Abstract
Left-handed Z-DNA has fascinated biological scientists for decades by its extraordinary structure and potential involvement in biological phenomena. Despite its instability relative to B-DNA, Z-DNA is stabilized in vivo by negative supercoiling. A detailed understanding of Z-DNA formation is, however, still lacking. In this study, we have examined the B-Z transition in a short guanine/cytosine (GC) repeat in the presence of controlled tension and superhelicity via a hybrid technique of single-molecule FRET and magnetic tweezers. The hybrid scheme enabled us to identify the states of the specific GC region under mechanical control and trace conformational changes synchronously at local and global scales. Intriguingly, minute negative superhelicity can facilitate the B-Z transition at low tension, indicating that tension, as well as torsion, plays a pivotal role in the transition. Dynamic interconversions between the states at elevated temperatures yielded thermodynamic and kinetic constants of the transition. Our single-molecule studies shed light on the understanding of Z-DNA formation by highlighting the highly cooperative and dynamic nature of the B-Z transition.
Keywords: magnetic tweezers, single-molecule FRET, Z-DNA
DNA is capable of adopting various conformations in addition to the conventional right-handed B-DNA double helix (1, 2). One of the most dramatic examples is Z-DNA, which is a left-handed helical form with a zigzag backbone (3, 4). Since its discovery, Z-DNA has drawn considerable attention from a broad range of research areas, including physical biochemistry, structural biology, and molecular biology, because of its intriguing and unusual structural features (3). The bases in Z-DNA alternate in syn- and anti-conformations while all the bases in B-DNA adopt the anti-conformation. Because the syn conformation is more stable for purines (Pu) than for pyrimidines (Py), alternating Pu and Py sequences readily adopt the Z-DNA conformation. Another striking feature of Z-DNA is the overturning of base pairs, posing a steric dilemma known as the chain sense paradox (3, 5).
Although Z-DNA is less stable than B-DNA at physiological ionic conditions, Z-DNA exists stably under certain conditions such as high ionic strength or negative supercoiling (4, 6–8). In the presence of a high-salt solution, the electrostatic repulsion between the phosphate backbones, which are closer in Z-DNA than in B-DNA, is reduced, stabilizing the Z-DNA conformation. Negative supercoiling induces the Z-DNA conformation even in physiological salt conditions because formation of Z-DNA significantly relieves torsional stress. Z-DNA has also been shown to exist stably in vivo in the presence of physiological negative supercoiling (9, 10).
Although there were first doubts as to whether Z-DNA plays any biological role, considerable experimental evidence for biological functions of Z-DNA has accumulated over the past two decades (11). In addition to the discovery of antibodies and proteins that bind selectively to Z-DNA (12, 13), Z-DNA formation has been strongly correlated with transcriptional activity. Transcription can induce Z-DNA formation, and the Z-DNA structure formed at or near the promoter region of genes regulates their transcription in vivo (14, 15). Z-DNA has also been implicated in various human diseases such as blood cancers, autoimmune diseases, and Alzheimer’s disease (16, 17).
Although the biological significance of Z-DNA has begun to be appreciated, our fundamental understanding of Z-DNA is far from complete (5). Despite the recent success in determining the structure of a B-Z-DNA junction (18), the thermodynamic and energetic details available for Z-DNA are still lacking. Additionally, the molecular mechanism behind the B-Z transition is not yet fully understood. Traditional methods, which have been widely used to study Z-DNA, such as x-ray crystallography, CD spectroscopy, electrophoresis, and antibody binding assays (3–8, 12, 19, 20), are not able to capture the dynamic features of the DNA molecule because of their limited spatial and temporal resolution. Moreover, the aforementioned techniques are not suitable for directly probing conformational changes in short, localized regions embedded in a long DNA molecule.
Hybrid schemes combining various single-molecule techniques (21, 22) have proven to be useful for measuring minute conformational changes under various mechanical conditions. Here, we utilized torquemetric fluorescence microscopy, our hybrid technique that combines single-molecule FRET (23, 24) and magnetic tweezers (24, 25) in order to gain direct and quantitative information regarding the thermodynamic parameters and dynamics of Z-DNA formation. This technique enables us to examine in real time conformational dynamics of a specific, localized DNA sequence under precisely controlled tension and superhelical density. By tracking single-molecule FRET signals from the donor and acceptor dyes flanking the guanine/cytosine (G/C) repeat sequence (Table S1) undergoing the B-Z transition (Fig. 1), we clearly identified two FRET states corresponding to the B- and Z-DNA conformations and found that the tension as well as the torsion applied to a DNA molecule is a critical factor in stabilizing Z-DNA. Contrary to the current knowledge, the superhelical density that is required to trigger the B-Z transition in the presence of approximately one picoNewton tension is remarkably small, at least fivefold smaller than the previously accepted values (8, 19). In addition, the supercoiling-induced B-Z transition was directly verified by observing a synchronous unbuckling of the DNA molecule. We also observed dynamic interconversions between the two states at elevated temperatures, which allowed us to evaluate thermodynamic and kinetic constants for the B-Z transition directly. The results from our single-molecule study revealed the highly cooperative and dynamic nature of the B-Z transition.
