Abstract
Ras and its effector Raf are key mediators of the Ras/Raf/MEK/ERK signal transduction pathway. Mutants of residue Q61 impair the GTPase activity of Ras and are found prominently in human cancers. Yet the mechanism through which Q61 contributes to catalysis has been elusive. It is thought to position the catalytic water molecule for nucleophilic attack on the γ-phosphate of GTP. However, we previously solved the structure of Ras from crystals with symmetry of the space group R32 in which switch II is disordered and found that the catalytic water molecule is present. Here we present a structure of wild-type Ras with calcium acetate from the crystallization mother liquor bound at a site remote from the active site and likely near the membrane. This results in a shift in helix 3/loop 7 and a network of H-bonding interactions that propagates across the molecule, culminating in the ordering of switch II and placement of Q61 in the active site in a previously unobserved conformation. This structure suggests a direct catalytic role for Q61 where it interacts with a water molecule that bridges one of the γ-phosphate oxygen atoms to the hydroxyl group of Y32 to stabilize the transition state of the hydrolysis reaction. We propose that Raf together with the binding of Ca2+ and a negatively charged group mimicked in our structure by the acetate molecule induces the ordering of switch I and switch II to complete the active site of Ras.
Keywords: GTPase, Raf, calcium, intrinsic hydrolysis
The Ras/Raf/MEK/ERK signaling pathway is the most well studied of five known mitogen activated protein kinase (MAPK) cascades involved in the mediation and timing of signaling events in the cell (1). This pathway is activated by Ras GTPase in response to extracellular signals and is involved in the control of cell proliferation, differentiation, and survival (2). In its resting state Ras is bound to GDP and is in a conformation in which it does not interact with Raf or other effector proteins (3). Guanine nucleotide exchange factors facilitate the release of GDP (4). Once the more abundant GTP binds, the Ras switch I (residues 30–40) and switch II (residues 60–76) regions become poised for interaction with effector proteins, leading to the propagation of signal transduction cascades. Ras has a low intrinsic rate of GTPase activity that is enhanced by 3–5 orders of magnitude in the presence of GTPase activating proteins (GAPs), resulting in depletion of Ras-GTP as the switch is turned off (5).
The biochemical properties of Ras and its oncogenic mutants have been well characterized in the absence of Raf or other factors (6, 7), and numerous structures of wild-type and oncogenic Ras mutants have been used to study the possible mechanisms through which Ras becomes defective in its ability to hydrolyze GTP (8–12). The switch regions in these structures, solved from the crystal form with symmetry of space group P3221, are modulated by crystal contacts to resemble the switch I and switch II conformations found in the Ras/RasGAP complex (5, 13). Since the structure of this complex has elucidated the mechanism through which GAP enhances the GTPase activity of Ras (5), it seems reasonable, given its similarity to the canonical structure of the uncomplexed form, that it could also serve as a framework for the mechanism of intrinsic hydrolysis in Ras (14, 15). Based on this assumption, a mechanism for intrinsic hydrolysis has been proposed with a two-water model: The γ-phosphate of GTP abstracts a proton from W189, which in turn activates the catalytic W175 for nucleophilic attack on the γ-phosphate during the hydrolysis reaction (16).
More recently, we proposed an alternative mechanism based on the structure of Ras from crystals with symmetry of space group R32 (17), where switch I and water molecules in the active site are as observed in the Raps*/Raf complex (18) and switch II is unhindered by crystal contacts. It is clear that the R32 crystal form is an excellent mimic for the Ras active site in the complex with Raf (17) and could model events that may occur in this complex in the absence of GAPs. Raf interacts with Ras-GTP through two domains: the Ras-binding domain (RBD) and the cystein-rich domain (CRD) (19). The crystal structure of Raf-RBD in complex with Raps-GppNHp shows the conserved switch I residue Y32 with its hydroxyl group interacting with a γ-phosphate oxygen atom through a bridging water molecule (20), exactly as we see in our structure (17). In our proposed mechanism a proton from the catalytic water molecule is shuttled via the γ-phosphate to the water molecule bridging the γ-phosphate to Y32 (which we have also named W189 due to its proximity to the γ-phosphate, although it does not overlap with the position of W189 in the P3221 crystal form) and is eventually delivered to the GDP leaving group. The fact that switch II is disordered with no electron density for Q61 means that a key catalytic residue was left out of this mechanism. In the present paper, we resolve this issue with analysis of a crystal structure of wild-type Ras in which switch II is ordered through an allosteric switch. We show that the remote allosteric site binds Ca2+ but not Mg2+ in the crystals. The result is a more complete picture of our proposed mechanism of intrinsic hydrolysis in Ras, and a paradigm shift for future studies of regulation in the Ras/Raf//MEK/ERK pathway.
