Abstract
The stochasticity of chromosome organization was investigated by fluorescently labeling genetic loci in live Escherichia coli cells. In spite of the common assumption that the chromosome is well modeled by an unstructured polymer, measurements of the locus distributions reveal that the E. coli chromosome is precisely organized into a nucleoid filament with a linear order. Loci in the body of the nucleoid show a precision of positioning within the cell of better than 10% of the cell length. The precision of interlocus distance of genomically-proximate loci was better than 4% of the cell length. The measured dependence of the precision of interlocus distance on genomic distance singles out intranucleoid interactions as the mechanism responsible for chromosome organization. From the magnitude of the variance, we infer the existence of an as-yet uncharacterized higher-order DNA organization in bacteria. We demonstrate that both the stochastic and average structure of the nucleoid is captured by a fluctuating elastic filament model.
Keywords: chromosome segregation, chromosome structure, nucleoid structure, polymer physics
Prokaryotic chromosomes are organized into a compact DNA-protein complex called the nucleoid (1, 2). The physical structure of chromosomes has functional consequences, for example it affects gene regulation from the simplest prokaryotes (1) to multicellular organisms (3). Nucleoid organization and condensation also appear to play a central role in chromosome segregation: Mutants with defective chromosome segregation are typically accompanied by abnormal nucleoid organization or condensation (2). Although a significant number of such genes have been identified by genetic screens, the mechanism by which these molecular players effect the cellular-scale nucleoid structure is not yet understood (2). Similarly, the mechanism by which prokaryotic chromosomes are segregated is still hotly debated (2, 4, 5). The apparent dispensability of a mitotic-spindle-like mechanism in chromosome segregation in Escherichia coli (2, 4) has led to speculation that nucleoid organization and segregation may be the result of several redundant mechanisms, including polymer physics—embodied by the combined effects of entropy, confinement, and excluded volume—rather than resulting from the action of dedicated cellular machinery alone (2, 6, 7).
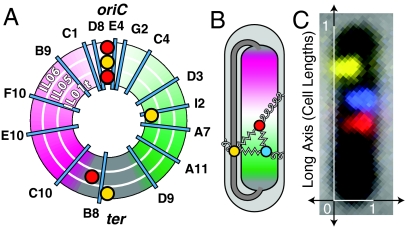
This paper complements earlier work by focusing on the measurement and theoretical interpretation of two classes of statistical measures of chromosome organization: (i) the distributions of the positions of individual loci within the cell; and (ii) the distributions of displacements between pairs of genetic loci. We argue that the measurement and analysis of these distributions sheds light on the mechanisms of chromosomal positioning that have not been revealed in earlier measurements. The cellular-scale structure of the circular Caulobacter crescentus chromosome has already been shown to be linearly organized between replication cycles, with the origin of replication at one pole and the terminus (ter) at the other (8). A model based on qualitative measurements of the circular E. coli chromosome structure suggests that the E. coli nucleoid has a similar structure with the origin at midcell (6, 9, 10, 11). (See Fig. 1A, B for schematic illustration of the E. coli nucleoid organization.) Our quantitative measurements of E. coli nucleoid structure confirm this qualitative picture: The body of the nucleoid is linearly organized along the long-axis of the cell, implying that the nucleoid has a nearly-constant linear packing density, except for a short region, genomically ter-proximate, which connects the two arms of the chromosome (11). But in spite of the observation of a linear chromosome organization in both E. coli and C. crescentus, the mechanism which gives rise to this characteristic structure is unknown.
Fig. 1.
Experimental schematic: The E. coli fiducial strains (IL01t, IL05, IL06) each carry two fiducial fluorescent loci (red and yellow foci) in addition to a probe locus (cyan lines). (A) A schematic map of the genomic location of the probes by strain. The three concentric rings represent the three fiducial strains. (B) The left and right arms of the E. coli chromosome are positioned on opposite sides of the nucleoid, with the origin at midcell. The terminus-proximate loci are positioned at both ends of the nucleoid, as well as in a crossing region that bridges the two poles of the nucleoid. External positioning mechanisms, which position a locus directly relative to the cell, are represented by curly springs. Internal positioning mechanisms, which position loci relative to one another, are represented by zigzag springs. (C) A typical composite image for the “IL06 I2” strain.