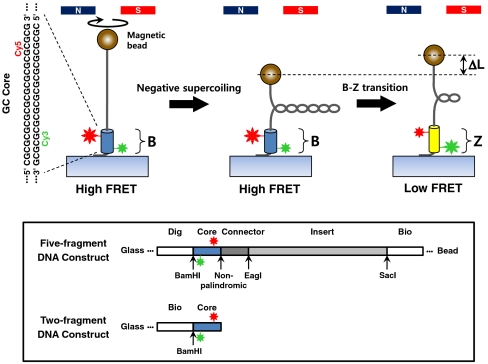
Fig. 1.
Negative supercoiling induces the B-Z transition. When the DNA tether containing the GC core DNA flanked by donor (Green) and acceptor (Red) dyes is underwound by magnetic tweezers (Left), the FRET efficiency changes from a high to a low value, indicating the transition from the B-form (Blue) to the Z-form (Yellow) (Right). When the supercoiled DNA is buckled at low tension (Middle), the B-Z transition removes some plectonemes, increasing its extension by ΔL (Right). Inset: DNA constructs. A five-fragment DNA construct containing the donor and acceptor dyes in the middle and two distinct attachment sites (Dig & Bio) at the ends was used in the hybrid scheme. The dimer construct bearing the dye pair was used for FRET measurements.
Results
Assignment of Conformational States Based on FRET Efficiency Measurements.
In order to assign the conformational states of the alternating GC repeat sequence (GC core, ∼7.1 nm and ∼8.2 nm long in the B- and Z-form, respectively), we measured the FRET efficiency (FE) from the GC core at various salt concentrations in the absence of external tension and torsion (Fig. 2). At the lowest concentration for each salt, this mechanically unconstrained core DNA revealed a peak FE of ∼0.50 corresponding to B-DNA, in addition to a peak FE of ∼0 arising from donor-only molecules. Because Z-DNA is favored under high-salt conditions (4, 6), the new peak (FE = 0.12) detected in high-salt conditions can be attributed to Z-DNA. According to these measurements, FEs from 0.03 to 0.21 and from 0.42 to 0.58 were assigned to Z- and B-DNA, respectively. The methylated GC core (Gm5C core) shows a similar tendency, with the exception that the transition occurs at lower salt concentrations (Fig. S1). The methyl groups in (Gm5C)n strengthen hydrophobic interactions, thus stabilizing Z-DNA at lower salt concentrations (6, 26). All the FE results from unconstrained core DNAs are listed in Table S2.
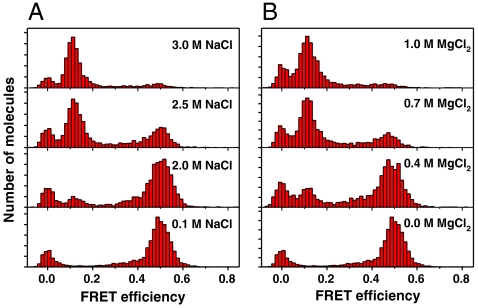
Fig. 2.
Single-molecule FRET histograms of the GC core for various concentrations of (A) NaCl and (B) MgCl2.
The salt conditions at which B- and Z-DNA are equally probable (midpoint conditions) were deduced from our FE histograms and are consistent with previous experimental data (26), with the exception of the Gm5C core in the presence of MgCl2, in which the midpoint is 0.6 mM MgCl2. The discrepancy found for Gm5C is likely due to imperfect enzymatic methylation of cytosine. To further support the structural assignment shown above, we measured the CD spectra of the GC core at 0.1 M and 3.0 M NaCl (Fig. S2). The CD spectra obtained at the low- and high-salt conditions are similar to the known spectra of B- and Z-DNA (27), respectively. The substantial inversion of the CD spectrum suggests that the transition occurs over the entire (GC)11 sequence. Considering these results, the internal dye labeling for fluorescence measurement has little effect on the B-Z transition of the (GC)11 sequence.
Tension As Well As Torsion Facilitates the B-to-Z Transition.
To study the effects of tension and torsion on the B-Z transition of a core DNA, we monitored the change in FEs at room temperature as a function of force and superhelical density (σ ≡ (Lk - Lk0)/Lk0, where Lk is the linking number of DNA, Lk0 is the Lk of relaxed DNA, and ΔLk = Lk - Lk0 is the linking difference) as shown in Fig. 3A–C and Fig. S3A and B. Time traces of the FE (Bottom) obtained for the GC core for varying σ values (Top) are plotted together with the fluorescence intensities (FIs; Middle) of the donor and acceptor dyes (Fig. 3A and B and Fig. S3A and B). As seen in these traces, a molecule initially adopting the B-form undergoes the transition to the Z-form by negative supercoiling. All the transitions were a one-step process with no detectable intermediate state. Spontaneous conformational switching could not be observed at any σ value during FRET measurements, the duration of which is limited by dye photobleaching. These observations are compatible with previous antibody binding assays performed at room temperature (28, 29), in which the transition rate was very slow (∼1 h) even at the midpoint σ value where the two states are equally probable. However, as soon as the DNA returned to σ = 0, the FE quickly returns to the value for the B-form, indicating that the transition is reversible. The Gm5C core behaved similarly while a control random core showed no change in FE under negative supercoiling (Fig. S4).
