Abstract
Telomerase is a ribonucleoprotein complex consisting of a protein reverse transcriptase (TERT) and an RNA subunit (TR). Telomerase normally adds telomeric DNA repeats to chromosome ends. Here, we engineer human and Tetrahymena cis-telomerase RNAs, each having a DNA primer covalently linked to its 3′ end. We find that cis-telomerase synthesizes DNA with increased repeat addition processivity (RAP) but does not completely rescue the RAP defect of the L14A mutant of Tetrahymena TERT. This supports the conclusion that L14 has a function beyond binding the DNA primer and preventing dissociation during multiple rounds of repeat addition. By comparing cis-telomerases with various linker lengths, we find that a 5 nt linker gives near-optimal activity, indicating that the distance between the 3′ end of the telomerase RNA pseudoknot region and the 5′ end of the DNA primer is ∼33 Å. Even a 2 nt linker (∼14 Å) gives some activity, indicating a high degree of conformational flexibility in this ribonucleoprotein complex. More generally, the cis system will allow structure-function relationships of each RNA molecule to be read directly through the reaction that it performs on itself.
Keywords: ribonucleoprotein complex, processivity, reverse transcriptase, telomerase
Natural RNAs involved in biocatalysis act either “in cis” (intra-molecular reactions) or “in trans” (intermolecular reactions). Cis-acting RNAs are exemplified by the self-splicing group I and group II introns and the self-cleaving hammerhead and hairpin ribozymes (1, 2). Trans-acting RNAs include: the ribosomal RNAs that use aminoacylated tRNAs as substrates (3), the spliceosomal small nuclear RNAs that use pre-mRNAs as substrates (4), and the RNase P ribozyme that uses tRNA precursors containing 5′-leader sequences as substrates (5). In a number of cases, cis-acting ribozymes have been converted into trans-acting RNA enzymes that splice or cleave exogenous RNA substrates. The incentive for engineering these trans-acting ribozymes was initially to demonstrate that they were true catalysts, capable of multiple-turnover reactions. A subsequent rationale was to create RNA-cleaving agents for biotechnology. Transformation in the other direction, from trans-acting to cis-acting RNAs, has been accomplished for RNase P. By engineering a chimeric RNA that joins the RNase P RNA to its pre-tRNA substrate, pre-tRNA-RNase P RNA conjugates were designed to undergo accurate and efficient self-cleavage (6, 7). In the present work, we describe the conversion of the telomerase RNA-TERT complex, which normally acts in trans to extend telomeric DNA primers, into a cis-acting system that extends its own 3′ end. Thus, a trans-acting RNA can be converted into a cis-acting RNA even in the context of a complex ribonucleoprotein (RNP).
The telomerase RNP uses its reverse transcriptase subunit (TERT) to copy a template sequence within its intrinsic RNA subunit (TR), thereby adding telomeric DNA repeats to the ends of chromosomes (8, 9). Although TERT has clear sequence conservation among divergent organisms, the RNA subunit shows little primary sequence conservation even between closely related species. More surprisingly, the length of telomerase RNA varies widely among eukaryotes; it ranges from 150–200 nt for ciliates (10, 11) and 300–500 nt for vertebrates (12) to more than 1 kb for budding and fission yeasts (13–15).
Despite the apparent lack of sequence conservation among species, recent studies revealed several conserved secondary structure elements in telomerase RNAs (16–20). In addition to providing a template for the reverse transcriptase domain of TERT, telomerase RNA contains non-template regions that also play important roles in telomerase catalysis, such as binding TERT to form the RNP (21), contributing to telomerase repeat-addition processivity (RAP) (22), and orienting the DNA substrate to the active site of the enzyme (20). Thus, telomerase RNA is an essential component of the telomerase RNP enzyme, actively participating in many aspects of catalysis (23). Consistent with this proposal is the fact that multiple mutations in the non-template region of human TR have been found in patients with autosomal dominant dyskeratosis congenita, a progressive syndrome characterized by abnormal skin manifestations, nail dystrophy, and bone marrow failure. Some of these TR mutations have been shown to severely impair telomerase activity and result in shorter telomeres (24–26).