To probe the mechanism of nucleoid organization, we measure and analyze the position fluctuations of single loci and the correlation between the fluctuations of locus pairs within a unique conceptual framework. We consider locus positioning mechanisms which fall into two generic classes: external and internal. An external mechanism positions a locus directly, without acting through any other genetic loci in the nucleoid. For instance, a protein factor like FtsK, which is positioned at the septum and binds a ter-proximate locus, positions that locus by an external mechanism. An internal mechanism positions a locus relative to another locus in the nucleoid. For instance, proteins like H-NS, which is a DNA bridging protein, and MukBEF, mediate DNA-DNA interactions and therefore structure the nucleoid by an internal mechanism (12). To differentiate between these two modes of positioning, we independently measure the single-locus position and the interlocus distance distributions. Loci that are strongly coupled by intranucleoidal interactions are expected to exhibit correlated fluctuations from their mean positions, which is not the case if external positioning mechanisms are dominant*.
To quantitatively interpret these correlations and the locus positioning distributions more generally, we introduce a model of nucleoid structure: the Fluctuating Filament Model; see Fig. 4. This model includes intranucleoid interactions as well as two explicit mechanisms of external positioning: (i) cellular tethering of loci; and (ii) cellular confinement of the chromosome. The analysis of the experimentally determined locus distributions in the context of this model reveals that the nucleoid positioning in slow growing E. coli cells in G1 phase (before the initiation of segregation) is dominated by strong intranucleoid interactions. Strong external positioning occurs only at the poles. The precision of this positioning, its dependence on cell length, and the fine-scale features of the locus position distributions of pole-proximate loci, all strongly suggest that cellular confinement plays the dominant role in external positioning in G1 phase. The observed high degree of internal ordering has implications for chromosome segregation and suggests that the E. coli chromosome is folded into a prokaryotic chromosome fiber.
Fig. 4.
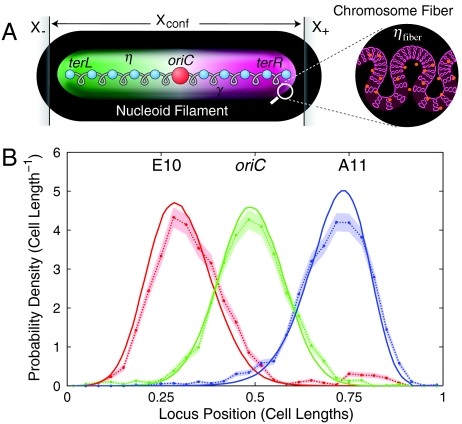
(A) The Fluctuating Filament Model. The nucleoid is represented by an elastic body with effective elasticity γ. The genome is condensed (from ter-left to ter-right) to form a nucleoid filament with constant linear DNA packing density η. Confinement of the nucleoid between positions X- and X+ acts to position the poles of the nucleoid. The chromosome fiber is folded to form the nucleoid filament. The packing density of the chromosome fiber is ηfiber. (B) Predicted singe-locus position distributions for three loci. The observed distributions (points and error regions) show both quantitative and qualitative agreement with the predicted distributions (solid curves). In particular, the predicted distributions reproduce the confinement-induced asymmetry for oriC-distant loci.
Results
Locus Distributions.
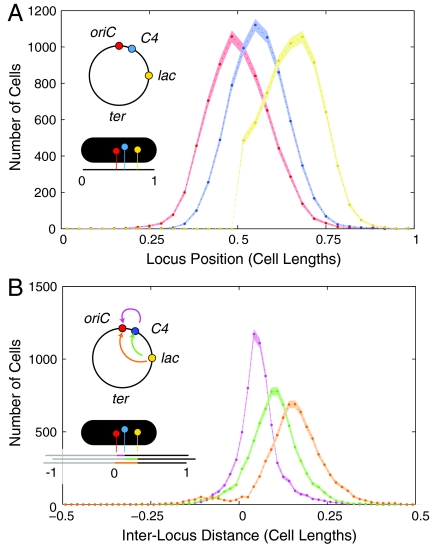
Single-locus position and interlocus distance distribution functions were generated from measurements of the locus positions. The distributions for the “IL01t C4” strain are shown in Fig. 2A, B. The distribution of loci positions in the body of the nucleoid (ter-distant) were found to be well-approximated by a Gaussian. To characterize the dependence of the distributions on genomic position, we fit the experimentally determined distributions to Gaussian distributions to determine the mean and variance as best-fit values. oriC-distant loci were characterized by asymmetric Gaussian-like distributions which decayed more steeply on the side closer to the cell pole. (See lac distribution in Fig. 2A.) For ter-proximate loci, cells showed two behaviors: Loci were either positioned at one of the poles of the nucleoid, or in an unpositioned state that was distributed uniformly between the poles of the nucleoid. (Please see SI Text.) The structural significance of this crossing region is clear from the schematic drawing (Fig. 1B): The two poles of the nucleoid are connected by a low-packing-density fiber that stretches between them. We estimate the crossing region consists of just 8% of the genome† and therefore has a packing density of just 1/10 that of the body of the nucleoid. This low packing density marks the crossing region as structurally distinct and therefore a potential target of remodeling. Although the genomic location of this region is always ter-proximate, it is stochastic.