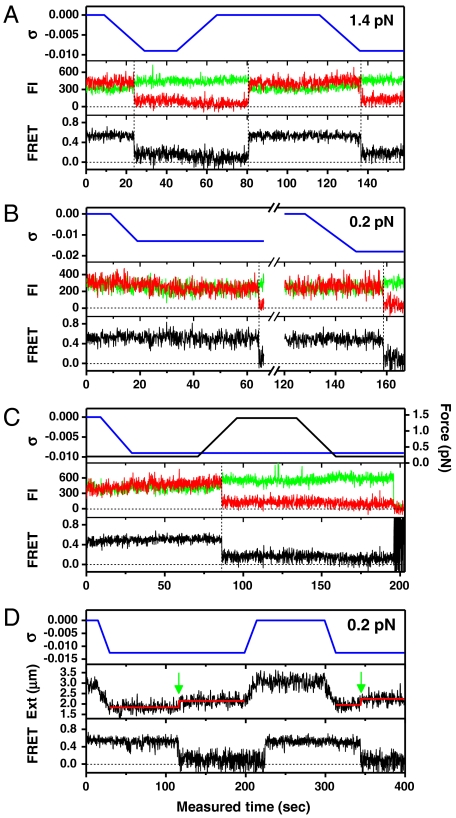
Fig. 3.
Single-molecule time traces for donor and acceptor fluorescence intensities (FI) (Middle) and FRET (Bottom) for the GC core in the presence of torsion (Top, Blue) at (A) 1.4 pN, (B) 0.2 pN, and (C) varying force (Top, Black). (D) Time trace of FRET at 0.2 pN (Bottom) under varying torsion (Top) shown together with the extension of the DNA molecule (Middle). The Red Lines indicate the average extensions of the DNA molecule at σ = -0.013 before and after the B-Z transition detected by FRET measurement (Green Arrows).
The inversion of helical sense by Z-DNA formation absorbs some negative linking difference, and, thus, reduces the effective negative linking difference on the rest of the molecule. In order to monitor such a topological effect, we measured the FE from the core DNA and the extension of the entire molecule at 0.2 pN simultaneously (Fig. 3D). As seen in Fig. 3D and illustrated in Fig. 1, the height of a DNA-bound bead decreased gradually as the DNA was being unwound because most of the linking difference is distributed to writhe at 0.2 pN, and the DNA was extended by ∼300 nm (ΔL) at the moment the core was converted to the Z-form. According to the available structure data (3, 19), the B-to-Z conversion of a (GC)11 segment results in a loss of approximately four negative superhelical turns. Since the length of one plectoneme is estimated to be ∼70 nm from the slope of the extension-vs.-ΔLk curve (30), and also from the top and middle panels of Fig. 3D, the steps of ∼300 nm (Green Arrows in Fig. 3D) agree well with the loss of ∼4 plectonemes. This result confirms that the decrease in FE originates from supercoiling-induced Z-DNA formation and that the presence of dyes does not block propagation of the Z-form toward the ends of the (GC)11 repeat.
In order to systematically analyze the effect of σ on the B- and Z-DNA states, all the relevant FRET data from torsionally stressed DNA molecules were summarized (Fig. 4 and Fig. S3C–E). The FEs of the GC and methylated GC cores, given the indicated mechanical parameters, are plotted in Fig. 4A and B and Fig. S3C. The conformational states adopted by the DNA molecules can be clearly assigned to either B- or Z-forms according to their FEs. The fraction of the Z-form from Fig. 4A and B and Fig. S3C is shown in Fig. 4C and D and Fig. S3D, respectively. The population of Z-form DNA increased rapidly as the DNA became more negatively supercoiled. The population distribution is inverted in a narrow range of σ, which is characteristic of a cooperative transition (4, 19, 29). At F = 0.2 pN, the transition midpoint is located at -σ ≈ 0.013, which is much smaller than -σ ≈ 0.045 for the transition midpoint reported from bulk assays on supercoiled plasmids (8, 19). Tension, missing in assays with plasmids, must be responsible for the large discrepancy. Since the tension to a DNA molecule modulates the distribution of its linking difference between twist and writhe (31), tension as well as torsion is expected to play an important role in the B-Z transition. The midpoint of the transition at a higher tension (F = 1.4 pN) was observed at σ values as low as σ = -0.006. As shown in Fig. 3C, a molecule in the B-form at σ = -0.009 and 0.2 pN isomerized quickly to the Z-form while the force was elevated to 1.4 pN. The results obtained for other forces also support the idea that Z-DNA is formed at smaller -σ for higher force (Fig. S3). For F = 0.2 pN, we only used -σ ≤ 0.018 because larger negative supercoiling brought the bead down into the evanescent field region, generating an enormous auto-fluorescence signal from the bead. Similarly, we could not obtain data at forces lower than 0.2 pN because the strong auto-fluorescence signal from a bead bound to a slightly underwound DNA molecule precluded FRET measurements.
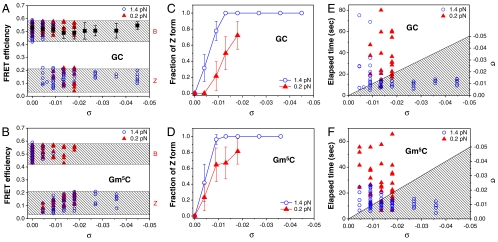
Fig. 4.
