Abstract
Many well-known immune-related C-type lectin-like receptors (CTLRs) such as NKG2D, CD69, and the Ly49 receptors are encoded in the natural killer gene complex (NKC). Recently, we characterized the orphan NKC gene CLEC2A encoding for KACL, a further member of the human CLEC2 family of CTLRs. In contrast to the other CLEC2 family members AICL, CD69, and LLT1, KACL expression is mostly restricted to skin. Here we show that KACL is a non–disulfide-linked homodimeric surface receptor and stimulates cytotoxicity by human NK92MI cells. We identified the corresponding activating receptor on NK92MI cells that is encoded adjacently to the CLEC2A locus and binds KACL with high affinity. This CTLR, termed NKp65, stimulates NK cytotoxicity and release of proinflammatory cytokines upon engagement of cell-bound KACL. NKp65, a distant relative of the human activating NK receptor NKp80, possesses an amino-terminal hemITAM that is required for NKp65-mediated cytotoxicity. Finally, we show that KACL expression is mainly restricted to keratinocytes. Freshly isolated keratinocytes express KACL and are capable of stimulating NKp65-expressing cells in a KACL-dependent manner. Thus, we report a unique NKC-encoded receptor-ligand system that may fulfill a dedicated function in the immunobiology of human skin.
Keywords: CLEC2 family, natural killer cells, NKRP1 receptors, skin
Natural killer (NK) cells express a diverse array of activating and inhibitory receptors regulating NK activity depending on their respective environmental “niche” (1, 2). Major inhibitory NK receptors are the human killer Ig–like receptors, the mouse Ly49 receptors, and CD94/NKG2A which all bind MHC class I molecules and avert cytolysis of MHC class I–proficient cells (3). NKG2D, in contrast, is an activating NK receptor stimulating NK cytotoxicity upon ligation of diverse stress-induced MHC class I–related molecules thereby promoting NK-mediated elimination of infected or malignant cells (4–6). NKG2D, CD94/NKG2A, and Ly49 molecules all are dimeric type II transmembrane receptors with a single C-type lectin-like domain (CTLD) (7) and are encoded in the NKC on human chromosome 12 (7, 8). The human NKC contains approximately 30 genes encoding for C-type lectin-like receptor (CTLR) expressed on NK cells, T cells, myeloid cells, and other immune-related cells as well as some poorly defined orphan genes.
Similar to human killer Ig–like receptor and Ly49 receptor families, the NKC-encoded NKRP1 family comprises both inhibitory and activating NK receptors (9). However, in contrast to the former, NKRP1 receptors do not engage MHC class I molecules. Rather, at least some mouse NKRP1 receptors ligate members of the CLEC2 family with NKRP1 and CLEC2 genes interspersed in a distinct subregion of the NKC establishing a unique system of genetically linked C-type lectin-like receptor–ligand pairs. For example, the inhibitory Nkrp1d and the activating Nkrp1f receptors bind the CLEC2 family members Clr-b and Clr-g, respectively (9, 10). However, as expression of most mouse Clr proteins is poorly characterized and ligands of other Nkrp1 receptors such as NK1.1 remain unknown, the immunobiology of these NKC-based receptor-ligand systems is far from being understood. Also in humans, corresponding NKRP1–CLEC2 receptor–ligand pairs have recently been characterized: LLT1 (encoded by CLEC2D) was identified as a ligand of the inhibitory NK receptor NKR-P1A/CD161 (11, 12) and another member of the human CLEC2 family, AICL (CLEC2B), as a ligand of the activating NK receptor NKp80 (13). LLT1 is expressed by activated DC and B cells and was shown to inhibit NK cytotoxicity, whereas AICL on myeloid cells stimulates NK cytotoxicity as well as a cross-talk between NK cells and monocytes (13, 14).