Fig. 2.
(A) Histogram of long-axis locus position for IL01t C4 cells. The genomic locus positions are shown schematically in the inset. (B) Histogram of interlocus long-axis distance between loci. The interlocus distance distributions for oriC-C4 (magenta), C4-lac (green), and lac-oriC (orange) reveal that the variance of the distribution increases with the mean distance.
Mean Locus Position.
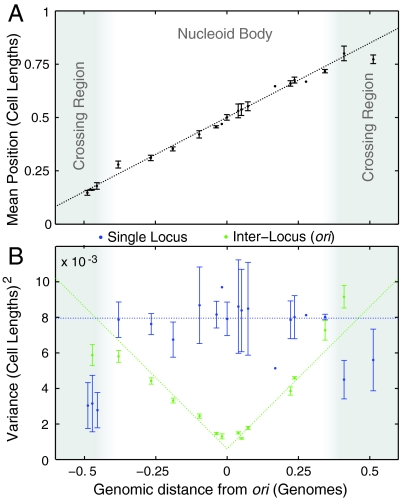
The mean position of loci is shown as a function of genomic position in Fig. 3A. As described previously, oriC is positioned at midcell and the left and right arms of the chromosome are positioned on opposing sides of the cell (11, 13). The most striking feature of this plot is that the mean position is linear in the genomic position, interpolating smoothly from ter-left at the left pole, through the origin at midcell, to ter-right at the right pole. This linear dependence implies that the nucleoid is linearly organized along the long-axis and has an approximately constant linear packing density of 1.6 Mb/μm. This nucleoid structure is strikingly reminiscent of C. crescentus, where a similar linear organization was observed although with ori and ter positioned at opposing poles (8).
Fig. 3.
(A) The nucleoid is linearly organized. The mean long-axis position of probe loci is plotted as a function of genomic distance from oriC. (The error bars represent day-to-day variation.) (B) The precision of locus positioning. The variance, σ2, of locus position measures the precision of locus positioning. The single-locus variance (blue) is roughly constant throughout the nucleoid body (excluding the crossing region). (The error bars represent day-to-day variation.) The oriC-probe interlocus variance (green) measures the precision of probe-locus position relative to the position of oriC.
Locus Variance.
The notion of locus positioning implies more than a mean value; it implies a precision. A locus uniformly distributed throughout the cell would have a mean position at midcell, but would not be considered “well-positioned.” The precision of positioning is quantified by the variance (σ2) of the locus position distribution. In Fig. 3B, we plot the single-locus and the oriC-probe interlocus variance as a function of genomic position. The single-locus variance (blue) is roughly constant throughout the body of the nucleoid‡. All loci in the body are positioned with an accuracy of σ ∼ 9% of the cell length. In Fig. 3B, we plot the interlocus variance with respect to oriC (green). In marked contrast to the single-locus variance, which is approximately constant throughout the body of the nucleoid, the interlocus variance is strongly dependent on the interlocus genomic distance between loci. The oriC-proximate loci are positioned with much greater precision (that is, smaller variance) with respect to oriC than with respect to the cell. The positions of oriC and oriC-proximate loci are therefore highly correlated. Although this result is not completely unexpected, our measurements reveal that the variance increases linearly with interlocus genomic distance:
| [1] |
This coupling is not unique to oriC, but most likely a general feature of nucleoid structure§. All loci in the body of the nucleoid appear to be positioned with greater precision relative to their neighbors than with respect to the cell, suggesting that intranucleoidal interactions play a central role in shaping nucleoid structure.
