-tagged XPA gene. Cell extract was prepared from Flag-XPA expressing cells and XPA was immunoprecipitated with anti-Flag agarose resin. After extensive washing, the bound proteins were eluted with Flag peptide and separated by SDS–PAGE (Fig. 4A). The proteins that were specifically retained by the Flag-XPA resin were identified by mass spectrometry. The highest molecular weight protein identified in our screen was HERC2. This is a putative HECT E3 ligase and a member of a six-member HERC family that has been implicated in a number of cellular processes including G-protein signaling and membrane trafficking (23). Naturally occurring Herc2 mutations cause the so-called rjs (runty, jerky, sterile) phenotype in mice (27), but there have been no cellular or biochemical studies reported on HERC2 indicative of its E3 ligase activity. Hence, we proceeded to look at the XPA-HERC2 interaction in more detail and to determine the effect of HERC2 on XPA modification and stability.
Fig. 4.
Interaction of XPA with the putative ubiquitin E3 ligase HERC2. (A) HEK293 cells stably expressing either Flag-tagged XPA or Flag-tagged RAD1 (control) were induced to express the corresponding proteins. The cell lysates were then mixed with anti-Flag agarose beads and the bound proteins were visualized by SDS-PAGE and SYPRO Ruby staining. The XPA-interacting proteins were identified by mass spectroscopy. For clarity, we indicate only the bands corresponding to XPA, HERC2, and DNA-PKcs (which migrates like a doublet of HERC2). (B) XPA was immunoprecipitated from extract of either Flag-XPA or mock (control) expressing cells with anti-Flag beads and the unbound and bound fractions were probed with the appropriate antibodies. The input lanes contain 5% of the input protein. (C) Two cell lines were fractionated into “cytosol,” “nuclear,” and “chromatin” fractions and the distribution of XPA, HERC2, HERC1, and markers for the three subcellular fractions GAPDH (cytosolic), ORC2 (nuclear), H2A.X (chromatin) were analyzed by immunoblotting. (D) 293T and A549 cells were permeablized, fixed, stained with XPA and either HERC1 or HERC2 antibodies (Fig. S6), and imaged using a fluorescent microscope.
First, we investigated the XPA-HERC2 interaction specificity by immunoprecipitating XPA and probing the immunoprecipitate for HERC2, as well as for proteins known to interact with XPA (RPA70 and RPA32) (28) and proteins that are not expected to interact with XPA (HERC1, MDM2, and ACTIN). The results are shown in Fig. 4B. As expected, XPA binds tightly to RPA such that under the conditions of the experiment, where XPA is overproduced 22-fold over the endogenous levels, nearly 90% of RPA is in a complex with XPA (compare lanes 3 and 4). Significantly, while neither MDM2 nor ACTIN is detected in the XPA immunoprecipitate (lane 6), about 40% of HERC2 is immunoprecipitated with anti-Flag antibodies (compare lanes 3, 4, and 6). We further confirmed the XPA-HERC2 interaction by coimmunoprecipitating endogenous HERC2 with anti-XPA antibodies in A549 cells (Fig. S3).
Second, because the other members of the HERC family of putative E3 ligases that have been investigated so far are associated with membrane or cytoplasmic organelles (29), and XPA is primarily located in the nucleus (30), we wished to ascertain whether XPA and HERC2 have similar subcellular localization to allow interaction between the two and ubiquitination of XPA. We analyzed the subcellular localization of the two proteins by biochemical and cell biological methods using two human cell lines. Fig. 4C shows the results of the subcellular fractionation. In agreement with previous reports (30), XPA is found mostly in the nuclear fraction (Fig. 4C, lanes 2 and 5). The small fraction of XPA that is found in the cytosol is most likely the result of incidental permeabilization of the nuclear membrane during fractionation. Importantly, while HERC1, which is known to be associated with the Golgi complex and endoplasmic reticulum (29) exhibits exclusively cytosolic localization, HERC2 shows nearly equal distribution between the cytosol and the nuclear fractions (Fig. 4C, lanes 1 and 2, and 4 and 5). These conclusions were confirmed by immunostaining. In agreement with the subcellular fractionation data, HERC1 is exclusively cytoplasmic, XPA is exclusively nuclear, and HERC2 exhibits both cytoplasmic and nuclear localization (Fig. 4D). Taken together, the data presented so far show that XPA is an unstable protein, colocalizes with a putative ubiquitin ligase, and appears to directly interact with this ligase, suggesting that HERC2 may be responsible for directing the proteolysis of XPA and thus participating in its circadian oscillatory expression pattern.