Recently, we reported that the orphan gene CLEC2A encodes a fourth member of the human CLEC2 family of NKC-encoded CTLR (15) that we now term KACL (keratinocyte-associated C-type lectin). KACL transcripts were almost exclusively detected in human skin, clearly contrasting the broad presence of transcripts of other CLEC2 family member in hematopoietic cells (15–17). In the present study, we addressed expression and a potential immune-related function of KACL. We find KACL specifically expressed on keratinocytes stimulating NK cytotoxicity by engaging a hitherto unknown activating CTLR, thereby indicating that this receptor–ligand pair may specifically contribute to the immunosurveillance of human skin.
Results
Ectopic KACL Stimulates NK Cytotoxicity.
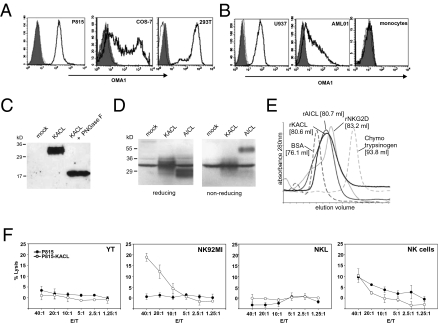
In a previous study, we characterized the NKC-resident orphan gene CLEC2A encoding for a CTLR that we have now termed KACL. Skin-associated mRNA expression clearly distinguished KACL from the other human CLEC2 family members AICL, LLT, and CD69 (15). To analyze protein expression and function of KACL, we generated the mAb OMA1 that specifically binds to soluble KACL ectodomains (rKACL) and to KACL ectopically expressed on mouse and primate cell lines, but not to the KACL relatives AICL and LLT1 (Fig. 1A and Fig. S1). In a screen of a panel of human cell lines, only a few cell lines of myeloid origin specifically bound OMA1, with U937 expressing highest levels of KACL (Fig. 1B). This is in line with our previously reported mRNA expression profile and paralleled expression of AICL on cell lines (13, 15). Next, we assessed KACL expression by human peripheral blood mononuclear cells (PBMCs) from healthy donors. As previously reported (15), no significant levels of KACL transcripts were detected in freshly isolated human PBMCs or in PBMCs after treatment with potent stimulatory compounds such as phythemagglutinine or phorbol myristate acetate. Accordingly, we failed to detect surface KACL on PBMC including T cells using OMA1 (Fig. S2). This was unexpected, because a previous study reported KACL (also known as PILAR) expression by activated T cells (18). Immunoblot analyses of KACL-transfected COS-7 revealed that KACL is a glycoprotein of approximately 32 kDa (Fig. 1C). Removal of N-linked glycosylation resulted in a protein of approximately 20 kDa well in accord with the predicted molecular mass of KACL monomers and the presence of three potential N-glycosylation sites. In contrast to AICL, which forms disulfide-linked homodimers, KACL runs as a monomer under both reducing and nonreducing conditions (Fig. 1D). However, in gel filtration, rKACL eluted at a similar volume as did ectodomains of NKG2D (rNKG2D) or AICL (rAICL) which are of similar molecular mass and known to occur as homodimers (Fig. 1E). This strongly suggested that KACL occurs as a non–disulfide-linked homodimer in contrast to AICL, LLT1, and CD69, which all form disulfide-bonded homodimers (19). Having established that KACL is a bona fide cell surface CTLR, we addressed its potential immune-related function. As the KACL relatives AICL and LLT1 modulate NK cytotoxicity by engaging NK receptors NKp80 and NKR-P1A, respectively, we tested various human NK cell lines for cytotoxicity against P815 ectopically expressing KACL. No significantly increased cytotoxicity of YT or NKL cells toward P815-KACL transfectants was observed, and NK cells isolated from healthy donors did not specifically react against P815-KACL (Fig. 1F). In contrast, NK92MI cells exhibited a pronounced cytotoxic response against P815-KACL cells, but did not lyse P815-neo control transfectants, suggesting that NK92MI cells bear an activating receptor stimulating NK cytotoxicity upon KACL engagement (Fig. 1F).
Fig. 1.