Fluctuating Filament Model.
The qualitative conclusions about the role of intranucleoidal interactions in shaping nucleoid structure can be explored quantitatively in the context of an effective physical model. We introduce the Fluctuating Filament Model which represents the nucleoid as a fluctuating elastic filament, rather than resolving the detailed DNA conformation. (The Fluctuating Filament Model is shown schematically in Fig. 4A and is discussed in detail in SI Text, The Fluctuating Spring Model). The filament is assumed to have a constant genome packing density η (expressed in units of base pairs per micron) between ter-left and ter-right¶. The undeformed length of the nucleoid is Xnucl = genome length/η. The fluctuations of the nucleoid are described by an effective elastic modulus, γ, expressed in units of base pairs per micron squared∥. η and γ are assumed to completely characterize the intranucleoid interactions. In addition to internal organization, we confine the nucleoid to a region length Xconf by forbidding DNA loci outside the region X- to X+. We then compute the probability of different conformations assuming a Boltzmann Law: The probability of a given configuration is p ∝ exp(-G), where G is the total elastic energy of the Fluctuating Filament Model expressed in thermal units, kBT. The model statistics are calculated by summing over nucleoid configurations, which is performed by Metropolis Monte Carlo Integration.
Model Results.
To illustrate the role of the positioning mechanisms in shaping chromosome structure, we have computed the distribution functions for an expository nucleoid structure that is organized by both tethering interactions and confinement, as well as intranucleoid interactions. These calculation are described in detail in SI Text, The Fluctuating Spring Model. We summarize the general features of the model in this section.
(i) The Fluctuating Filament Model generically recovers the linear dependence of the interlocus distance variance on genomic distance (L) for proximate loci 1. The linear dependence of the interlocus variance is a generic signature of positioning by uniform nearest-neighbor interactions**.
(ii) The single-locus variance is shaped by both external and internal interactions. At genomic positions that are tethered to the cell, the variance has a local minimum††. The nucleoid elasticity determines the depth of these tethering-induced minima relative to the variance in the positioning of proximate loci.
(iii) Confined nucleoids (Xnucl≥Xconf) can result in nearly-constant single-locus variance‡‡ but, confinement manifests itself in the shape of the position distribution functions, which display a characteristic asymmetry: the distribution function decays more steeply on the side directly affected by confinement§§.
Discussion
Intranucleoidal Interactions.
The observed linear dependence of the interlocus variance on interlocus genomic distance strongly suggests that intranucleoid interactions play a dominant role in shaping the E. coli nucleoid. This measurement alone argues strongly against multiple individually addressed (externally positioned) loci (1, 8), unless these tethered loci are all located at the poles of the nucleoid. Likewise, the single-locus variance appears to be at a minimum only at the cell poles, further supporting the argument that no loci in the body of the nucleoid are strongly positioned by tethering interactions. If loci in the body of the nucleoid are tethered, these interactions must either result in very weak positioning or be short-lived. These results do not imply that DNA tethering does not play a significant role in structuring the nucleoid at other steps in the cell cycle¶¶.
The Case for Confinement.
Two mechanisms of external positioning—polar tethering and confinement—could be acting at the poles of the nucleoid to reproduce the observed variance data. A number of arguments favor the confinement model: (i) The nucleoid is known to be confined by the cell and the size of the variance is commensurate with what is expected from confinement-induced positioning; (ii) If our analysis is repeated for cell populations grouped by length, the precision of cellular positioning decays with cell length while the precision of relative intranucleoid positioning increases with cell length, as predicted in a confinement-based model***. Furthermore, in very long cells, where replication initiation has been arrested, the nucleoid does not continue to grow with the cell and is not positioned at the poles of the cell, as would be predicted by polar tethering. Analogously, reducing the length of the nucleoid by condensation also leads to the loss of nucleoid positioning (14); (iii) The distribution of pole-proximate loci exhibit the characteristic asymmetry predicted by confinement-induced positioning. (See the observed distributions in Fig. 4B and in SI Text, Quantitative analysis of locus histograms.) In short, there is no compelling argument to propose the existence of a yet-to-be-identified factor to position the terminus when cellular confinement is sufficient to position the nucleoid.