HERC2 is an Ubiquitin Ligase for XPA and Promotes its Proteolysis.
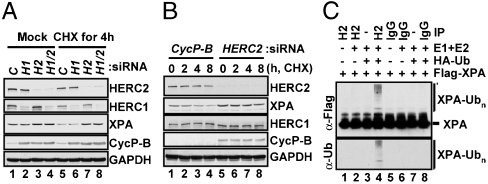
To determine whether HERC2 influences the stability of XPA, we downregulated HERC2 or HERC1 with siRNA and measured the levels of XPA in the siRNA treated cells. As is seen in Fig. 5A, downregulation of HERC2 (lane 7), but not HERC1 (lane 6), stabilizes XPA after cycloheximide (CHX) treatment. Fig. 5B shows the XPA degradation kinetics under conditions of CHX treatment in the presence or absence of HERC2 siRNA. In the absence of HERC2 siRNA, XPA is degraded with kinetics similar to that shown in Fig. 3, whereas in the presence of HERC2 siRNA and CHX, the XPA levels essentially remain constant for the duration of the experiment (8 h). Because XPA is not the only limiting factor in nucleotide excision repair (31), we also analyzed the effect of HERC2 downregulation on the other core repair factors and found that these proteins were not affected by CHX treatment or by CHX treatment combined with HERC2 siRNA over a period of 8 h (Fig. S4A). We also tested for the effect of HERC2 on some of the key DNA damage checkpoint proteins because they also affect the cellular response to genotoxic agents and again, did not see an effect of HERC2 downregulation on these proteins either (Fig. S4B). Thus, it appears that, of all the excision repair and checkpoint proteins tested, HERC2 plays a role only in proteolysis of XPA and as a consequence in circadian oscillation of excision repair activity.
Fig. 5.
Identification of HERC2 as an XPA ubiquitin ligase. (A) 293T cells were transfected with control Cyclophilin-B siRNA (C), HERC1 siRNA (H1), HERC2 siRNA (H2), or both HERC1 and HERC2 siRNAs (H1/2), and then incubated with cycloheximide (CHX) where indicated and the levels of the target proteins as well as of XPA and GAPDH (loading control) were determined by immunoblotting. (B) Control CycP-B or HERC2 siRNA treated cells were incubated with CHX for the indicated times and the levels of the HERC2, XPA, control proteins, and GAPDH were determined by immunoblots. (C) A549 cell extract was incubated with Protein A agarose and either HERC2 antibodies or control rabbit IgG. The beads were collected and incubated with the indicated proteins at 30 ºC for 30 minutes followed by separation on SDS–PAGE and analysis by western blotting with either Flag antibodies for XPA (Top) or with ubiquitin antibodies (Bottom).
The data presented so far indicate that HERC2 may ubiquitinate XPA and thus target it for proteolytic degradation, but there are no reports on the E3 ligase activity of HERC2. Therefore, to test our model, we conducted an ubiquitination assay with HERC2 using XPA as a substrate. HERC2 was immunoprecipitated from A549 cells and, after extensive washing, the immunoprecipitate was supplemented with E1, E2, HA-tagged ubiquitin (HA-Ub) and Flag-XPA under appropriate reaction conditions. The products were separated by SDS–PAGE and probed with either Flag or ubiquitin antibodies. As seen in Fig. 5C, when supplemented with E1 and E2, the immunoprecipitated HERC2 does ubiquitinate XPA (lane 4) and hence it is possibly a bona fide E3 ubiquitin ligase. HERC2 also ubiquitinates itself or another interacting protein in these reactions (Fig. S5).
Downregulation of HERC2 Improves Cisplatin-(GpG) Repair Efficiency.