Results
H-Ras with 23 residues truncated at the C-terminus is used for the structural studies and includes the entire catalytic domain, residues 1–166 (referred to as Ras throughout the paper). The RasY32F mutant is similarly truncated. The structure of Ras in which switch II is disordered was obtained from crystals grown in 200 mM calcium chloride, with symmetry of the space group R32 (17). More recently we found that the crystals grew larger if calcium acetate was used instead of calcium chloride, and the resulting model revealed structural features that are explored here. Wild-type Ras-GppNHp was solved in the presence of calcium acetate to 1.3 Å resolution from a crystal with R32 symmetry. Crystals removed from the mother liquor and soaked either in 200 mM calcium acetate or 200 mM magnesium acetate yielded structures solved to 1.85 and 1.86 Å resolution, respectively, and crystals transferred to calcium acetate after a magnesium acetate soak resulted in a 2.1 Å resolution structure. The RasY32F-GppNHp structure was solved from a crystal with symmetry of the space group P212121 to 1.8 Å resolution. Data collection and refinement statistics for the five structures are presented in Table S1.
Structure of Wild-Type Ras in the Presence of Calcium Acetate Reveals Allosteric Modulation of Switch II.
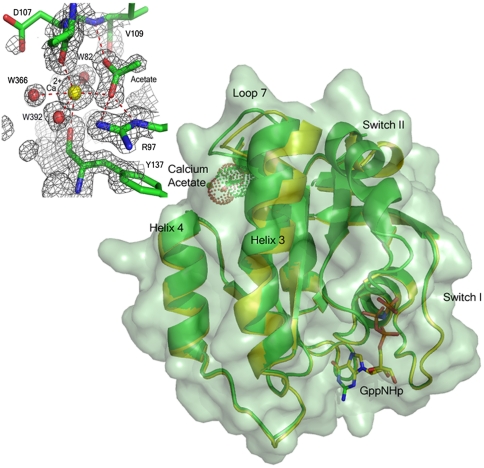
The overall structure of wild-type Ras in the presence of 200 mM calcium acetate is similar to that in the presence of 200 mM calcium chloride (PBD ID code 2RGE), with one striking exception: There is a shift of loop7, helix 3, and the C-terminal end of switch II, which culminates in the ordering of the N-terminal portion of switch II and placement of Q61 in the active site near the bridging water molecule that interacts with Y32 and the O1G atom of GppNHp (Fig. 1). This shift is modulated by the binding of calcium acetate at a site remote from the catalytic center. The structural elements involved in the shift include the C-terminal end of helix 3 (residues 98–103) and the sequentially adjacent loop 7 (residues 104–108). Anchoring residues on either end of this shifted region interact directly with the bound acetate molecule: The side chain of R97 interacts with one of the acetate oxygen atoms and the main chain amide of V109 donates an H-bond to the other (Fig. 1). Accompanying the shift in residues 98–108 is also a shift in residues 69–75 comprising the C-terminal portion of switch II, with R68 nestled between helix 3 and residues 60–67 at the N-terminal portion of the switch.
Fig. 1.
The allosteric switch in Ras. Ribbon diagram of Ras-GppNHp showing the shift in helix 3 and loop 7 due to the binding of calcium acetate in the allosteric site. Ras bound to calcium acetate is shown in green with a semitransparent surface. Note the presence of switch II in the model. Ras in the absence of calcium acetate is in yellow (PBD ID code 2RGE). The GppNHp and acetate (with van der Waals dot surface) are shown in stick. (Upper Left) Details of the calcium acetate binding site with a 2Fo-Fc electron density map contoured at the 1σ level. Dashed lines indicate H bonds and calcium ion coordination. All figures were generated in PyMol.
The Ca2+ ion binds between helix 4 and loop 7. It is hexacoordinated, interacting closely with one of the acetate oxygen atoms, three water molecules, W82, W392, W366, and the main chain carbonyl oxygen atoms of D107 and Y137, which are opposite each other in the coordination sphere (Fig. 1). The observed shift appears to be a result of a decrease in distance between these two residues due to their involvement in coordinating the Ca2+ ion. While the carbonyl of Y137 on helix 4 changes positions only slightly, D107 shifts significantly toward Y137, bringing with it loop 7 and the C-terminal portion of helix 3. The shift creates a set of conditions leading to formation of two connected H-bonding networks, network 1 and network 2 described below, that have the effect of ordering switch II and placing Q61 in what we propose is its precatalytic conformation at the beginning of the hydrolysis reaction. These H-bonding networks comprise the elements of an allosteric switch, which in the present structure is in the “on” conformation, and which is “off” in our previously published structures of wild-type Ras and of the RasQ61L oncogenic mutant (17).
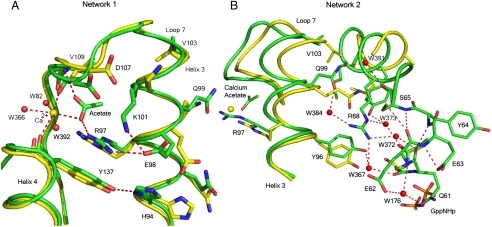
Network 1 includes the calcium acetate with R97 at the center. It involves H94, R97, E98, and K101 (on the side of helix 3 facing away from switch II), the side chains of D107 (loop 7), and Y137 (helix 4) (Fig. 2A). This network, consisting entirely of protein atoms and the calcium acetate, clusters in a space that is largely occupied by crystallographic water molecules in the structure of Ras in the presence of calcium chloride. With the helix 3 residues that face helix 4 placed within network 1, the helix 3 residues Y96, Q99, and R102 facing switch II are in position to contribute, along with switch II residues M72, Y71, D69, R68, S65, E62, and several water molecules, to network 2 centered around R68 (Fig. 2B). This leads to an extension of the α-helix found in the C-terminal half of switch II.