Distribution of FEs as a function of σ for (A) GC and (B) Gm5C cores. The top and bottom shaded regions show the ranges of FE for B- and Z-DNA, respectively. The FE of the random core is independent of σ at 1.4 pN (Black Square). Relative populations of the FRET states for (C) GC and (D) Gm5C cores as a function of σ. The elapsed time from the onset of DNA twisting (t = 0) until the transition was observed is displayed for (E) GC and (F) Gm5C cores. In the shaded regions below the line showing completion of DNA twisting in (E) and (F), DNA tethers were being twisted. For data points in the shaded region, the B-Z transition occurred during the twisting. Due to the constant rate of DNA twisting, at any time (Left Axis) during the twisting, σ can be calculated (Right Axis). Data were collected for two different forces (Blue Circle, 1.4 pN; Red Triangle, 0.2 pN). The error bars in (A) are given by standard deviations of the data from the random core, and those in (C) and (D) are the standard errors of the sample proportions.
The elapsed time from the start of DNA twisting (t = 0) until the transition is displayed in Fig. 4E and F and Fig. S3E. These results also reveal that tension as well as negative supercoiling facilitates the B-to-Z transition. At F = 1.4 pN, the DNA molecules tested were converted to the Z-form at nearly the same threshold σ ≈ -0.010, regardless of the final σ. This result is again consistent with the observations that in the presence of tension, the B-Z transition occurs at a -σ much smaller than previously observed values (8, 19). The Gm5C core undergoes the B-Z transition at a -σ even smaller than the GC core at 0.2 pN, while both cores exhibit similar σ dependence at 1.4 pN. This result implies that tension is a more important factor in the B-Z transition than the methyl modifications.
Dynamic Interconversions Between the B- and Z-DNA States Under Specific Torsion and Temperature Conditions Reveal Highly Torsion-Dependent Rate Constants.
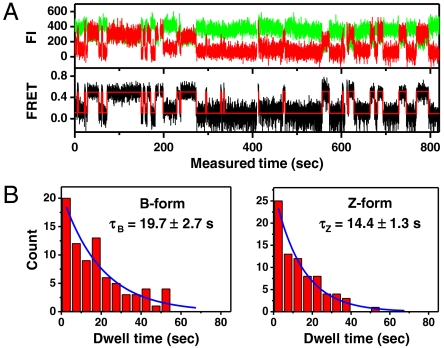
To address the dynamics of the B-Z transition, we performed experiments similar to those above at elevated temperatures. Fig. 5A shows an FE time trace from the GC core at 37 °C, F = 1.4 pN, and σ = -0.013 (ΔLk = -15) together with its idealized FRET trajectory (Red Lines) generated by hidden Markov modeling (32). By attenuating the excitation power, we managed to obtain long-time FE traces (∼3000 sec) without photobleaching of dyes. In such long-time traces, we were able to observe that DNA molecules switched back and forth between the two FE states. The histograms of the dwell times for the B- and Z-states shown in Fig. 5B are well fitted to a single exponential decay equation, which yields the rate constants of the forward (kBZ = 1/τB = 0.051 sec-1) and reverse (kZB = 1/τZ = 0.070 sec-1) reactions. From the rate constants, we calculated the equilibrium constant of the reaction (Keq = kBZ/kZB = 0.73), which agrees well with the ratio of the occupation times of the two states (ttotal,Z/ttotal,B = 0.73), read directly from Fig. 5A. The data fit well to a single exponential decay equation, indicating that the B-Z transition follows first-order kinetics involving no measurable intermediate state and that the GC core can be regarded as a two-state system (4, 7). The average dwell times (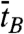 and
and  ) are equivalent to τB and τZ provided that the dwell time histogram is described by a single exponential decay. Since FE traces are often not long enough to get a good fit, we use
) are equivalent to τB and τZ provided that the dwell time histogram is described by a single exponential decay. Since FE traces are often not long enough to get a good fit, we use 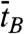 and
and 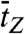 instead of τB and τZ.
instead of τB and τZ.
Fig. 5.
(A) Single-molecule time traces for donor and acceptor FIs and FEs for the GC core at 37 °C, 1.4 pN, and σ = -0.013 (ΔLk = -15). (B) Dwell time histograms of the B-to-Z and Z-to-B transitions. Each histogram is well fitted to a single exponential curve (Blue) with a time constant τ.