KACL is a non–disulfide-linked homodimeric CTLR and stimulates NK cytotoxicity. (A) MAb OMA1 specifically detects ectopic KACL on mouse and primate cells. P815 stably transfected with KACL (solid), but not mock-transfected P815 (dotted), were stained with OMA1. COS-7 transiently and 293T stably transfected with KACL (solid), but not the respective AICL transfectants (dotted) were stained by OMA1. Shaded histograms are isotype controls. (B) KACL is expressed on some myeloid cell lines, but not on monocytes. U937, AML01, and monocytes isolated from PBMCs were stained with OMA1 (solid). Pretreatment of OMA1 with rKACL (dotted) reduced stainings to isotype levels (shaded). (C) Monomeric KACL is a glycoprotein of approximately 32 kD. Lysates of KACL-transfected COS-7 (with or without PNGase F treatment) analyzed by immunoblotting with anti–histidine-tag antibody. (D) Lysates of COS-7 transfected with myc-tagged KACL and FLAG-tagged AICL cDNA separated by SDS/PAGE were analyzed by immunoblotting with anti-c-myc/anti-FLAG mAbs. KACL migrates similarly to a cross-reactive protein also apparent in mock transfectants. (E) Sizing of rKACL and rAICL by gel filtration compared with BSA (66 kDa), rNKG2D (41 kDa), and chymotrypsinogen (25 kDa). (F) NK92MI, but not YT, NKL, or freshly isolated NK cells, exhibit an enhanced cytolysis of P815-KACL.
NKp65 Is a CTLR Encoded Adjacent to KACL in the NKC.
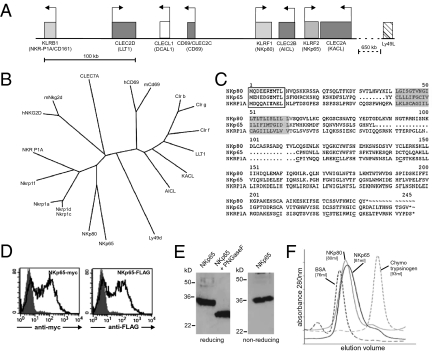
To characterize the receptor on NK92MI cells specifically interacting with KACL, we assessed binding of ectodomains of known activating NK receptors to 293T or COS-7 cells ectopically expressing KACL (Fig. 1A). Neither the ectodomains of NKC-encoded C-type lectin-like NK receptors such as NKp80 or NKG2D nor the ectodomains of the Ig-like receptors NKp30, NK44, NKp46, 2B4, and DNAM-1 bound to KACL-expressing cells (Fig. S3). Further, experiments forcing ligand-induced down-regulation did not provide any evidence for KACL serving as a ligand for NKp80 or NKR-P1A (Fig. S3). As we failed to identify a receptor for KACL among known activating NK receptors, we considered the possibility that KACL may interact with a yet undescribed receptor encoded in the NKC in proximity to the CLEC2A locus based on the genetic linkage of the KACL relatives AICL and LLT1 with their respective receptors. Computational analysis of the genomic region flanked by the genes CLEC2B and CLEC2A identified putative exons encoding for a CTLD. Employing exon-spanning oligonucleotides, we amplified a corresponding partial transcript from activated human NK cells. Subsequently, we obtained the sequence of the complete ORF by RACE-PCR. This ORF is encoded by a gene, termed KLRF2, consisting of six exons extending over 15 kb, and positioned just approximately 3 kb telomeric from the CLEC2A gene in a tail-to-tail orientation (Fig. 2A). The ORF predicted a typical C-type lectin-like type II transmembrane glycoprotein of 207 amino acids sharing the key features of a CTLD, such as six conserved cysteines and the WIGL motif (Fig. 2C) (7). Comparative sequence analyses of the CTLD with CTLD of other NKC-encoded human CTLR identified NKp80 as closest relative and placed this CTLR into a subgroup of related CTLRs comprising human NKp80 and NKR-P1A as well as the mouse Nkrp1 receptors, which we suggest to term NKRP1 subfamily (Fig. 2B). We named this CTLR NKp65 in view of its molecular mass and homodimeric occurrence (as detailed later), and to indicate its similarity to