Fig. 2.
H-bonding networks 1 and 2 linking the allosteric site to Q61. The structures in the presence and absence of calcium acetate are shown in green and yellow, respectively. Dashed lines indicate H-bonding interactions involved in the network. (A) Network 1 centered on R97. Shift in helix 3 not visible from this orientation. (B) Network 2 centered on R68, showing the interactions that stabilize switch II. In this orientation the shift in helix 3 is clearly visible. Note the steric overlap between the ordered switch II in the calcium acetate bound structure (green) with helix 3 residues in the structure with an empty allosteric site (yellow). R68 is connected through single water molecules to Y96 (W367) and Q99 (W384) on helix 3, and the carbonyl group of Gly60 (W373) and Q61 (W372) on switch II. All four water molecules are unique to the calcium acetate-bound structure.
The extended switch II helical structure places key elements important in ordering the N-terminal half of the switch, from which the side chain of catalytic residue Q61 extends. For example, S65 in the ordered switch II conformation makes a good H bond with the main chain carbonyl group of E62, while its carbonyl group connects to Q99 on helix 3 through water molecule W391 (Fig. 2B). Q99 in turn links to R68 through W384. Surprisingly, the side chain of E62, which is disordered in all other available structures of Ras, is very well ordered and interacts through water molecule W176 with the side chain oxygen atom of Q61. Further details are given in SI Text and Fig. S1. While the side chain of Q61 is involved in this network, its carbonyl group is bridged to the side chain of R68 through water molecule W372. The net effect of all these interactions is that Q61 is positioned through both backbone and side chain H bonds that place it in the active site and that connect it through networks 1 and 2 to the remote allosteric site, occupied in our structure by the calcium acetate as a mimic to a yet undiscovered natural modulator.
The active site features reported for the wild-type Ras structure solved in the presence of calcium chloride (17) are also observed in the calcium acetate bound structure, with the added feature of the ordered switch II with placement of the Q61 near the bridging water molecule W189. Thus, in both structures, all of switch I is in the same conformation as found in the Raps/Raf-RBD complex with Y32 closed over the nucleotide and interacting with it through the bridging water molecule. Two water molecules interact closely with the Y32 side chain oxygen atom: On one side is W396, which links to N86 at the N-terminal end of helix 3 through an H-bonding network involving W162 and W101, and on the other is the bridging water molecule, W189, which in turn H-bonds to the O1G atom of the γ-phosphate of GppNHp (Fig. S1). The T35 residue maintains its well-known interactions within the active site: Its side-chain hydroxyl group coordinates the Mg2+ ion and its main-chain carbonyl oxygen atom makes a good H bond to the catalytic water molecule (W175). In addition to donating an H bond to the carbonyl oxygen atom of T35 and to the O1G atom of the nucleotide analogue as observed in the calcium chloride condition, in the presence of calcium acetate the catalytic water molecule also accepts an H bond from the backbone N atom of the Q61 residue, which in our previously published structure (PBD ID code 2RGE) is disordered.
Ras Binds Ca2+ but not Mg2+ at the Allosteric Site.
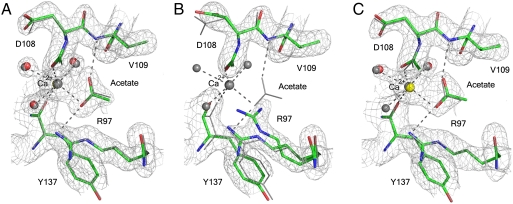
As an initial test of the specificity of the allosteric site for Ca2+, crystals grown in the presence of calcium acetate were removed from the mother liquor and soaked in stabilization solutions containing either 200 mM calcium acetate or 200 mM magnesium acetate. The soak in 200 mM calcium acetate is a control to make sure that a transfer to stabilization solution in itself does not remove the Ca2+ ion and that comparisons are made between calcium acetate and magnesium acetate solutions that are otherwise identical. Fig. 3A and B shows the allosteric binding site in the two resulting structures. The structure soaked in calcium acetate is the same as that described above from crystals taken directly from the mother liquor, with clear electron density for the calcium acetate in the allosteric site. The soak in magnesium acetate, however, resulted in a structure with an “empty” allosteric site and with an alternate conformation for R97 typical of the off state of the allosteric switch, overlapping with the acetate binding site as seen in crystals grown in calcium chloride. Although there are other features of the off state, such as an alternate position of Y96 relative to R68 and a small shift of loop 7 toward the off state, the full shift is not made, probably due to constraints of the crystal packing. Crystals soaked in magnesium acetate and then transferred back to calcium acetate show a bound Ca2+ ion and acetate molecule as seen in Fig. 3C, regaining all of the features of the on state of the allosteric switch.
Fig. 3.
Allosteric site in structures resulting from soaked crystals. (A) Soak in calcium acetate. (B) Soak in magnesium acetate. (C) Backsoak from magnesium acetate to calcium acetate. The three structures are superimposed on the Ras structure from crystals taken directly from the mother liquor containing calcium acetate, depicted in gray. The 2Fo-Fc electron density maps were contoured at the 1σ level.
The Y32 Hydroxyl Group Stabilizes Switch I.