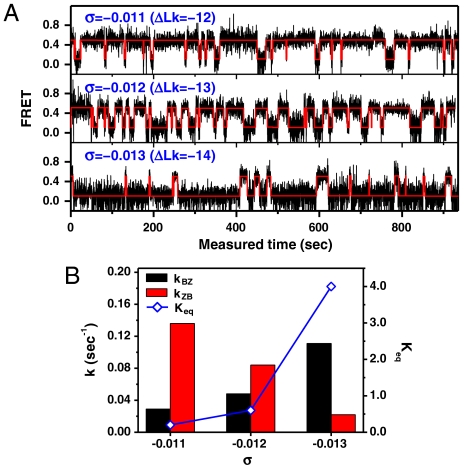
Even at elevated temperatures, rapid conformational switching like that shown in Fig. 5A was only observed around the midpoint σ. Traces of the GC core obtained for various σ values at 40 °C and 1.4 pN are shown in Fig. 6 together with the values of kBZ, kZB, and Keq. The larger -σ is, the larger the probability of adopting the Z-form, analogous to the result in Fig. 4C. Since the fraction of Z-form molecules, which was 0.2 at σ = -0.011 (ΔLk = -12), increased sharply to 0.8 at σ = -0.013 (ΔLk = -14) in Fig. 6A, the σ window for the transition is much narrower than in Fig. 4C. This feature may reflect the fact that in Fig. 6A we acquired an entire dataset for different σ values from one molecule, which has a unique Lk0, whereas the results in Fig. 4C exhibit a broadening of the Lk0 by collecting data from many different molecules (33). The rate constants are sensitive to changes in σ as well. The forward (kBZ) rate increases rapidly with increasing -σ while kZB showed the opposite trend. No spontaneous switching in either direction was observed when ΔLk was changed only by ± 2 turns from the midpoint.
Fig. 6.
(A) Single-molecule FE time traces for the GC core at 40 °C and 1.4 pN with σ = -0.011, -0.012, and -0.013 (ΔLk = -12,-13,-14). (B) Rate constants and equilibrium constants of the transition.
Interestingly, the midpoint -σ in Fig. 6 (-σ = 0.012, ΔLk = -13) is larger than the midpoint -σ at room temperature (-σ = 0.006, ΔLk = -7). The helical twist (θ) per base in B-DNA changes linearly with temperature, and its temperature derivative (dθ/dT) is -0.012°/K·bp (33). For our DNA tether (∼12 kbp), the temperature derivative of the cumulative helical twist of the entire molecule is -144°/K (ΔLk0 = -0.40 turn/K). Because Lk0 is reduced by 6.0 turns at 40 °C, compared to room temperature, the DNA tether should be further underwound by ΔLk = -6.0 in order to maintain the same torsional stress, assuming that the torsional modulus of B-DNA remains the same.
A similar temperature dependence of the B-Z transition was observed by varying the temperature for a fixed force and a fixed σ (Fig. S5). According to the above relation, ΔLk0 is decreased by 1.2 turn per 3 °C increment. Thus, the equilibrium of the B-Z transition shifts toward the B-DNA state as the temperature is raised. Unfortunately, this additional effect of temperature on the helical twist makes it difficult to determine the activation energy because the rate constants are extremely sensitive to such changes of ΔLk and thus σ.
Discussion
The most striking finding presented here is that the B-Z transition occurs at σ values as low as -0.006 (ΔLk = -7) at F = 1.4 pN and room temperature. Although the midpoint -σ at the lower forces is somewhat larger (Fig. S3), it still remains considerably smaller than the value reported in literature under force-free condition. For the sake of simplicity and concreteness, we only consider the case of F = 1.4 pN. Because neither a plectoneme nor a bubble is formed at such small -σ values, the linking number change only appears as twist (ΔLk = Tw) (31, 34). The energy by twist is given as follows (31, 34):
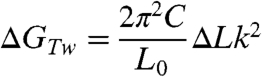 |
[1] |
where C is the torsional modulus (C = 86 nm kBT) (31, 35) and L0 the contour length of the DNA tether (L0 = ∼ 3800 nm), giving GTw ∼ 0.26ΔLk2 kcal/mol. Z-DNA formation relieves the torsional stress of negatively supercoiled DNA by absorbing twist. When a (GC)i repeat is flipped from the B- to Z-form, the accompanying twist change, according to the conventional zipper model (19), is:
| [2] |
where a is the twist change when a (GC)1 dinucleotide unit flips from a B-helix (10.5 bp/turn) to a Z-helix (-12 bp/turn), a = 2 × [(1/10.5) + (1/12)], and b is the twist allocated to a B-Z junction, which was determined to be 0.4 turn from the zipper model. Because i = 11 for our GC core, ΔTw = -4.7 turns, meaning that 4.7 turns are released upon forming Z-DNA. At ΔLk = -7, the twist energy is 12.9 kcal/mol if the whole molecule is in B-form, whereas the twist energy is reduced to 1.4 kcal/mol when the core adopts the Z-form. Because the energies of the two states should be the same at this midpoint ΔLk, the difference between the twist energies of B- and Z-forms should be equal to the formation energy of Z-DNA (ΔGZ). Considering the error in estimating the midpoint ΔLk, ΔGZ becomes 11.5 ± 2.5 kcal/mol, which is somewhat lower than the free energy calculated by Peck et al. (19) using the parameters determined from 2D gel assays (ΔGZ,Zipper = 17.3 kcal/mol). In the case of the Gm5C core, the transition midpoint is σ = -0.005 (ΔLk = -6). Thus, ΔGZ for (Gm5C)11 is determined to be 9.1 ± 2.5 kcal/mol by similar energy arguments.
ΔGZ estimated in this work depends on what value of torsional modulus (C) is used. Unfortunately, the correct value for C is not fully established (30, 31, 34) and depends on the environmental conditions. In our estimation, we used the most widely used value (86 nm kBT). The discrepancy in ΔGZ can be attributed to different experimental conditions (tension, temperature, etc.) between 2D gel assays and single-molecule manipulation techniques.