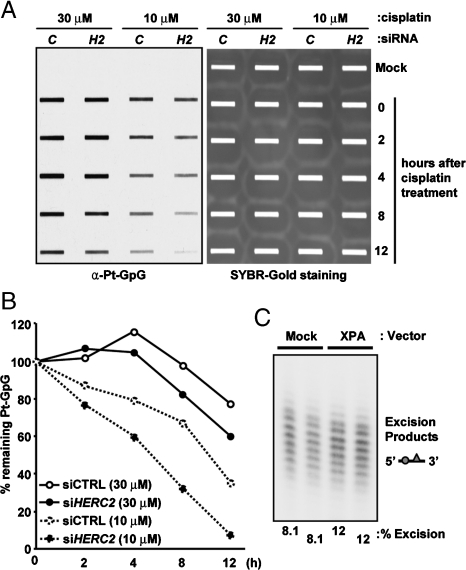
Our data indicate that XPA is a limiting factor in nucleotide excision repair in vivo, and therefore we wished to know whether increasing XPA levels by inhibition of ubiquitination and proteolytic degradation of XPA would manifest in a faster rate of repair of cisplatin–DNA adducts. HEK293 cells that were transfected with either control siRNA or HERC2 siRNA were incubated with either 10 μM or 30 μM cisplatin for 3 h, and then the drug was removed from the medium and the Pt-(GpG) excision kinetics were determined by DNA slot blot using monoclonal antibodies specific for this adduct. As seen in Fig. 6A and B, downregulation of HERC2 results in a faster rate of Pt-(GpG) repair, supporting the conclusion that in tissues or cell lines with a circadian clock, the rate of nucleotide excision repair is controlled by ubiquitination of XPA by HERC2 E3 ligase. In line with this conclusion, even in a cell culture where each cell has a circadian rhythm but the culture on the whole is without a circadian rhythm because the circadian phases of the cells are out of synchrony (2), overexpression of XPA even in the presence of HERC2 leads to a modest but significant increase in excision activity (Fig. 6C) because apparently under these conditions the rate of XPA synthesis far exceeds the rate of ubiquitination by HERC2.
Fig. 6.
Downregulation of HERC2 improves cisplatin-(GpG) repair efficiency. (A) A549 cells in which either Cyclophilin-B (C) or HERC2 (H2) were downregulated by siRNA were treated with the indicated doses of cisplatin for 3 hours, washed, and then incubated in fresh medium for the indicated times before harvesting. The Pt-(GpG) diadduct levels were analyzed by immunoblotting (Left) and SYBR-Gold staining of the membrane showing total DNA in the slot blot (Right). (B) Quantitative analysis of the data from Panel A. (C) Extracts were made from circadian asynchronous cultures of HEK293 cells or HEK293 cells overexpressing Flag-XPA 22-fold higher than the endogenous protein levels and used in excision assays with cisplatin-(GpTpG) substrate. Each lane represents an independent experiment. The effect on excision repair is not proportional to the level of XPA overexpression because under these conditions other excision repair factors become rate-limiting.
Discussion
Our findings, in light of current knowledge about the molecular mechanism of the circadian clock, are schematically summarized in Fig. S8. In wild-type mice, oscillatory transcription of the clock genes (Crys and Pers) and the clock-controlled gene Xpa, combined with the action of E3 ligases specific for the respective proteins, engender the core circadian clock and the oscillation of one of the clock output pathways, nucleotide excision repair, respectively. In the absence of Crys, there is no circadian clock, XPA is expressed at constitutively high levels, and the excision repair activity is elevated and ceases to oscillate. The data presented in this paper raise some important points relevant to the regulation of nucleotide excision repair in mammals and the evolution of the circadian clock, and are of potential significance to rational approaches to chronochemotherapy.
Regulation of Nucleotide Excision Repair.
Our study has defined a relatively direct pathway for regulation of nucleotide excision repair in mice and humans. To recapitulate briefly, we find that XPA is regulated in a circadian manner in all mouse tissues tested, with the exception of testis. We show that oscillation of the XPA protein is accomplished by two mechanisms: 1) transcriptional regulation by cryptochrome and other core clock proteins, and 2) proteolysis by the ubiquitin (HERC2 E3 ligase)-proteasome pathway. Our findings of circadian clock control of XPA, and of excision repair activity in brain and liver, constitute the most direct evidence to date of any form of control of nucleotide excision repair activity. Although numerous studies have shown that both the core excision repair factor XPC and the accessory factor XPE (DDB2) are ubiquitinated by the CUL4A-ROC1-DDB1 E3 ubiquitin ligase following DNA damage (32–34), at present it is unclear what effects these posttranslational modifications have on excision repair (35). The ubiquitination of XPC does not lead to its proteolytic degradation, and it is unclear whether it affects its function (36). Ubiquitination of DDB2 leads to its eventual proteolytic degradation (33). However, it is a matter of ongoing debate whether DDB2 plays a direct role in repair or influences cell survival directly or indirectly by participating in apoptosis and cell cycle regulation (34, 37–40). In contrast, the findings reported in this paper clearly show that XPA is controlled by transcriptional and posttranslational mechanisms, and that the oscillatory rhythm of the protein resulting from these regulatory mechanisms has a drastic effect on the repair capacity of the cell. Hence, in studies on excision repair capacity in animals and even mammalian cell lines, with the aim of determining the effects of various parameters, such as genetic background, race, gender, and age, on excision repair rate, the time of day (or circadian time) of sampling or assay should be considered as an important variable.