The G1 structure of the nucleoid is predicted by a simple model dominated by: (i) intranucleoid interactions that position the vast majority of loci; and (ii) cellular confinement which positions the poles of the nucleoid. The model is specified by just two parameters characterizing the nucleoid: the positioning modulus (γ = 57 ± 1 genomes/cell length2) characterizes the intranucleoid interactions, and the packing density (η = 1.16 ± 0.01 genomes/cell length). Two additional parameters, limiting coordinates of the nucleoid-accessible cytoplasm, describe the cell. (A cytoplasmic region length Xconf = 0.88 ± 0.01 cell lengths is accessible to the nucleoid.) The model parameters were determined by a chi-squared fit to the means and variances of the locus distributions†††. The model not only recapitulates the observed dependence of the mean and variance on locus position as a function of genomic position (Fig. 3), but predicts the entire single-locus and interlocus distribution functions for all loci in the body of the nucleoid. Fig. 4B compares the observed and predicted distributions functions for three loci. A particularly significant feature of this plot is the prediction of a fine-scale feature of the distribution: the confinement-induced asymmetry characterized by a steep decay on the confinement-proximate side of the distribution (left side of E10 and right side of A11). This feature is not fit in the determination of the optimal model parameters but is rather a prediction of the model.
Higher-Order DNA Structure.
Although we have argued that nucleoid positioning—for instance, the positioning of oriC at midcell—is predicted by the Fluctuating Filament Model, the two key structural features of this model (constant linear packing density and the intranucleoidal interactions) have not been derived from a microscopic model that explicitly treats the polymeric nature of the chromosome. To interpret the significance of our results, we now explicitly consider the predictions of an equilibrium polymer model. In a concentrated polymer solution, the Flory theorem predicts that the effects of excluded volume on the equilibrium polymer conformation can be ignored (15). In a polymer model, the effective elasticity is
 |
[2] |
where ηfiber is the basepair packing density of the chromosome fiber and ℓp is the bend persistence length‡‡‡. Higher-order structure (or lengthwise condensation (18)) condenses the DNA locally via genomically-proximate interactions (as opposed to long-ranged interactions which bridge genomically-distant sequences) to form a chromosome fiber. These models are still polymer models, but the chromosome fiber has a larger basepair packing density per unit fiber length (ηfiber) than double-stranded DNA (dsDNA). For instance, in eukaryotes 10 and 30 nm fiber are both examples of higher-order DNA structure. In the absence of higher-order structure, we expect the effective elasticity to be approximately that observed for unstructured dsDNA: γ = 90 kb/μm2, which is between two and three orders of magnitude smaller than the observed effective elasticity (γ = 3 × 104 kb/μm2). To match the observed γ, the persistence length of DNA would have to be reduced by a factor of 300 to half a base pair! Neither the addition of a large number of nucleoid associated proteins (NAPs) which bridge and bend the DNA (12) nor supercoiling can explain such a large discrepancy in γ, if the polymer structure is equilibrated. The large effective elasticity is therefore evidence for higher-order structure.
One possible structural realization of a prokaryotic chromosome fiber is an ordered stack of plectonemic loops, drawn schematically in the inset of Fig. 4A. It is assumed that the genomic positions of the plectonemes are essentially frozen in and cannot equilibrate. If these plectonemic loops corresponded to the 10 kb topological domains reported to structure the E. coli chromosome (19) and the plectoneme loop stems are spaced by 100 base pairs, the resulting chromosome fiber would have 100-fold larger fiber packing density (ηfiber) than dsDNA and therefore a 100-fold increase in the effective elasticity (γ), approaching the observed value. Intriguingly, a back-of-the-envelope estimate of the width of such a fiber suggests that it is on order 0.5 μm, roughly the same width as the cell and therefore the nucleoid filament itself§§§. This short, thick fiber is poorly approximated by Eq. 2: the nucleoid is better modeled by an elastic filament model than a polymer model.
The success of the Fluctuating Filament Model in describing the structure of the E. coli nucleoid suggests that the nucleoid may be more analogous to the precisely structured eukaryotic mitotic chromosome than to a eukaryotic interphase chromosome. The ordered structure of the nucleoid between replication cycles may have important implications for understanding chromosome segregation. Since the polymer model fails to describe the G1 structure of prokaryotic chromosomes, it is important to reconsider our assumptions about the nature of chromosome segregation in light of our new structural insights into chromosome structure.