Tyrosine 32 is a well-conserved residue in the Ras GTPase superfamily (21). In Ras-GTP it samples different conformations in solution, including a state where it is closed over the nucleotide, as is found in the complex with Raf, and a range of open conformations where Y32 is away from the nucleotide, as exemplified in the Ras/RasGAP complex (22). These changes are local to Y32 and appear to happen within the confines of a switch I anchored by the γ-phosphate of GTP. In Ras-GDP switch I is disordered in solution (23), although in the crystal structure (PBD ID code 4Q21) Y32 is stabilized in a conformation away from the active site and occupying the Raf-RBD binding pocket (24).
In order to further investigate the role of the Y32 hydroxyl group, this residue was mutated to F32 and the structure of RasY32F-GppNHp was solved from crystals with symmetry of space group P212121 containing three molecules in the asymmetric unit. In all three molecules switch I occludes the Raf-RBD binding pocket with F32 away from the active site. Switch II is disordered to various degrees, and electron density for the side chain of Q61 is absent (SI Text and Fig. S2). In one of the monomers, the catalytic and bridging water molecules are present and switch I differs from the wild-type conformation primarily in the vicinity of F32. In the other two monomers both water molecules are absent. The allosteric switch is off in all three monomers with no bound Ca2+ ion or acetate molecule in the allosteric site, in spite of the presence of 200 mM calcium acetate in the crystallization conditions. This provides a clue that Y32 may be an important element of the allosteric switch.
Having shown that Raf-RBD-CRD (a construct of Raf that includes both Ras-binding domains) binds RasY32F in a nucleotide-dependent manner (SI Text and Fig. S3), we then examined the effect of the mutation on the intrinsic hydrolysis rate of the free mutant relative to wild-type Ras and the effect of binding Raf-RBD-CRD on the hydrolysis rate of the mutant (details in SI Text and Fig. S4). The rate of 0.016/ min for wild-type Ras is as previously observed (25). The presence of Raf-RBD-CRD has no effect on this rate. The RasY32F mutant has a hydrolysis rate of 0.006/ min, a greater than 2-fold decrease relative to wild type, which is rescued to a wild-type rate of 0.014/ min in the presence of Raf-RBD-CRD. These results, combined with the structures of RasY32F-GppNHp, indicate that the destabilization of switch I with an accompanying decrease in occupancy of the active site water molecules results in the slower intrinsic hydrolysis rates. The binding of Raf occurs at switch I and in the adjacent pocket that is at least partially occluded in RasY32F-GppNHp (Fig. S4). Thus, in order for the complex to form, F32 must be closed over the nucleotide with switch I adopting the conformation observed in the wild-type complex. In spite of switch I adopting several Ras-GDP-like conformations in free RasY32F-GppNHp, the presence of the γ-phosphate provides a scaffold that facilitates a shift to the active switch I conformation in the presence of Raf-RBD-CRD that cannot be achieved with RasY32F-GDP. As indicated by the wild-type rate of intrinsic hydrolysis in the mutant complex, this conformation is sufficient to stabilize all of the elements of the intrinsically slow hydrolysis reaction that are so far understood from previous biochemical and structural work on Ras: the coordination of Mg2+ by T35 and the positioning of the active site water molecules. The hydroxyl group of Y32 clearly plays an important role in stabilizing these elements in free Ras-GppNHp, and it appears to be an important link in the allosteric switch. If Y32 contributes directly to catalysis, this contribution is not rate limiting, and possible effects due to the removal of the hydroxyl group may be masked by an even slower step in the hydrolysis reaction in the absence of the allosteric switch: placement of the catalytic Q61 in the active site of Ras. In order to measure the contribution of Y32 to catalysis, it will be necessary to activate the allosteric switch in solution so that we can measure the reaction rates in a situation where Q61 is in place, a feat that we have not succeeded in doing by simply adding calcium acetate either in the absence or presence of Raf-RBD-CRD.
Discussion
Proposed Mechanism of Intrinsic Hydrolysis.
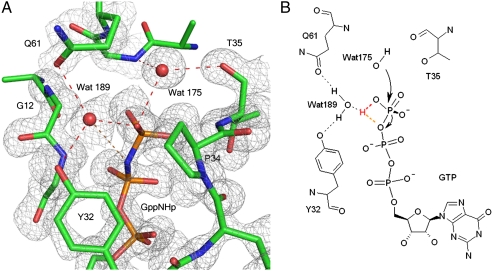
It is now established that intrinsic hydrolysis in Ras occurs through a loose, dissociative-like transition state (TS) with significant concerted character, as does the GAP-catalyzed reaction (26). This does not rule out a mechanism in which the GTP itself receives a proton from the catalytic water molecule (27). Within these parameters, we propose a general outline for catalysis deduced from our structure of the ground state of Ras with an activated allosteric switch, augmenting the mechanism we previously published (17). As the reaction proceeds, a proton is shuttled from the catalytic water molecule via the O1G atom of the γ-phosphate of GTP to the bridging water molecule, which is particularly prone to accept an H bond, as it can donate H bonds to both Y32 and Q61 (Fig. 4). This would promote development of a partial positive charge in the active site near the β-γ bridging oxygen atom of GTP to stabilize a dissociative-like TS creating a counterpart, albeit weaker, to the arginine finger provided by GAPs. This is the key element lacking in our previously published mechanism. It is consistent with kinetic isotope effect experiments in which it was concluded that Q61 must mediate stabilization of negative charges on the leaving group oxygen during the reaction (26). We envision that as the reaction proceeds, W189 moves toward the β-γ bridging oxygen atom of GTP and away from the Q61 side chain, which can then flip to interact directly with the γ-phosphate in a manner analogous to that observed in the Ras/RasGAP structure (5). Indeed the ordered structure of switch II in our Ras-GppNHp model is not so different from that observed in the complex with GAP, and a flip of the Q61 side chain would put it within reach of the γ-phosphate. This process would release the allosteric switch, resulting in a disordered switch II, ultimately leaving more room for the release of the inorganic phosphate product at the end of the reaction. The allosteric switch that we propose leads to enhanced catalysis is consistent with recently published molecular dynamics simulations where correlated motions are observed between switch II residues 66–74 and helix 3/loop 7 residues 93–110 in Ras-GTP, but not in Ras-GDP (28). Correlated or “breathing” motions involving switch II and helix 3 have also been observed experimentally by NMR (29).