Even if the possible inaccuracy in C is taken into account, the midpoint -σ we obtained is still much smaller than previously reported values. To explain this discrepancy, we considered the energy of supercoiling in the absence of tension. The energy of a supercoiled DNA at a given ΔLk is (33, 36):
 |
[3] |
where N is the total number of base pairs (∼12 kbp), giving ΔG ∼ 0.05ΔLk kcal/mol. According to Eqs. 1 and 3, a much larger -ΔLk is necessary in the absence of tension than in its presence in order to overcome the energy cost of Z-DNA formation. This discrepancy can be also explained by the following qualitative argument. Since the ratio of writhe to twist in plasmid is known to be approximately 3∶1 and torque induced by twist is responsible for the B-Z transition, the onset σ at zero tension would be roughly four times the onset σ at 1.4 pN (∼-0.01 at 37 °C) in which almost all linking difference is distributed to twist. Then, the onset σ at zero tension (∼-0.04) is in the range known from literature (σ < -0.03). Thus, our result is consistent with the previous results obtained in bulk.
The difference in twist energy before and after the B-Z transition (ΔGTw,B - ΔGTw,Z) varies linearly with ΔLk, assuming that the twist absorbed by the Z segment is constant. The difference in total energy also varies linearly, assuming that ΔGZ is constant. The energy difference in the absence and presence of the Z-form per unit ΔLk (ΔΔLk = 1), calculated from Eq. 1, is ∼2.5 kcal/mol·turn. Thus, even a small change in ΔLk can alter the relative populations of the B and Z states, as well as the rate constants of the B-Z transition significantly (Figs. 4 and 6).
A variety of base-flipping models, which can explain the chain sense paradox, have been proposed. In one model, breaking of the hydrogen bonds between pairing bases is required (4, 19). In another model, flipping of the base pairs occurs through a series of correlated internal motions of backbones and glycosidic bonds without breaking hydrogen bonds (37). Thus, the possibility of bubble formation as an initial step of the B-Z transition is a key issue in the mechanism of the B-Z transition (5). According to our results, the B-Z transition occurs at a much smaller -σ than required for DNA denaturation. At 1.4 pN, DNA denaturation occurs at approximately σd = -0.014 (ΔLk = -15 for 12-kbp DNA) (31, 34). Even if the denaturation is initiated at σ≥σd, it will be preferentially in AT-rich regions. The probability of forming a bubble instead of Z-DNA in the GC core would be extremely small at our midpoint σ. Recent structural data has indicated that base stacking is maintained even between B- and Z-segments across a B-Z junction (18). An intermediate may not be a simple bubble where a few hydrogen bonds are broken and base stacking interactions are disrupted over a few bases, but a symmetric “double-bulge” with two extruded bases whose two adjacent base pairs retain base stacking interactions.
Our results also show unambiguously that the torsion-absorbing, non-B-DNA structure in the alternating GC sequence cannot be a cruciform, because such a structure would yield a higher FRET efficiency than the B-DNA state (38), contrary to our present observations.
The results obtained at low forces are more complex to analyze than at high force because a given ΔLk is distributed to both twist and writhe. Accumulation of twist is, however, certainly important for the B-Z transition as the larger -σ is required for the transition at low forces. It should be also noted that the midpoint -σs for 0.3 and 0.4 pN are closer to the midpoint -σ for 1.4 pN than for 0.2 pN as seen in Fig. S3D. According to the σ-extension curve in ref. 31, writhe starts to be formed as soon as DNA is unwound at 0.2 pN, whereas writhe is not formed at forces above 0.3 pN until a certain onset σ is reached. Thus, twist energy can be accumulated more easily at these forces than 0.2 pN by unwinding DNA. Once the twist energy exceeds the energy required for Z-DNA formation, the transition occurs regardless of subsequent writhe formation. This observation supports again the importance of twist for the B-Z transition and the critical role of force therein from the fact that it modulates the partition of ΔLk between twist and writhe.
Conclusion
We have presented previously undescribed experimental results characterizing the thermodynamics and dynamic aspects of the Z-DNA formation in a (GC)11 sequence in the presence of precisely controlled tension and superhelicity. The formation of Z-DNA was governed by torsional stress in the DNA molecule. The tension to the DNA was also found to be a critical factor in facilitating the formation of Z-DNA, particularly at very small -σ values. The rate constants kBZ and kZB were directly measured by real-time monitoring of the interconversions between the B- and Z-DNA states. Our straightforward measurement provides a reliable way to determine the rate constants of the B-Z transition, avoiding ambiguity and technical difficulties associated with antibody binding and control of supercoiling by DNA intercalators. Taken together, our findings also provide some clues to the transition mechanism.
Z-DNA is believed to play important biological roles in living cells (11). Z-DNA forming sequences such as (GC)n and (GT)n are known to exist commonly at promoter regions (39). Our results suggest that Z-DNA can be formed in vivo more easily than expected from previous results, especially in the presence of tension. Such a sequence is a very effective regulator of torsional stress during gene expression because it is transformed to Z-DNA beyond the onset σ value for the B-Z transition while it is rapidly recovered to B-DNA after torsional stress is removed. A preferred conformation for a given σ value is quickly reached and held after a brief conformation search at the onset σ, which is the hallmark of the cooperative B-Z transition. Our hybrid single-molecule approach, which enables us to elucidate the underlying details of Z-DNA formation, sheds light on the intriguing phenomena of the B-Z transition.