Cryptochrome, “Escape from Light,” and Evolution of the Circadian Clock.
Cryptochrome, which is a key component of the circadian clock, phylogenetically is related to DNA photolyase, which repairs UV-induced pyrimidine photodimers using blue light as the energy source (41). According to the modern version of the “Escape from Light” hypothesis for the evolution of the circadian clock (41, 42), an ancient aquatic organism employed the photolyase/cryptochrome ancestor as a blue-light sensor (cryptochrome) to regulate its diel vertical movements so as to optimize nutrient availability and minimize exposure to genotoxic UV light, and as a blue-light activated repair enzyme (photolyase) that repairs UV damage that inevitably occurs despite the presence of an escape from light mechanism. Subsequently, during evolution, this protoenzyme gave rise to the present day cryptochrome, which is strictly a clock protein, and photolyase, which is strictly a repair enzyme. It is rather remarkable, therefore, that cryptochrome still participates in repair, albeit indirectly, by regulating the nucleotide excision repair activity in mice and humans. In these organisms, which lack photolyase, nucleotide excision repair is the sole repair mechanism for removing UV-induced dipyrimidine photoproducts that can also be repaired by the phylogenetic relative of cryptochrome, photolyase, in other organisms such as fruit flies (41).
Whether circadian control of nucleotide excision repair confers a selective advantage or is an evolutionary relic cannot be answered based on available data. It is conceivable that even nocturnal animals such as mice are exposed to sunlight that produces DNA damage, and therefore the continuous rise in excision repair activity during the day serves a useful purpose. Conversely, it might be advantageous to reduce the excision repair activity at night, when it apparently is not needed, so as to reduce the risk of attack of normal DNA by this repair system (“gratuitous repair”) that may cause deleterious mutations (43, 44). In this regard, it is interesting to note that UvrA, the key damage recognition protein in E. coli nucleotide excision repair, in a manner similar to XPA, is also tightly regulated by proteolysis so as to avoid gratuitous repair and harmful mutagenesis (45).
Chronochemotherapy.
The daily oscillation of excision repair activity is likely to affect the efficacy of chemotherapy with certain drugs. Cisplatin and its second and third generation derivatives are among the most commonly prescribed anticancer drugs (3). With the development of new derivatives that have better uptake, activation, and DNA binding properties, the range of cancers that are treated with cisplatin is continuously expanding (3). However, despite the many improvements in cancer treatment regimens that have increased the efficacy and decreased the side effects of the drug, the time of administration, which may significantly affect both the efficacy and the side effects (toxicity), has not become a commonly used variable in clinical practice (2). This is due in part to the lack of a mechanistic foundation for the roles of circadian factors in chemotherapy.
The cellular response to genotoxic agents is dictated by several factors (46, 47), including the cell cycle phase, DNA repair capacity, and potential for apoptosis. Hence, in designing a chronochemotheraphy regimen, all these factors must be taken into account. Here we show that in mouse liver, excision repair of cisplatin adducts exhibits a robust circadian oscillatory pattern, with a maximum at ∼5 pm and a minimum at ∼5 am. These findings should be useful in designing mechanism-based chronochemotherapy trials in mice and eventually in humans.
Materials and Methods
Organ Harvesting and Preparation of Cell-Free Extracts.
Cell-free extracts (CFEs) from C57BL/6J mice and Cry1-/-Cry2-/- mice in the same background (26) were prepared as described previously (10) with modifications. See SI Text for details.
Excision Assay.