Fig. 4.
Proposed mechanism of intrinsic hydrolysis in Ras. (A) Ras-GppNHp active site showing Y32, Q61, the catalytic (W175) and bridging (W189) water molecules near the nucleotide. Electron density (gray) is from a 2Fo-Fc map contoured at the 1σ level. H-bonding interactions involving these residues in the ground state are shown in red dashed lines. The orange dashed line indicates the H bond between W189 and the β-γ oxygen of GTP that we propose helps stabilize the transition state of the reaction. (B) Schematics of the reaction mechanism leading to the transition state. Hydrogen atoms are not shown except those for W175 and W189 and hydrogen atoms that interact directly with them. The flexibility in directionality of H bonds may be important.
Reconsidering the Relevance of Intrinsic Hydrolysis in Ras.
Ras is a protein that has long been considered to undergo conformational changes associated with regulation strictly due to the state of the bound nucleotide, where it can interact with effectors in the GTP-bound but not in the GDP-bound state. We have uncovered an additional level of modulation of switch II (and along with it catalytic residue Q61) for Ras in the GTP-bound state that may influence its ability to hydrolyze GTP and therefore the timing of interaction with Raf. We propose that the allosteric switch is not general to all effectors, but is activated by Raf, which interacts with Ras through switch I (20, 30). In addition to leaving a free switch II in contrast to RasGAP and other effectors that bind both switches, Raf is unique in that its 3.5 nM affinity for Ras (31) is 3 orders of magnitude stronger than the micromolar affinity observed for Ras/RasGAP (32) and for Ras interaction with effectors such as PI3K (33) and RalGDS (34). Given that GAPs and effectors have overlapping binding sites (35), GAPs must displace the effectors in order to turn off the GTPase signaling and directly control the timing of the Ras/effector interaction. Since RasGAP, PI3K and RalGDS have similar affinities to Ras, it is likely that GAP-catalyzed hydrolysis is the primary mechanism of negative regulation of pathways involving these effectors. However, it is unlikely that GAPs can outcompete Raf at the concentrations found in cells† (36). It has been suggested that the role of RasGAP in regulating the Ras/Raf pathway could be to deplete the pool of available free Ras and that Raf could somehow increase the GTP hydrolysis rate (35, 37). However this has not been part of the discourse in the literature for the last 15 years, given the slow intrinsic hydrolysis rate of Ras and the lack of rate enhancement observed in vitro in the presence of Raf. Given our discovery of the allosteric switch, this possibility must be seriously reconsidered and investigated further. We propose a model in which the binding of Raf is controlled by the state of the bound nucleotide through interaction of the RBD, but the timing of interaction is modulated allosterically at the remote site by Ca2+ in conjunction with another cellular factor containing a negatively charged group, mimicked by the acetate in our structure.
The discovery of an allosteric switch in Ras provides insight into the mystery associated with the function of Q61 in intrinsic catalysis and the slow rate of hydrolysis observed for Ras in the absence of GAPs. It suggests a unique mechanism by which the binding of Raf and simultaneous association of a negatively charged ligand at the remote allosteric site could increase hydrolysis rates to biologically relevant levels in the presence of Ca2+, promoting attenuation of the signaling in Ras/Raf-associated pathways. It appears that in order to have a fully formed active site for intrinsic hydrolysis of GTP, Ras needs to have both switch I and switch II in highly ordered conformations. Full stabilization of switch I can be achieved with the binding of Raf. Both in free Ras and in the complex with Raf, however, switch II is likely disordered in the absence of the allosteric switch (29, 30). In this situation the correct placement of Q61 for catalysis would be expected to be a slow, rate-determining step in the hydrolysis reaction, leading to the previously observed hydrolysis rates, where the binding of Raf alone would not be expected to have an effect. However, the simultaneous binding of Raf at switch I, placing the hydroxyl group of Y32 in position to interact with the bridging water molecule and enable the allosteric switch, and a natural ligand at the remote allosteric site (perhaps a membrane phospholipid or carboxylate group) in the presence of Ca2+ could stabilize switch II through the mechanism we described above involving H-bonding networks 1 and 2. The completion of the active site would then no longer be a rate-determining step in the reaction, leading to an increase in intrinsic hydrolysis rate. Calcium has been shown to be one of the activators of the Ras/Raf/MEK/ERK pathway, promoting increases in Ras-GTP levels leading to the recruitment of Raf to the membrane (38). Furthermore, we show here that the allosteric site binds Ca2+, but not Mg2+, supporting a specific role for calcium as we propose. It is conceivable that the timing of the Ras/Raf interaction is affected by the higher Ca2+ levels with binding of a membrane negatively charged group to the allosteric site once the complex with Raf is in place. An increase in Ca2+ concentration has already been shown to inactivate the Ras/Raf/MEK/ERK pathway in at least two different instances (39, 40). Although the balance between the activating and inactivating roles of Ca2+ with respect to the Ras/Raf pathway is still not well understood, it is possible that the allosteric switch may be one of the means through which this balance is achieved by direct action of Ca2+ on Raf-bound Ras at the membrane.