RAP is a unique feature of telomerase; after one round of nucleotide addition, the same primer can translocate, realign to the RNA template, and undergo another round of nucleotide addition. A recent study of the Telomerase Essential N-terminal (TEN) domain of the Tetrahymena telomerase protein catalytic subunit showed that Leu14 in this domain is essential for RAP, as substitution of L14 with various other amino acids, even with isoleucine, largely eliminated primer extension products corresponding to the addition of multiple repeats (27). A simple explanation for the loss of RAP in L14 mutants might have been that L14 is important in mediating telomerase-DNA primer interaction; a weakened interaction could cause rapid dissociation of the product after one round of nucleotide addition, thereby impairing primer translocation. However, DNA photo-crosslinking and primer challenge experiments suggested that the L14A mutant retains the primer anchor site and recycles primers at a rate similar to WT telomerase (27).
In this work, we construct a cis-telomerase by linking the 3′-end of the telomerase RNA pseudoknot region to the 5′ end of the DNA primer. This allows an independent test of whether L14 in the TEN domain of TERT functions to bind to the DNA primer and, therefore, help primer translocation in RAP. In addition, the linker that joins the RNA with the DNA primer in cis-telomerase can be used as an indicator of an important distance constraint within the active RNP enzyme—the distance between the pseudoknot domain and the template region. More generally, cis-telomerase should facilitate future structure-function analysis of telomerase RNAs, because the structure-function relationships of the RNA component of the RNP enzyme can be read directly through the reaction that it catalyzes on itself.
Results
Construction of Human Cis-Telomerase.
We transcribed the pseudoknot domain (hTR44-184) with T7 RNA polymerase and then used splinted ligation (28) to join 5′ mono-phosphorylated DNA primers with telomeric sequence to the RNA 3′ end (Fig. 1A and Fig. S1). Cis-telomerase RNAs with DNA linkers of two different lengths, 17 and 13 nt, were initially constructed. The resulting hTR44-184/telomeric DNA primer conjugates were refolded, mixed with the CR4-CR5 RNA domain (the other RNA domain required for telomerase activity in addition to the pseudoknot domain; refs. 12 and 29), and assembled with HA-tagged hTERT translated in rabbit reticulocyte lysates (RRLs). The assembled cis-telomerase RNP complex was then immuno-purified from the RRL and the telomerase reaction was initiated by adding dNTPs to the reaction buffer. Both cis-telomerases were active (Fig. 1B).
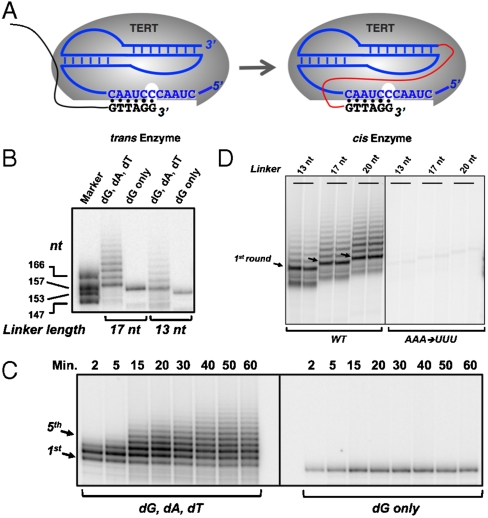
Fig. 1.
Design and characterization of human cis-telomerase. (A) Telomerase is composed of a protein subunit (Gray Oval) and an RNA subunit (Secondary Structure, Blue). The cis-telomerase is constructed by linking the 3′ end of the telomerase RNA pseudoknot region to the 5′ end of the DNA primer with a flexible linker (Red), such that the 3′ end of the RNA/DNA chimera can align to the template region of the telomerase RNA (Blue Letters) and be elongated. (B) Telomerase assays of cis-telomerases with linkers of lengths 17 nt and 13 nt. (Linker sequences in Table S1; 13 nt #2 used here.) In both versions, human telomeric sequence GTTAGG is at the very 3′ end of the DNA/RNA chimera. Lanes 2 and 4, dATP, dTTP, and dGTP were added to provide all the deoxynucleotide substrates needed for the synthesis of the human telomeric sequence. Each band represents one round of telomeric DNA addition with a spacing of 6 nt between the bands. Lanes 3 and 5, dGTP was the only nucleotide added. Marker, mixture of four cis-telomerase RNAs of the lengths indicated, each 3′-end labeled. (C) Time course of cis-telomerase reactions with either dG, dA, and dT provided or only dG provided. (D) Assays of cis-telomerases with a telomerase RNA triple-helix disrupting mutation. In the AAA → UUU mutants, nucleotides 174-AAA-176 in the human telomerase RNA were mutated to UUU.