Conclusion.
In this paper we characterize the stochasticity of the E. coli nucleoid structure between replication cycles. We measured both single-locus position and interlocus distance distribution. Analysis of single-locus position fluctuations revealed that all loci in the body of the nucleoid are positioned with a precision of roughly 10% of cell length with respect to the cell. The analysis of the coupling of genomically proximate loci reveals that loci are precisely positioned relative to their neighbors (< 4% cell length), suggesting that the nucleoid organization is dominated by intranucleoidal interactions. In particular, we demonstrate that the variance of the interlocus distance distribution grows linearly with their genomic separation, which is the signature dependence of a conformation structured by nearest-neighbor interactions. To investigate the role of these internal interactions, we introduce the Fluctuating Filament Model which approximates the E. coli nucleoid as a confined, elastic filament with a constant DNA packing density. The nucleoid is characterized by just two parameters; the linear packing density which governs the mean position of loci, and an effective elasticity which characterizes the fluctuations. The quantitative agreement of nucleoid structure with the Fluctuating Filament Model suggests that the most natural structural analogue of the nucleoid maybe the mitotic rather than the interphase eukaryotic chromosome. The precise internal order, relative to that predicted by polymer models, is strong evidence for higher-order DNA structure. We anticipate that these unique insights into the structure of the nucleoid between replication cycles will have important implications for our understanding of prokaryotic chromosome segregation.
Methods
The methods are described in detail in SI Text, Experimental Methods. To probe the nucleoid structure in live cells, we adapted both the Fluorescent Repressor Operator System (FROS) (20) and the ParB-parS system (21) to visualize three genetic loci concurrently. Both labeling technologies allow genomic loci to be visualized by the aggregation of fluorescently labeled proteins at an ectopic sequence introduced at the locus of interest. Our fiducial strains carry two fiducial labels, one at oriC and the other at an oriC-distant locus. The number of oriC loci is used to determine whether the cells have initiated the segregation process (20, 21). The second fiducial label is used to determine the left-right polarity of the cell (11, 13). (See Fig. 1 for schematic maps of the fiducial strains.) From the fiducial strains, we generated a collection of probe strains by inserting the parS sequence at a random, probe locus using a mariner-based transposon (8, 22). The snapshot technique was used to generate a population-based distribution of locus position. The microscopy is described in more detail in SI Text, Microscopy and growth conditions. Cells are segmented from a phase contrast image, and loci in the three fluorescence channels (RFP, YFP, and CFP) are identified, counted, and their position determined along the long and short axes of the cell by custom MATLAB software. Each cell is oriented along the long-axis using the position of a fudicial label on either the left or the right arm of the chromosome. Fig. 1C shows the image of a typical E. coli cell with all three fluorescent labels. We analyze locus positioning in cells in which we observe a single oriC locus. Since the loci undergo a period of sister cohesion after replication, but before segregation, the population we study is most likely a mix of cells in G1 and early S phase (6). The strains, plasmids, growth conditions, and imaging protocols are described at length in SI Text.
Supplementary Material
Acknowledgments.
The authors would like to thank Hernan Garcia, Dieter Heermann, Suckjoon Jun, and Rob Phillips for their comments and suggestions. The authors are also especially grateful to Aude Bourniquel and Nancy Kleckner, whose preliminary data and insights into nucleoid structure influenced our own interpretation of the results presented in this paper. P.A.W. and R.L. were supported by National Science Foundation Grant PHY-084845 and the Whitehead Institute for Biomedical Research. J.K., J.M., and K.C. were supported in part by the National Science Foundation, DMR-0706458 and MRSEC-0820492.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912062107/DCSupplemental.
*Elaborate models can be proposed that cannot be simply characterized in this framework. For instance, positioning mechanisms can be proposed in which the position of the external chromosome tethers are correlated.
†See SI Text The crossing region.
‡The interpretation of the variation in the crossing region is more subtle since these distributions are not well-approximated by a Gaussian distribution. Please see the discussion in SI Text, The crossing region.
§The probe-lac interlocus variance shows the same trend. Please see SI Text, Intranucleoidal interactions are not unique to oriC.
¶The crossing region is not included in the model.