The idea that a molecule at or near the membrane may act in conjunction with Ca2+ to activate the allosteric switch is supported by molecular dynamics simulations of full length lipid-modified Ras in a 1,2-dimyristoylglycero-3-phosphocholine bilayer, showing that in Ras-GTP the catalytic domain makes more extensive interactions with the membrane phospholipids than in Ras-GDP, and is positioned such that the allosteric site near loop 7 is adjacent to the membrane (41). The computational model shows that Ras-GTP interacts with phospholipid head groups at residues R128, R135, and Q165, anchoring helix 4 to the membrane. This is supported experimentally in cells by mutation of the two arginine residues to alanine, which results in a decrease in neurite outgrowth due to impaired signal transduction (41). In our crystal structures of Ras-GppNHp, helix 4 is nestled against extensive crystal contacts, with R135 making a salt bridge with the terminal carboxylate group of a symmetry-related molecule. Thus, the stabilization of helix 4 by the membrane appears to be mimicked to some extent in crystals with symmetry R32. This may be an important element of the allosteric switch, as helix 4 contains Y137 (only two residues away from R135) that coordinates to the Ca2+ ion through its carbonyl group. Lack of this stabilizing interaction in solution may prevent the activation of the allosteric switch, accounting for the fact that we have been unable to measure an increase in the intrinsic hydrolysis rate by adding calcium acetate to the solutions in which the hydrolysis experiments are performed in vitro. If our assessment is correct that the crystal contacts serve a similar function to that of the membrane as far as stabilizing Y137 on helix 4, it appears that the structural components that lead to a highly ordered switch II and placement of Q61 in the active site are sensitive to both placement of Ras against the membrane in a manner that happens only in the GTP-bound form (41) and to the binding of Raf, which is necessary for the stabilization of Y32. In this situation Y32 could serve as a venue through which the binding of Raf is imposed as a requirement in the activation of Ras in lieu of activation by GAPs, ensuring that the membrane-bound Ras-GTP does not engage in futile hydrolysis of GTP to GDP before it has a chance to recruit Raf from the cytoplasm and activate the signal transduction cascade.
Materials and Methods
Ras Mutagenesis, Expression, and Purification.
The mutant RasY32F was made by site-directed mutagenesis of wild-type Ras 1-166, using the Quick Change II kit from Stratagene, following the manufacturers instructions. Both wild-type Ras and RasY32F expression, purification, storage, and GppNHp exchange were as previously published (13).
Ras-GppNHp Crystallization.
The Ras-GppNHp protein solution consisted of 20 mg/mL protein in a stabilization buffer (20 mM Hepes pH 7.5, 50 mM NaCl, 20 mM MgCl2, and 10 mM dithioerythritol). Crystals were grown at 18 °C for 1 week using sitting drop crystallization trays containing 5 μL protein solution and 5 μL reservoir solution. The reservoir solution was composed of 500 μL of 200 mM calcium acetate hydrate, 20% w/v PEG 3350, and 0.05% n-Octyl-β-D-glucopyranoside diluted with 50 μL of stabilization buffer. Synchrotron data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamline at the Advanced Photon Source (APS).
Soaks in Calcium Acetate and Magnesium Acetate Solutions.
For the calcium acetate and magnesium acetate soaks, Ras-GppNHp crystals grown as above were transferred to a solution containing 10 mM Hepes pH 7.5, 30% w/v PEG 3350, 10 mM MgCl2, 25 mM NaCl, and either 200 mM Ca(OAc)2 or 200 mM Mg(OAc)2 and flash frozen after 1 or 2 h for data collection. After soaking in Mg(OAc)2 for 3 days, a crystal was then transferred back to the solution containing 200 mM Ca(OAc)2 and soaked for 24 h before data collection. Datasets for the soaked crystals were collected on our home MAR345 area detector mounted on a Rigaku RuH3R rotating anode generator.
RasY32F-GppNHp Crystallization.
RasY32F-GppNHp was concentrated to 9–12 mg/mL in a stabilization buffer (above). Crystals were grown from hanging drops at 18 °C, consisting of 3 μL protein solution and 3 μL reservoir solution: 200 mM Ca(OAc)2, 5 mM MgCl2, 10 mM DTT, and either 18–20% PEG 3350 or 20% PEG 6000. Synchrotron data were collected at the SER-CAT beamline at APS.