One hallmark of the human telomerase reaction is that telomerase can copy the same small stretch of its internal RNA template multiple times on the same DNA primer; therefore, a ladder with a 6 nt interval is observed in a sequencing gel. Interestingly, in the cis-telomerase reaction, we found that multiple copies of the telomeric DNA were also added to the same DNA primer when dATP, dTTP, and dGTP were provided to allow the telomerase to finish a full round of nucleotide addition (Fig. 1C Left), even though in this case the DNA primer was covalently linked to the telomerase RNA. When only dGTP was provided, a single product was observed (Fig. 1C Right), consistent with the expectation that extension would stop after addition of a single dG (note the template-primer alignment in Fig. 1A).
Time course experiments of cis-telomerase reactions showed that the first round of nucleotide addition reached its maximum after 15 min; however, adding additional repeats took longer, reaching a maximum at around 1 h (Fig. 1C). We then tested whether mutations that impair native telomerase catalysis also affect the cis-telomerase. A triple-helix structure in the telomerase RNA has been shown to be important for catalysis (18, 20). For human and yeast telomerases, substitutions of As in the triple-helix with Us completely abolish the hydrogen-bonding network, therefore disrupting the structure and greatly decreasing telomerase activity (18, 20). When we introduced these triple-helix disrupting mutations (174AAA176 → UUU) into three versions of cis-telomerase with 13 nt, 17 nt, and 20 nt linker lengths, we found that they lost more than 95% of their activity (Fig. 1D). This experiment further assures that linking the DNA primer to the end of the telomerase RNA pseudoknot domain doesn’t affect the way that telomerase carries out its catalysis.
Intramolecular Versus Intermolecular Primer Elongation.
If the cis-telomerase is really carrying out an intramolecular reaction, the RNA that is directly linked to the DNA primer must also be the template provider for reverse transcription. Alternatively, the same profile of reaction products might be generated if the covalently linked DNA primer of one telomerase RNA were to flip from its own RNA to base pair with another telomerase RNA, thereby forming an intermolecular telomerase RNA-DNA primer complex (Fig. 2A). To test whether such DNA primer flipping occurs, we carried out two sets of experiments.
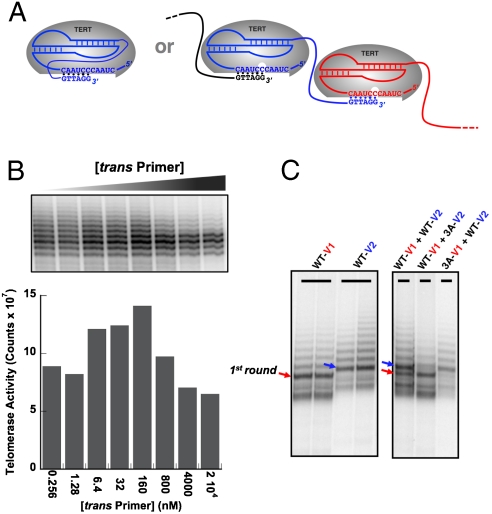
Fig. 2.
Cis-telomerase RNA is a real cis-system. (A) Diagram showing a hypothetical switch between two possible modes of RNA-conjugated primer elongation, intramolecular (Left) and intermolecular (Right). (B) Increasing amount of the trans primer was added to the cis-telomerase reactions to compete for the conjugated primer of the cis-telomerase. The concentrations of the trans primer are indicated below the bar graph. (C) Mixing assays to test for possible intermolecular primer elongation. Left, cis-telomerases with linkers of two different lengths (13 nt #2 linker for V1 and 17 nt linker for V2) produced bands with distinct mobility. Right, when wild type and inactive mutant telomerase RNAs were mixed, only the primers conjugated to the wild-type telomerase RNAs were elongated. Thus, there is no intermolecular exchange of conjugated DNA primers between telomerase RNAs. The 3A mutation is the triple-helix disrupting mutation (174AAA176 → UUU) used in Fig. 1D. The “1st Round” of telomerase extension was assigned by comparison with +1 markers made by elongation of cis-telomerase RNA by a reverse transcriptase in the presence of only dGTP. Bands below those marked 1st Round are due to an uncharacterized nuclease activity either intrinsic to telomerase or co-purified from the in vitro translation system.