Materials and Methods
Preparation of DNA Samples.
Sample preparation is described in detail in SI Methods and elsewhere (24). Our DNA tether consists of five parts: Insert DNA (∼12 kbp), core DNA, connector, Dig-linker, and Bio-linker (Fig. 1). The core DNA (38 bp), which contains a FRET dye pair, is of primary interest. The GC core DNA contains the (GpC)11 sequence in which dyes are separated by 14 bp (Fig. 1). In the Gm5C core, cytosine bases were methylated at C5 atoms. The random core bears a randomly arranged sequence. The sequences of core DNAs are given in Table S1. DNA tethers were attached to streptavidin-coated magnetic beads (MyOne, Dynal, Invitrogen) in phosphate buffered saline (PBS, pH 7.4). The DNA-bead complexes were injected into a sample chamber coated with anti-digoxigenin (Roche). After a 1–2 h incubation, the imaging buffer (10 mM Tris-HCl (pH 7.4), 100 mM NaCl, 2 mM Trolox, and oxygen scavenging solution) was injected into the chamber. A brief passivation step using sonicated DNA (sssDNA, Invitrogen) was used when needed to prevent nonspecific binding between the glass surface and beads. To measure the FEs of core DNAs free from tension and torsion, the core DNAs were ligated only to Bio-linkers (Fig. 1). These were immobilized on a neutravidin-coated glass surface. The FEs were measured in the imaging buffer containing various amounts of salts.
Single-Molecule FRET and Magnetic Tweezers Experiment.
An objective-type TIRF system equipped with the overhead magnetic tweezers was utilized. The force exerted to DNA was determined by utilizing the equipartition theorem and measuring the Brownian fluctuations of the DNA-bound bead. The diffraction pattern of a DNA-bound bead generated by illuminating it with near-IR light was used to measure the extension of the DNA molecule. Singly tethered and torsionally constrained DNA molecules were chosen for experiments by checking the σ-vs.-extension relationship at high and low forces (1.4 and 0.5 pN). Fluorescence intensities of the donor and acceptor dyes in the core DNAs were recorded to obtain the FE defined by IA/(IA + ID) where IA and ID are the acceptor and donor intensities, respectively. The temperature of the sample chamber was controlled as described previously (40). The objective lens beneath the chamber was warmed by surrounding the lens with a solenoidal tubing that circulates water from a temperature-controlled water bath and the chamber was also directly heated from above by Ohmic heat generated by a current-carrying ITO plate. A thermocouple inserted in the sample chamber was used to measure the temperature. The margin of error in temperature measurement was less than 1 °C. All experiments were performed at room temperature unless otherwise specified. For long-term measurements, the power of the excitation laser was significantly attenuated.
Supplementary Material
Acknowledgments.
This work was supported by the grants from the National Research Foundation of Korea (C00155) and the Seoul R&BD Program (NT080639).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911528107/DCSupplemental.
References
- 1.Rich A. DNA comes in many forms. Gene. 1993;135:99–109. doi: 10.1016/0378-1119(93)90054-7. [DOI] [PubMed] [Google Scholar]
- 2.Wells RD. Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci. 2007;32:271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Wang AH-J. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 4.Pohl FM, Jovin TM. Salt-induced co-operative conformational change of a synthetic DNA: Equilibrium and kinetics studies with poly(dG-dC) J Mol Biol. 1972;67:375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- 5.Fuertes MA, Cepeda V, Alonso C, Pérez JM. Molecular mechanisms for the B-Z transition in the example of poly[d(G-C)·d(G-C)] polymers. A critical review. Chem Rev. 2006;106:2045–2064. doi: 10.1021/cr050243f. [DOI] [PubMed] [Google Scholar]
- 6.Rich A, Nordheim A, Wang AH-J. The chemistry and biology of left-handed Z-DNA. Annu Rev Biochem. 1984;53:791–846. doi: 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- 7.Jovin TM, Soumpasis DM, McIntosh L. The transition between B-DNA and Z-DNA. Annu Rev Phys Chem. 1987;38:521–560. [Google Scholar]
- 8.Nordheim A, et al. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982;31:309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- 9.Rahmouni AR, Wells RD. Stabilization of Z-DNA in vivo by localized supercoiling. Science. 1989;246:358–363. doi: 10.1126/science.2678475. [DOI] [PubMed] [Google Scholar]