See SI Text and Table S1 for details on data collection and refinement statistics for the five crystal structures presented in this article.
Supplementary Material
Acknowledgments.
Senthil Kumar first grew crystals of RasG12V in the presence of calcium acetate in our lab, which prompted us to try similar conditions for wild-type Ras. Melissa Young grew crystals of the RasY32F mutant. Use of the Advanced Photon Source at the Argonne National Laboratory was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38. This research is supported by the NIH (R01 CA096867-01A1).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org, with the following accession codes: Ras grown in mother liquor with Ca(OAc)2 (PDB ID code 3K8Y); Ras soaked in Ca(OAc)2 (PDB ID code 3LBH); Ras soaked in Mg(OAc)2 (PDB ID code 3LBN); Ras soaked in Mg(OAc)2 and back soaked in Ca(OAc)2 (PDB ID code 3LBI); RasY32F (PDB ID code 3K9N).
This article contains supporting information online at www.pnas.org/cgi/content/full/0912226107/DCSupplemental.
*Raps refers to the Ras homologue Rap with the switch I mutations E30D,K31E that results in a switch I region identical in sequence to that of Ras.
†The concentration range is 0.1–1.6 μM for Ras and 3.0–500 nM for Raf.
References
- 1.Sacks DB. The role of scaffold proteins in MEK/ERK signalling. Biochem Soc Trans. 2006;34(5):833–836. doi: 10.1042/BST0340833. [DOI] [PubMed] [Google Scholar]
- 2.Torii S, Nakayama K, Yamamoto T, Nishida E. Regulatory mechanisms and function of ERK MAP kinases. J Biochem. 2004;136(5):557–561. doi: 10.1093/jb/mvh159. [DOI] [PubMed] [Google Scholar]
- 3.Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: Does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 14(11):639–647. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 4.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394(6691):337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 5.Scheffzek K, et al. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277(5324):333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- 6.John J, Schlichting I, Schiltz E, Rosch P, Wittinghofer A. C-terminal truncation of p21H preserves crucial kinetic and structural properties. J Biol Chem. 1989;264(22):13086–13092. [PubMed] [Google Scholar]
- 7.Neal SE, Eccleston JF, Hall A, Webb MR. Kinetic analysis of the hydrolysis of GTP by p21N-ras. The basal GTPase mechanism. J Biol Chem. 1988;263(36):19718–19722. [PubMed] [Google Scholar]
- 8.Franken SM, et al. Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H-ras. Biochemistry. 1993;32(33):8411–8420. doi: 10.1021/bi00084a005. [DOI] [PubMed] [Google Scholar]
- 9.Krengel U, et al. Three-dimensional structures of H-ras p21 mutants: Molecular basis for their inability to function as signal switch molecules. Cell. 1990;62(3):539–548. doi: 10.1016/0092-8674(90)90018-a. [DOI] [PubMed] [Google Scholar]
- 10.Tong LA, et al. Structural differences between a ras oncogene protein and the normal protein. Nature. 1989;337(6202):90–93. doi: 10.1038/337090a0. [DOI] [PubMed] [Google Scholar]
- 11.Tong LA, de Vos AM, Milburn MV, Kim SH. Crystal structures at 2.2 A resolution of the catalytic domains of normal ras protein and an oncogenic mutant complexed with GDP. J Mol Biol. 1991;217(3):503–516. doi: 10.1016/0022-2836(91)90753-s. [DOI] [PubMed] [Google Scholar]
- 12.Wittinghofer F, Krengel U, John J, Kabsch W, Pai EF. Three-dimensional structure of p21 in the active conformation and analysis of an oncogenic mutant. Environ Health Perspect. 1991;93:11–15. doi: 10.1289/ehp.919311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buhrman G, de Serrano V, Mattos C. Organic solvents order the dynamic switch II in Ras crystals. Structure. 2003;11(7):747–751. doi: 10.1016/s0969-2126(03)00128-x. [DOI] [PubMed] [Google Scholar]
- 14.Frech M, et al. Role of glutamine-61 in the hydrolysis of GTP by p21H-ras: An experimental and theoretical study. Biochemistry. 1994;33(11):3237–3244. doi: 10.1021/bi00177a014. [DOI] [PubMed] [Google Scholar]
- 15.Pai EF, et al. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: Implications for the mechanism of GTP hydrolysis. EMBO J. 1990;9(8):2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scheidig AJ, Burmester C, Goody RS. The pre-hydrolysis state of p21(ras) in complex with GTP: New insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins. Structure. 1999;7(11):1311–1324. doi: 10.1016/s0969-2126(00)80021-0. [DOI] [PubMed] [Google Scholar]
- 17.Buhrman G, Wink G, Mattos C. Transformation efficiency of RasQ61 mutants linked to structural features of the switch regions in the presence of Raf. Structure. 2007;15(12):1618–1629. doi: 10.1016/j.str.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nassar N, et al. Ras/Rap effector specificity determined by charge reversal. Nat Struct Biol. 1996;3(8):723–729. doi: 10.1038/nsb0896-723. [DOI] [PubMed] [Google Scholar]