First, we titrated the cis-telomerase reactions with various amounts of DNA primers as trans substrates with the final concentration of the trans DNA primer ranging from 0.256 nM to 2 × 104 nM. Because the concentration of the cis-telomerase is about 2 nM, an intermolecular reaction would be competed by relatively low concentrations of the trans primer. As shown in Fig. 2B, there is no competition up to 160 nM trans primer; even with the trans primer concentration as high as 2 × 104 nM, 10,000 times higher that the cis-telomerase concentration, at least half of the telomerase reaction still takes place in cis with the conjugated primer. Thus, cis-telomerase normally self-elongates, but the partial suppression of cis-activity and of RAP at very high trans-primer concentrations suggests that the tethered primer is in a bound-unbound equilibrium and can be competed by very high concentrations of exogenous primer, especially when the linker is becoming longer due to the telomerase elongation.
In an independent test of the intramolecularity of the reaction, we mixed a cis-telomerase bearing a triple-helix disrupting mutation in the pseudoknot region—a dead enzyme by itself—with wild-type cis-telomerases containing linkers of different length. If the above-mentioned intermolecular DNA primer/telomerase RNA interaction were responsible for the activity, then cis-telomerase with a triple-helix disrupting mutation would also be elongated. Different linker lengths allowed us to distinguish the two cis-telomerase products (wild type versus mutant) after telomerase reactions. As shown in Fig. 2C, the primers conjugated to the wild-type telomerase RNA were elongated, but not the ones attached to the triple-helix disrupting mutants. Therefore, we conclude that intermolecular exchange of conjugated DNA primers between telomerase RNAs is inconsequential, and each cis-telomerase is adding telomeric repeats to itself.
Effect of Linker Length.
One variable that we optimized during the cis-telomerase design was the length of the linker that joins the 3′ end of the pseudoknot domain of the telomerase RNA to the 5′ end of the DNA primer. If the linker were too short, the DNA primer might not be able to align to the RNA template region due to the length restraint; if the linker were too long, the extra single-stranded nucleic acid might base pair with a portion of the telomerase RNA and interfere with the telomerase reaction. We, therefore, engineered a range of linker lengths into the cis-telomerase system, from 2 nt to 26 nt with their linker sequences shown in Table S1. Surprisingly, even 2 nt and 4 nt linkers gave telomerase activity (Fig. 3A), and the activity was truly from cis elongation as shown by a trans primer challenge experiment (Fig. S2). There was a substantial increase in activity between 4 and 5 nt, after which activity showed only a weak dependence on linker length (Fig. 3A). Activity of telomerase RNAs with 22 nt, 23 nt, and 26 nt linkers (not shown) was similar to that of the 17 nt linker telomerase. In the case of the 13 nt linker, linker sequences containing A4 and T4 gave similar activity (Table S1).
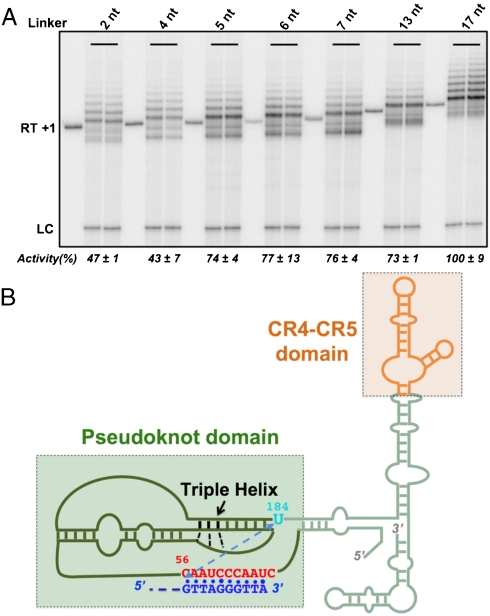
Fig. 3.
The effect of linker length on cis-telomerase activity. (A) Telomerase assays of cis-telomerases with linkers of various lengths, ranging from 2 to 17 nt. Lane RT and the First Lane of each set of three thereafter, each cis-telomerase extended by +1 nt with the reverse transcriptase (Superscript III reverse transcriptase from Invitrogen), thereby serving as a length marker for the telomerase assays in the next two lanes (duplicate samples). LC, loading control. Activity, average of four determinations, with +/- % S.E.M., relative to activity of the 17 nt linker cis-telomerase that had the highest activity of all cis-telomerases constructed (2–26 nt linkers). Cis-telomerase with 13 nt linker has sequence of 13 nt #2 shown in Table S1. (B) Secondary structure model of the human telomerase RNA (12), indicating the pseudoknot domain and the CR4-CR5 domain with bound DNA primer. Dashed Blue Line, distance spanned by linker in cis-telomerase.