∥If the nucleoid filament is in thermal equilibrium, γ is the elastic modulus of the filament divided by kT. Out of equilibrium, the interpretation of γ depends on the details of the active processes which structure the nucleoid. Please see the discussion in SI Text, Elastic constant need not be mechanical in nature for a detailed discussion.
**See SI Text, Elastic constant need not be mechanical in nature.
††See SI Text, The model results for the expository nucleoid structure..
‡‡See SI Text, The model results for the expository nucleoid structure.
§§See SI Text, Asymmetric distributions.
¶¶In fact, ter is known to be tethered at the z-ring in late S-C phase.
***See SI Text, The dependence of the model parameters on cell length.
†††See SI Text, The Fluctuating Spring Model fit to experimental data..
‡‡‡Recent studies have suggested that the Flory Theorem may fail to describe tightly confined polymers and that confinement may play a role in segregating and organizing the chromosome (7, 16). On the other hand, rigorous tests of the Flory Theorem have demonstrated that the theorem holds in a regime we believe to be relevant for describing unstructured DNA in the cell (17). As we consider larger fiber packing densities, these filaments are no longer described by Eq. 2 since the monomer size becomes significant in relation to the confinement radius of the nucleoid.
§§§Please see the estimate in SI Text, Polymer interpretation of the nucleoid stiffness.
References
- 1.Thanbichler Martin, Viollier PH, Shapiro Lucy. The structure and function of the bacterial chromosome. Curr Opin Genet Dev. 2005;15(2):153–162. doi: 10.1016/j.gde.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Reyes-Lamothe Rodrigo, Wang Xindan, Sherratt David. Escherichia coli and its chromosome. Trends Microbiol. 2008;16(5):238–245. doi: 10.1016/j.tim.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Dekker Job. Gene regulation in the third dimension. Science. 2008;319(5871):1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamaichi Yoshiharu, Niki Hironori. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 2004;23(1):221–233. doi: 10.1038/sj.emboj.7600028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toro Esteban, Hong Sun-Hae, McAdams HH, Shapiro Lucy. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA. 2008;105(40):15435–15440. doi: 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bates David, Kleckner Nancy. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121(6):899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jun Suckjoon, Mulder Bela. Entropy-driven spatial organization of highly confined polymers: Lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103(33):12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viollier PH, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101(25):9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14(2):212–223. [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Xindan, Possoz Christophe, Sherratt DJ. Dancing around the divisome: Asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005;19(19):2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Xindan, Liu Xun, Possoz Christophe, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20(13):1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luijsterburg MS, Noom MC, Wuite GJL, Dame RTH. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: A molecular perspective. J Struct Biol. 2006;156(2):262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen HJ, Ottesen JR, Youngren Brenda, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006;62(2):331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang Qinhong, Mordukhova EA, Edwards AL, Rybenkov VV. Chromosome condensation in the absence of the non-smc subunits of mukbef. J Bacteriol. 2006;188(12):4431–4441. doi: 10.1128/JB.00313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grosberg AY, Khokhlov AR. Statistical Physics of Macromolecules. New York: AIP Press; 1994. pp. 154–155. Ch. 4. [Google Scholar]
- 16.Jun Suckjoon, Thirumalai D, Ha B-Y. Compression and stretching of a self-avoiding chain in cylindrical nanopores. Phys Rev Lett. 2008;101(13):138101. doi: 10.1103/PhysRevLett.101.138101. [DOI] [PubMed] [Google Scholar]
- 17.Lua R, Borovinskiy AL, Grosberg AY. Fractal and statistical properties of large compact polymers: A computational study. Polymer. 2004;45:717–731. [Google Scholar]
- 18.Marko JF. Linking topology of tethered polymer rings with applications to chromosome segregation and estimation of the knotting length. Phys Rev E Statistical, Nonlinear, and Soft Matter Physics. 2009;79(5 Pt 1):051905. doi: 10.1103/PhysRevE.79.051905. [DOI] [PubMed] [Google Scholar]
- 19.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18(14):1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau IF, et al. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49(3):731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, Sergueev K, Austin S. The segregation of the Escherichia coli origin and terminus of replication. Mol Microbiol. 2002;46(4):985–995. doi: 10.1046/j.1365-2958.2002.03234.x. [DOI] [PubMed] [Google Scholar]
- 22.Chiang SL, Rubin EJ. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. 2002;296(1-2):179–185. doi: 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.