Linear (140 bp) or circular (2.9 kbp) substrates containing a single Pt-(GpTpG) adduct were prepared by established methods and the excision assay was carried out with 10 fmol of substrate and 100 μg of CFE in 25 μl excision buffer by incubating at 30 °C for 1 h. See SI Text for details.
In Vivo Excision Repair Assay.
A monoclonal antibody which recognizes Pt-(GpG) adducts (49) (Oncolyze) was used in a slot blot assay to quantify the adduct levels in the DNA of cisplatin treated A549 cells as a function of time. See SI Text for details.
Immunoblotting, RT-PCR, and siRNA.
See SI Methods for details.
Subcellular Fractionations and Immunofluorescence.
See SI Methods for details.
Proteomics and Ubiquitination Assay.
See SI Methods for details.
Supplementary Material
Acknowledgments.
We thank Michael Kemp for providing recombinant RPA and for critical reading of the manuscript, and Drs. Kenneth Kreuzer (Duke University), Bennett Van Houten (University of Pittsburgh), and Pengbo Zhou (Cornell University) for useful comments on the manuscript. This work was supported by National Institutes of Health Grants GM31082 and GM32833.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0915085107/DCSupplemental.
References
- 1.Kobayashi M, Wood PA, Hrushesky WJ. Circadian chemotherapy for gynecological and genitourinary cancers. Chronobiol Int. 2002;19:237–251. doi: 10.1081/cbi-120002600. [DOI] [PubMed] [Google Scholar]
- 2.Levi F, Schibler U. Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- 3.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 4.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107:1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 5.Hall MD, Okabe M, Shen DW, Liang XJ, Gottesman MM. The role of cellular accumulation in determining sensitivity to platinum-based chemotherapy. Annu Rev Pharmacol Toxicol. 2008;48:495–535. doi: 10.1146/annurev.pharmtox.48.080907.180426. [DOI] [PubMed] [Google Scholar]
- 6.Chaney SG, Sancar A. DNA repair: Enzymatic mechanisms and relevance to drug response. J Natl Cancer Inst. 1996;88:1346–1360. doi: 10.1093/jnci/88.19.1346. [DOI] [PubMed] [Google Scholar]
- 7.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 8.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 9.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997;272:23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 10.Kang TH, Reardon JT, Kemp M, Sancar A. Circadian oscillation of nucleotide excision repair in mammalian brain. Proc Natl Acad Sci USA. 2009;106:2864–2867. doi: 10.1073/pnas.0812638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang TH, Sancar A. Circadian regulation of DNA excision repair: implications for chrono-chemotherapy. Cell Cycle. 2009;8:1665–1667. doi: 10.4161/cc.8.11.8707. [DOI] [PubMed] [Google Scholar]
- 12.Johnsson A, et al. Pharmacokinetics and tissue distribution of cisplatin in nude mice: Platinum levels and cisplatin-DNA adducts. Cancer Chemother Pharmacol. 1995;37:23–31. doi: 10.1007/BF00685625. [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto Y, Sancar A. Circadian regulation of cryptochrome genes in the mouse. Brain Res Mol Brain Res. 1999;71:238–243. doi: 10.1016/s0169-328x(99)00192-8. [DOI] [PubMed] [Google Scholar]
- 14.Morse D, Cermakian N, Brancorsini S, Parvinen M, Sassone-Corsi P. No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse. Mol Endocrinol. 2003;17:141–151. doi: 10.1210/me.2002-0184. [DOI] [PubMed] [Google Scholar]
- 15.Lambert CM, Weaver DR. Peripheral gene expression rhythms in a diurnal rodent. J Biol Rhythms. 2006;21:77–79. doi: 10.1177/0748730405281843. [DOI] [PubMed] [Google Scholar]
- 16.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 17.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 18.Godinho SI, et al. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 19.Siepka SM, et al. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reischl S, et al. Beta-TrCP1-mediated degradation of PERIOD2 is essential for circadian dynamics. J Biol Rhythms. 2007;22:375–386. doi: 10.1177/0748730407303926. [DOI] [PubMed] [Google Scholar]