These observations suggest that 5 nt are sufficient to span the distance from the 3′ end of the telomerase RNA pseudoknot region to the 5′ end of the DNA primer in the active telomerase in its ground state (i.e., without distortion). These five nucleotides consist of two ribonucleotides and three deoxyribonucleotides. Taking 5.9 Å as the distance spanned by a ribonucleotide and 7.0 Å by a deoxyribonucleotide (30), this 5 nt linker is about 33 Å long. This length is somewhat shorter than that predicted from a previous computational model of the core region of the human telomerase RNA (44 Å in the model, shown in Fig. S3), based on distances measured by FRET (31).
Can Cis-Telomerase Rescue the RAP Defect in the Tetrahymena L14A Mutant?
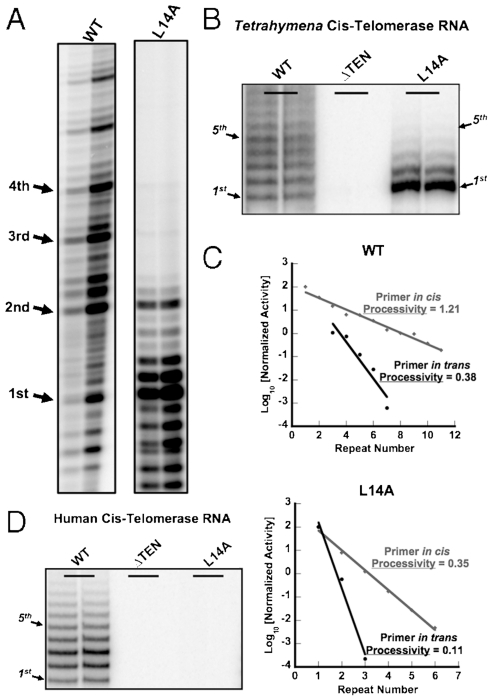
Leu14 in the TEN domain of Tetrahymena TERT has been shown to be essential for RAP (27) (Fig. 4A). The importance of Leu14 in RAP might have been that it mediated the interaction between the telomerase TEN domain and the DNA primer and held onto the primer during the telomerase reaction cycle. Weakened interaction in the mutant would then allow rapid dissociation of the product after one round of nucleotide addition, thereby impairing primer translocation. If this proposal were true, we would expect cis-telomerase to rescue the loss of RAP in the Leu14 mutant background because the primer is permanently linked to the telomerase. In the cis-telomerase system, the L14A mutant produced more discernible higher molecular weight bands corresponding to multiple rounds of telomeric DNA addition than the native L14A telomerase with the primer provided in trans (Fig. 4B). Quantification showed that cis-telomerase provided a 3-fold increase in RAP for the L14A mutant (Fig. 4C). However, RAP of the wild-type telomerase also increased about 3-fold in the cis-telomerase system (Fig. 4C). Therefore, we conclude that RAP defect in L14A mutant could not be rescued by attaching the DNA primer to the telomerase. This experiment suggests that the role of L14 is not simply to bind to the substrate and help it realign to the template that agrees with the results of previous photo-crosslinking and “primer challenge” experiments (27). The L14A mutation in the human TERT abolished most activity in the cis-telomerase system (Fig. 4D), just as it does in the native telomerase (27). Furthermore, when we deleted the whole TEN domains of Tetrahymena and human TERTs, neither of them was active in the cis-telomerase system (Fig. 4B and D). These results suggest that in addition to binding to the DNA primer, the TEN domain of TERT may have some other important functional roles in telomerase nucleotide addition and repeat addition, such as interacting with other domains of TERT or the telomerase RNA.
Fig. 4.
Cis-telomerase cannot rescue the RAP defect of the Tetrahymena TERT L14A mutant. (A) Direct telomerase activity assay with the native Tetrahymena telomerase, WT and L14A mutant. In the reactions, unligated DNA primer was provided in trans at 0.08 μM and 0.4 μM for the WT, and 0.4 μM and 2 μM for the L14A mutant. (B) Telomerase assays of the Tetrahymena cis-telomerase RNA with wild-type TERT, the TERT TEN domain deletion mutant (Tetrahymena TERT aa192-1117), and the TERT L14A mutant. (C) Measurement of RAP for the native telomerase (with the primer provided in trans) and the cis-telomerase, in either wild type or L14A TERT background. RAP is calculated from the slope of the line. Processivity = R1/2 = ln 2/(2.303k), where k is the slope and R1/2 is the number of repeats synthesized before half of the primers have dissociated, in analogy to t1/2 in radioactive decay. (D) Telomerase assays of human cis-telomerase RNA with wild-type TERT, the TERT TEN domain deletion mutant (human TERT aa197-1132), and the TERT L14A mutant.