- 10.Wittig B, Dorbic T, Rich A. The level of Z-DNA in metabolically active, permeabilized mammalian cell nuclei is regulated by torsional strain. J Cell Biol. 1989;108:755–764. doi: 10.1083/jcb.108.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rich A, Zhang S. Z-DNA: The long road to biological function. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 12.Lafer EM, Möller A, Nordheim A, Stollar BD, Rich A. Antibodies specific for left-handed Z-DNA. Proc Natl Acad Sci USA. 1981;78:3546–3550. doi: 10.1073/pnas.78.6.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herbert A, Lowenhaupt K, Spitzner J, Rich A. Chicken double-stranded RNA adenosine deaminase has apparent specificity for Z-DNA. Proc Natl Acad Sci USA. 1995;92:7550–7554. doi: 10.1073/pnas.92.16.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittig B, Wölfl S, Dorbic T, Vahrson W, Rich A. Transcription of human c-myc in permeabilized nuclei is associated with formation of Z-DNA in three discrete regions of the gene. EMBO J. 1992;11:4653–4663. doi: 10.1002/j.1460-2075.1992.tb05567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu R, et al. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell. 2001;106:309–318. doi: 10.1016/s0092-8674(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Vasquez KM. Z-DNA, an active element in the genome. Front Biosci. 2007;12:4424–4438. doi: 10.2741/2399. [DOI] [PubMed] [Google Scholar]
- 17.Anitha S, Rao KSJ, Latha KS, Viswamitra MA. First evidence to show the topological change of DNA from B-DNA and Z-DNA conformation in the hippocampus of Alzheimer’s brain. Neuromol Med. 2002;2:289–297. doi: 10.1385/nmm:2:3:289. [DOI] [PubMed] [Google Scholar]
- 18.Ha SC, Lowenhaupt K, Rich A, Kim Y-G, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 19.Peck LJ, Wang JC. Energetics of B-to-Z transition in DNA. Proc Natl Acad Sci USA. 1983;80:6206–6210. doi: 10.1073/pnas.80.20.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipps HJ, et al. Antibodies against Z DNA react with the macronucleus but not the micronucleus of the hypotrichous ciliate Stylonychia mytilus. Cell. 1983;32:435–441. doi: 10.1016/0092-8674(83)90463-4. [DOI] [PubMed] [Google Scholar]
- 21.Shroff H, et al. Biocompatible force sensor with optical readout and dimensions of 6 nm3. Nano Lett. 2005;5:1509–1514. doi: 10.1021/nl050875h. [DOI] [PubMed] [Google Scholar]
- 22.Hohng S, et al. Fluorescence-force spectroscopy maps two-dimensional reaction landscape of the Holliday junction. Science. 2007;318:279–283. doi: 10.1126/science.1146113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selvin PR, Ha T. Single-Molecule Techniques: A Laboratory Manual. New York: Cold Spring Harbor; 2008. [Google Scholar]
- 25.Nöllmann M, et al. Multiple modes of Escherichia coli DNA gyrase activity revealed by force and torque. Nat Struct Mol Biol. 2007;14:264–271. doi: 10.1038/nsmb1213. [DOI] [PubMed] [Google Scholar]
- 26.Behe M, Felsenfeld G. Effect of methylation on a synthetic polynucleotide: The B-Z transition in poly(dG-m5dC)·poly(dG-m5dC) Proc Natl Acad Sci USA. 1981;78:1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kypr J, Kejnovská I, Renčiuk D, Vorličková M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009;37:1713–1725. doi: 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pohl FM. Dynamics of the B-to-Z transition in supercoiled DNA. Proc Natl Acad Sci USA. 1986;83:4983–4987. doi: 10.1073/pnas.83.14.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peck LJ, Wang JC, Nordheim A, Rich A. Rate of B to Z structural transition of supercoiled DNA. J Mol Biol. 1986;190:125–127. doi: 10.1016/0022-2836(86)90082-3. [DOI] [PubMed] [Google Scholar]
- 30.Mosconi F, Allemand J-F, Bensimon D, Croquette V. Measurement of the torque on a single stretched and twisted DNA using magnetic tweezers. Phys Rev Lett. 2009;102:078301. doi: 10.1103/PhysRevLett.102.078301. [DOI] [PubMed] [Google Scholar]
- 31.Charvin G, Allemand J-F, Strick TR, Bensimon D, Croquette V. Twisting DNA: Single molecule studies. Contemp Phys. 2004;45:383–403. [Google Scholar]
- 32.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Depew RE, Wang JC. Conformational fluctuations of DNA helix. Proc Natl Acad Sci USA. 1975;72:4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marko JF. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys Rev E. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 35.Strick TR, Allemand J-F, Croquette V, Bensimon D. Twisting and stretching single DNA molecules. Prog Biophys Mol Biol. 2000;74:115–140. doi: 10.1016/s0079-6107(00)00018-3. [DOI] [PubMed] [Google Scholar]
- 36.Pulleyblank DE, Shure M, Tang D, Vinograd J, Vosberg H-P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: Formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci USA. 1975;72:4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harvey SC. DNA structural dynamics: Longitudinal breathing as a possible mechanism for the B↔Z transition. Nucleic Acids Res. 1983;11:4867–4878. doi: 10.1093/nar/11.14.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasnik I, et al. Branch migration enzymes as a Brownian ratchet. EMBO J. 2008;27:1727–1735. doi: 10.1038/emboj.2008.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schroth GP, Chou P-J, Ho PS. Mapping Z-DNA in the human genome. J Biol Chem. 1992;267:11846–11855. [PubMed] [Google Scholar]
- 40.Park J-S, Hyon J-Y, Lee KJ, Hong S-C. Temperature dependence of DNA elasticity and cisplatin activity studied with a temperature-controlled magnetic tweezers system. J Korean Phys Soc. 2008;52:1927–1931. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.