- 21.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 22.Hughes M, et al. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Sym. 2007;72:381–386. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 24.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc Natl Acad Sci USA. 1994;91:10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Selby CP, Thompson C, Schmitz TM, Van Gelder RN, Sancar A. Functional redundancy of cryptochromes and classical photoreceptors for nonvisual ocular photoreception in mice. Proc Natl Acad Sci USA. 2000;97:14697–14702. doi: 10.1073/pnas.260498597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehman AL, et al. A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc Natl Acad Sci USA. 1998;95:9436–9441. doi: 10.1073/pnas.95.16.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Gonzalo FR, Rosa JL. The HERC proteins: Functional and evolutionary insights. Cell Mol Life Sci. 2005;62:1826–1838. doi: 10.1007/s00018-005-5119-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyamoto I, Miura N, Niwa H, Miyazaki J, Tanaka K. Mutational analysis of the structure and function of the xeroderma pigmentosum group A complementing protein. Identification of essential domains for nuclear localization and DNA excision repair. J Biol Chem. 1992;267:12182–12187. [PubMed] [Google Scholar]
- 31.Kesseler KJ, Kaufmann WK, Reardon JT, Elston TC, Sancar A. A mathematical model for human nucleotide excision repair: Damage recognition by random order assembly and kinetic proofreading. J Theor Biol. 2007;249:361–375. doi: 10.1016/j.jtbi.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugasawa K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 33.El-Mahdy MA, et al. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. 2006;281:13404–13411. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, et al. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–467. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 36.Wang QE, et al. DNA repair factor XPC is modified by SUMO-1 and ubiquitin following UV irradiation. Nucleic Acids Res. 2005;33:4023–4034. doi: 10.1093/nar/gki684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itoh T, Cado D, Kamide R, Linn S. DDB2 gene disruption leads to skin tumors and resistance to apoptosis after exposure to ultraviolet light but not a chemical carcinogen. Proc Natl Acad Sci USA. 2004;101:2052–2057. doi: 10.1073/pnas.0306551101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 39.Hwang BJ, Ford JM, Hanawalt PC, Chu G. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc Natl Acad Sci USA. 1999;96:424–428. doi: 10.1073/pnas.96.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoyanova T, Roy N, Kopanja D, Bagchi S, Raychaudhuri P. DDB2 decides cell fate following DNA damage. Proc Natl Acad Sci USA. 2009;106:10690–10695. doi: 10.1073/pnas.0812254106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sancar A. Regulation of the mammalian circadian clock by cryptochrome. J Biol Chem. 2004;279:34079–34082. doi: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- 42.Gehring W, Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J Mol Evol. 2003;57(Suppl 1):S286–289. doi: 10.1007/s00239-003-0038-8. [DOI] [PubMed] [Google Scholar]
- 43.Branum ME, Reardon JT, Sancar A. DNA repair excision nuclease attacks undamaged DNA. A potential source of spontaneous mutations. J Biol Chem. 2001;276:25421–25426. doi: 10.1074/jbc.M101032200. [DOI] [PubMed] [Google Scholar]
- 44.Hasegawa K, Yoshiyama K, Maki H. Spontaneous mutagenesis associated with nucleotide excision repair in Escherichia coli. Genes Cells. 2008;13:459–469. doi: 10.1111/j.1365-2443.2008.01185.x. [DOI] [PubMed] [Google Scholar]
- 45.Pruteanu M, Baker TA. Controlled degradation by ClpXP protease tunes the levels of the excision repair protein UvrA to the extent of DNA damage. Mol Microbiol. 2009;71:912–924. doi: 10.1111/j.1365-2958.2008.06574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beland FA, Dooley KL, Sheldon WG, Delongchamp RR. Circadian variation in the induction of intestinal tumors by N-methyl-N-nitrosourea in male C57BL/6N mice. J Natl Cancer Inst. 1988;80:325–330. doi: 10.1093/jnci/80.5.325. [DOI] [PubMed] [Google Scholar]
- 47.Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 48.Reardon JT, Sancar A. Purification and characterization of Escherichia coli and human nucleotide excision repair enzyme systems. Methods Enzymol. 2006;408:189–213. doi: 10.1016/S0076-6879(06)08012-8. [DOI] [PubMed] [Google Scholar]
- 49.Liedert B, Pluim D, Schellens J, Thomale J. Adduct-specific monoclonal antibodies for the measurement of cisplatin-induced DNA lesions in individual cell nuclei. Nucleic Acids Res. 2006;34:e47. doi: 10.1093/nar/gkl051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.