Discussion
In this paper, we have described the design and characterization of a cis-telomerase RNA that, when reconstituted with TERT, can add telomeric repeats to itself. While conversion of a ribozyme from transacting to cis-acting had been achieved before with RNase P, to our knowledge this is a unique case for an RNP enzyme. We utilized the cis system to examine some structural and mechanistic aspects of the telomerase RNP enzyme.
Tethering the DNA primer to the telomerase RNA with nucleic acid linkers of various lengths produces cis-telomerases that can elongate the conjugated DNA primers. When the linker length is 5 nt or longer, cis-telomerase has reached its full activity. This fact implies that the critical length between the 3′ end of the telomerase RNA pseudoknot region and the alignment region of the RNA template is no greater than 33 Å in the active telomerase. This result suggests close proximity of the pseudoknot domain to the RNA template region, consistent with the previous chemical photo-crosslinking data (20). Furthermore, we find that cis-telomerase with even a 2 nt-long linker is partially active, implying that the RNP structure is flexible enough to assume an active structure even when portions of the RNA are unnaturally constrained. The Tetrahymena ribozyme (a 310 nt catalytic RNA molecule) undergoes long-range conformational dynamics of up to 50 Å, and its catalytic activity persists when sites separated by 50 Å in the ground-state structure are joined by a covalent crosslink (32). Like this pure-RNA enzyme, the telomerase RNP appears to have a great deal of conformational flexibility.
Because the DNA primer in the context of the cis-telomerase is attached to the telomerase by a flexible linker, the entropic cost of realigning the primer to the template after one round of DNA synthesis is expected to be reduced. We indeed observed increased RAP in the cis-telomerase. The fact that the RAP defect of the Tetrahymena L14A mutant TERT could not be rescued in the cis-telomerase supports the previous conclusion (27) that the major role of L14 in the TEN domain is not to mediate the telomerase-DNA primer interaction, but rather to facilitate primer-template translocation and realignment.
By covalently linking the DNA primer to the RNA subunit of telomerase, active RNP enzymes can be directly selected through the reaction that they catalyze on themselves. Therefore, the cis-telomerase provides a method to separate functional and nonfunctional pools of telomerase RNAs, and perhaps of bound TERT proteins as well. This ability will be particularly useful for applying a high-throughput chemical genetics method—Nucleotide Analog Interference Mapping (NAIM) (33)—to identify chemical groups in the telomerase RNA that are important for telomerase activity. In addition, cis-telomerase also provides a platform to select for active RNA molecules in a library of random mutants, or to select for second-site revertants of deleterious mutations.
Materials and Methods
In Vitro Transcription of Telomerase RNA.
Telomerase RNA fragments were prepared by run-off transcription in vitro with bacteriophage T7 RNA polymerase. Transcription reactions (1 mL) contained 0.05 mg DNA template produced from PCR, 1 X T7 transcription buffer (50 mM Tris-HCl pH 8.0, 10 mM spermidine, 25 mM DTT, 0.1% Triton X-100, 25 mM MgCl2), 4 mM each NTP, and 0.1 mg T7 RNA polymerase. The reaction was incubated at 37 °C for 2 h. In vitro transcribed RNAs were gel-purified and eluted into TE buffer (25 mM Tris-HCl pH 8.0 and 1 mM EDTA).
Construction of Cis-Telomerase RNAs.
For human cis-telomerase RNA, in vitro transcribed telomerase RNA pseudoknot domain (either acceptor RNA A or B shown in Fig. S1B and C, resp.), 5-′monophosphorylated DNA primer (Table S1), and a DNA splint oligo (Table S1) were mixed at 1∶3∶2.5 molar ratio. The mixture was annealed (80 °C for 4 min followed by 25 °C for 5 min) in the presence of 50 mM NaCl and 10 mM Tris-HCl pH 8.0. Ligation was initiated by the addition of 1 U/mL T4 RNA ligase 2 (from New England Biolabs) in the presence of the ligation buffer (35 mM NaCl, 50 mM Tris-HCl at pH 7.0, 2 mM MgCl2, 1 mM DTT, 0.5 mM ATP) (34). Reactions were complete in 1 h at 25 °C. The yield of the splinted ligation was around 70–80%. Ligation product was then gel purified, eluted in TE buffer, and concentrated by Centricon (Millipore). Depending on the linker length of the cis-telomerase RNA, either acceptor RNA A or B was used in the splint ligation, as indicated in Table S1. In the case of Tetrahymena cis-telomerase RNA, additional ribonucleotides (CCUUGC) were added to the 3′ end of the full-length telomerase RNA to facilitate annealing/splinted ligation; 5′-monophosphorylated CTCTCCTTCTCTTGGG was ligated to the RNA.