- 19.Bondeva T, Balla A, Varnai P, Balla T. Structural determinants of Ras-Raf interaction analyzed in live cells. Mol Biol Cell. 2002;13(7):2323–2333. doi: 10.1091/mbc.E02-01-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittinghofer A, Nassar N. How Ras-related proteins talk to their effectors. Trends Biochem Sci. 1996;21(12):488–491. doi: 10.1016/s0968-0004(96)10064-5. [DOI] [PubMed] [Google Scholar]
- 21.Valencia A, Chardin P, Wittinghofer A, Sander C. The ras protein family: Evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30(19):4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 22.Spoerner M, Herrmann C, Vetter IR, Kalbitzer HR, Wittinghofer A. Dynamic properties of the Ras switch I region and its importance for binding to effectors. Proc Natl Acad Sci USA. 2001;98(9):4944–4949. doi: 10.1073/pnas.081441398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraulis PJ, Domaille PJ, Campbell-Burk SL, Van Aken T, Laue ED. Solution structure and dynamics of ras p21.GDP determined by heteronuclear three- and four-dimensional NMR spectroscopy. Biochemistry. 1994;33(12):3515–3531. doi: 10.1021/bi00178a008. [DOI] [PubMed] [Google Scholar]
- 24.Milburn MV, et al. Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science. 1990;247(4945):939–945. doi: 10.1126/science.2406906. [DOI] [PubMed] [Google Scholar]
- 25.Cheng H, Sukal S, Deng H, Leyh TS, Callender R. Vibrational structure of GDP and GTP bound to RAS: An isotope-edited FTIR study. Biochemistry. 2001;40(13):4035–4043. doi: 10.1021/bi0021131. [DOI] [PubMed] [Google Scholar]
- 26.Du X, Sprang SR. Transition state structures and the roles of catalytic residues in GAP-facilitated GTPase of Ras as elucidated by (18)O kinetic isotope effects. Biochemistry. 2009;48(21):4538–4547. doi: 10.1021/bi802359b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosloff M, Selinger Z. Substrate assisted catalysis—application to G proteins. Trends Biochem Sci. 2001;26(3):161–166. doi: 10.1016/s0968-0004(00)01748-5. [DOI] [PubMed] [Google Scholar]
- 28.Grant BJ, Gorfe AA, McCammon JA. Ras conformational switching: Simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLOS Comput Biol. 2009;5(3):e1000325. doi: 10.1371/journal.pcbi.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Connor C, Kovrigin EL. Global conformational dynamics in ras. Biochemistry. 2008;47(39):10244–10246. doi: 10.1021/bi801076c. [DOI] [PubMed] [Google Scholar]
- 30.Thapar R, Williams JG, Campbell SL. NMR characterization of full-length farnesylated and non-farnesylated H-Ras and its implications for Raf activation. J Mol Biol. 2004;343(5):1391–1408. doi: 10.1016/j.jmb.2004.08.106. [DOI] [PubMed] [Google Scholar]
- 31.Minato T, et al. Quantitative analysis of mutually competitive binding of human Raf-1 and yeast adenylyl cyclase to Ras proteins. J Biol Chem. 1994;269(33):20845–20851. [PubMed] [Google Scholar]
- 32.Vogel US, et al. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature. 1988;335(6185):90–93. doi: 10.1038/335090a0. [DOI] [PubMed] [Google Scholar]
- 33.Pacold ME, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103(6):931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 34.Herrmann C, Horn G, Spaargaren M, Wittinghofer A. Differential interaction of the ras family GTP-binding proteins H-Ras, Rap1A, and R-Ras with the putative effector molecules Raf kinase and Ral-guanine nucleotide exchange factor. J Biol Chem. 1996;271(12):6794–6800. doi: 10.1074/jbc.271.12.6794. [DOI] [PubMed] [Google Scholar]
- 35.Moodie SA, et al. Different structural requirements within the switch II region of the Ras protein for interactions with specific downstream targets. Oncogene. 1995;11(3):447–454. [PubMed] [Google Scholar]
- 36.Fujioka A, et al. Dynamics of the Ras/ERK MAPK cascade as monitored by fluorescent probes. J Biol Chem. 2006;281(13):8917–8926. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- 37.Warne PH, Viciana PR, Downward J. Direct interaction of Ras and the amino-terminal region of Raf-1 in vitro. Nature. 1993;364(6435):352–355. doi: 10.1038/364352a0. [DOI] [PubMed] [Google Scholar]
- 38.Cullen PJ, Lockyer PJ. Integration of calcium and Ras signalling. Nat Rev Mol Cell Biol. 2002;3(5):339–348. doi: 10.1038/nrm808. [DOI] [PubMed] [Google Scholar]
- 39.Choi J, et al. T-type calcium channel trigger p21ras signaling pathway to ERK in Cav3.1-expressed HEK293 cells. Brain Res. 2005;1054:22–29. doi: 10.1016/j.brainres.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Lockyer PJ, Kupzig S, Cullen PJ. CAPRI regulates Ca2+-dependent inactivation of the Ras-MAPK pathway. Curr Biol. 2001;11(12):981–986. doi: 10.1016/s0960-9822(01)00261-5. [DOI] [PubMed] [Google Scholar]
- 41.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3-phosphocholine bilayer. J Med Chem. 2007;50(4):674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.