In Vitro Telomerase Activity Assay.
The human and Tetrahymena telomerase activity assays followed the direct assay protocols of Chen and Greider (35) and Zaug et al. (27), resp. Briefly, epitope-tagged human and Tetrahymena TERT proteins were expressed from phTERT-HA2 and pET28a-T7-tTERT by using the TNT quick-coupled transcription/translation system (Promega). Each 50 μL reaction contained 40 μL TNT-master mix, 2 μL PCR enhancer, 1 μL 1 mM methionine, 7 μL water, and 0.5 μg plasmid DNA. After incubation at 30 °C for 2 h, 0.5 μg of cis-telomerase RNA (and CR4-CR5 domain for human telomerase assays) was added and incubated at 30 °C for 0.5 h. Lysate was diluted 2-fold with immunoprecipitation (IP) buffer (10 mM HEPES pH 7.5, 100 mM potassium glutamate, 1 mM MgCl2, 1 mM DTT and 10% glycerol). Assembled telomerase complexes were affinity-purified by using the C-terminal HA tag of recombinant human TERT protein with anti-HA agarose beads (Santa Cruz Biotechnology) or using the N-terminal T7 tag of recombinant Tetrahymena TERT with anti-T7·Tag agarose beads (Novagen). Each lysate was diluted to 100 μL with IP buffer and 15 μL of anti-HA agarose beads was added for immunoprecipitation at 4 °C for 3–4 h or overnight. Beads were washed with IP buffer three times and then resuspended in 1X telomerase assay buffer. The reaction mixture (20 μL each) contained 1X telomerase assay buffer (50 mM Tris-HCl pH 8.0, 50 mM KCl, 1 mM MgCl2, 5 mM β-mercaptoethanol and 1 mM spermidine), 5 mM dATP, 5 mM dTTP, 0.05 mM dGTP and 0.03 mM [α-32P]dGTP (3000 Ci/mmol) with 6 μL immuno-purified telomerase complex. The reaction was incubated at 30 °C for 30 min and the products were precipitated with the addition of 100 μL 3.6 M NH4OAc, 1 mg glycogen and 450 μL ethanol. The reaction mixture was incubated at -80 °C for 1 h followed by centrifugation at 4 °C for 30 min; the pellet was then washed with 75% cold ethanol and resuspended in 16 μL 1X RNA loading buffer (40% formamide, 10 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.05% xylene cyanol). The denatured samples were loaded onto a 10% polyacrylamide/TBE/7 M urea denaturing gel for electrophoresis. After electrophoresis, the gel was dried and exposed to a phosphorimager screen (Amersham Biosciences). Telomerase processivity was quantitated as described previously (36). In brief, the intensity of each major repeat band was measured, and normalized to the number of 32P-labeled nucleotides incorporated. The log of the normalized intensities was then plotted versus the repeat number.
Supplementary Material
Acknowledgments.
We thank Arthur Zaug, Elaine Podell, and Jayakrishnan Nandakumar (all at the University of Colorado-Boulder) for materials and helpful discussions. F.Q. was a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. This work was supported in part by National institute of Health Grant R01 GM28039.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909366107/DCSupplemental.
References
- 1.Cech TR. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 2.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 3.Steitz TA, Moore PB. RNA, the first macromolecular catalyst: The ribosome is a ribozyme. Trends Biochem Sci. 2003;28:411–418. doi: 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- 4.Madhani HD, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 5.Evans D, Marquez SM, Pace NR. RNase P: Interface of the RNA and protein worlds. Trends Biochem Sci. 2006;31:333–341. doi: 10.1016/j.tibs.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Kikuchi Y, Sasaki-Tozawa N, Suzuki K. Artificial self-cleaving molecules consisting of a tRNA precursor and the catalytic RNA of RNase P. Nucleic Acids Res. 1993;21:4685–4689. doi: 10.1093/nar/21.20.4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank DN, Harris ME, Pace NR. Rational design of self-cleaving pre-tRNA-ribonuclease P RNA conjugates. Biochemistry. 1994;33:10800–10808. doi: 10.1021/bi00201a030. [DOI] [PubMed] [Google Scholar]
- 8.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu Rev Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 9.Collins K. The biogenesis and regulation of telomerase holoenzymes. Nat Rev Mol Cell Biol. 2006;7:484–494. doi: 10.1038/nrm1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- 11.Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- 12.Chen JL, Blasco MA, Greider CW. Secondary structure of vertebrate telomerase RNA. Cell. 2000;100:503–514. doi: 10.1016/s0092-8674(00)80687-x. [DOI] [PubMed] [Google Scholar]
- 13.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 14.Leonardi J, Box JA, Bunch JT, Baumann P. TER1, the RNA subunit of fission yeast telomerase. Nat Struct Mol Biol. 2008;15:26–33. doi: 10.1038/nsmb1343. [DOI] [PubMed] [Google Scholar]
- 15.Webb CJ, Zakian VA. Identification and characterization of the Schizosaccharomyces pombe TER1 telomerase RNA. Nat Struct Mol Biol. 2008;15:34–42. doi: 10.1038/nsmb1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen JL, Greider CW. An emerging consensus for telomerase RNA structure. P Natl Acad Sci USA . 2004;101:14683–14684. doi: 10.1073/pnas.0406204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin J, et al. A universal telomerase RNA core structure includes structured motifs required for binding the telomerase reverse transcriptase protein. P Natl Acad Sci USA. 2004;101:14713–14718. doi: 10.1073/pnas.0405879101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theimer CA, Blois CA, Feigon J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol Cell. 2005;17:671–682. doi: 10.1016/j.molcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 19.Shefer K, et al. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol Cell Biol. 2007;27:2130–2143. doi: 10.1128/MCB.01826-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat Struct Mol Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chappell AS, Lundblad V. Structural elements required for association of the Saccharomyces cerevisiae telomerase RNA with the Est2 reverse transcriptase. Mol Cell Biol. 2004;24:7720–7736. doi: 10.1128/MCB.24.17.7720-7736.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller MC, Collins K. Telomerase recognizes its template by using an adjacent RNA motif. P Natl Acad Sci USA. 2002;99:6585–6590. doi: 10.1073/pnas.102024699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cech TR. Crawling out of the RNA world. Cell. 2009;136:599–602. doi: 10.1016/j.cell.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Armanios MY, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 25.Chen JL, Greider CW. Telomerase RNA structure and function: Implications for dyskeratosis congenita. Trends Biochem Sci. 2004;29:183–192. doi: 10.1016/j.tibs.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Vulliamy T, et al. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 27.Zaug AJ, Podell ER, Cech TR. Mutation in TERT separates processivity from anchor-site function. Nat Struct Mol Biol. 2008;15:870–872. doi: 10.1038/nsmb.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore MJ, Query CC. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- 29.Tesmer VM, et al. Two inactive fragments of the integral RNA cooperate to assemble active telomerase with the human protein catalytic subunit (hTERT) in vitro. Mol Cell Biol. 1999;19:6207–6216. doi: 10.1128/mcb.19.9.6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saenger W. New York: Springer-Verlag; 1984. Principles of nucleic acid structure. [Google Scholar]
- 31.Gavory G, et al. Structural analysis of the catalytic core of human telomerase RNA by FRET and molecular modeling. Biochemistry. 2006;45:13304–13311. doi: 10.1021/bi061150a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cohen SB, Cech TR. Dynamics of thermal motions within a large catalytic RNA investigated by cross-linking with thiol-disulfide interchange. J Am Chem Soc. 1997;119:6259–6268. [Google Scholar]
- 33.Strobel SA, Shetty K. Defining the chemical groups essential for Tetrahymena group I intron function by nucleotide analog interference mapping. P Natl Acad Sci USA. 1997;94:2903–2908. doi: 10.1073/pnas.94.7.2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nandakumar J, Ho CK, Lima CD, Shuman S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J Biol Chem. 2004;279:31337–31347. doi: 10.1074/jbc.M402394200. [DOI] [PubMed] [Google Scholar]
- 35.Chen JL, Greider CW. Functional analysis of the pseudoknot structure in human telomerase RNA. Proc Natl Acad Sci USA. 2005;102:8080–8085i. doi: 10.1073/pnas.0502259102. discussion 8077–8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F, et al. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature. 2007;445:506–510. doi: 10.1038/nature05454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.