NKp80. The stalk region connecting the CTLD to the cell membrane is fairly short in NKp65 (24 aa vs. 52 aa in NKp80), whereas the amino-terminal cytoplasmic sequence is again homologous to NKp80 (Fig. 2C). Interestingly, a tyrosine at position 7 of the cytoplasmic domain is present in all three receptors, including NKR-P1A. In NKp65, tyrosine 7 is embedded in a sequence identical to the hemITAM motif described for some myeloid-specific CTLR such as Dectin-1 and Clec1b (21, 22). We next tested whether NKp65 is a bona fide cell surface CTLR. Surface expression of NKp65 was demonstrated on COS-7 cells after transfection of a carboxy-terminally tagged NKp65 cDNA (Fig. 2D). Immunoblot analysis of NKp65-transfectants revealed that NKp65 is a glycoprotein of approximately 32 kDa under reducing and nonreducing conditions that was reduced to approximately 24 kDa after removal of N-linked carbohydrates (Fig. 2E) in line with the predicted molecular mass and two N-linked glycosylation sites flanking the CTLD. Purified ectodomains of NKp65 (rNKp65) produced in 293T cells were subjected to gel filtration and eluted at a similar volume as rNKp80 (Fig. 2F), suggesting that NKp65 forms non–disulfide-linked homodimers, whereas NKp80 and NKR-P1A form disulfide-linked homodimers (23, 24).
Fig. 2.
NKp65 is an NKC-encoded CTLR distantly related to NKp80. (A) NKp65 is encoded in the NKC by KLRF2 flanked by CLEC2A and CLEC2B. Schematic representation of a subregion of the human NKC. Boxes and arrows represent CLEC2 (dark gray) and NKRP1 genes (light gray) and transcriptional orientation, respectively. (B) Phylogenetic analysis of CTLD of NKRP1 and CLEC2 receptors of man and mouse. NKG2D, CLEC7A, and Ly49D are included for comparison. The phylogenetic tree was generated using the PHYLIP program. (C) Sequences of human NKRP1 receptors NKp80, NKR-P1A, and NKp65. Tyrosine-based motifs in the cytoplasmic domain are boxed and predicted transmembrane domains are shaded. Conserved cysteines of the CTLD are underlined. (D) NKp65 is a cell surface CTLR. Myc- or FLAG-tagged NKp65 was transfected into COS-7 and surface expression detected by tag-specific mAb. (E) NKp65 is a glycoprotein. Immunoblot analysis of lysates from transfected COS-7 with an anti–histidine-tag mAb. (F) Sizing of rNKp65 by gel filtration compared with BSA (66 kDa), rNKp80 (approximately 60 kDa), and chymotrypsinogen (25 kDa).
NKp65 Is a High-Affinity Receptor for KACL.
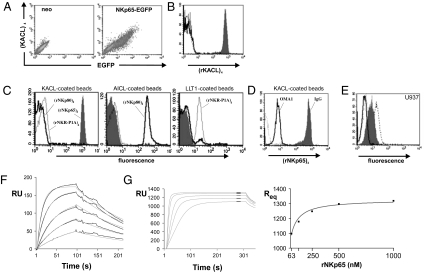
To address whether NKp65 is a receptor for KACL, we assessed binding of KACL tetramers to cells ectopically expressing NKp65. Only NKp65-expressing cells, but not controls or NKp80- or NKR-P1A-expressing cells, were stained by KACL tetramers (Fig. 3A and Fig. S3). We further investigated specific binding of NKp65 to KACL by using the purified, biotinylated ectodomains of these and related CTLR described here or elsewhere (13). In line with results described already, KACL tetramers strongly bound to rNKp65 immobilized on streptavidin-coated microspheres (imNKp65), but not to imNKp80 or imNKR-P1A (Fig. 3B) at variance to a previous study (18). Conversely, NKp65 tetramers also bound imKACL, but not to imAICL or imLLT1 (Fig. 3C). Further, NKp80 and NKR-P1A tetramers bound only to their known ligands imAICL and imLLT1, respectively, demonstrating that there is no significant cross-reactivity between these cognate and genetically coupled receptor-ligand pairs (Fig. 3C). Pretreatment of imKACL with mAb OMA1 largely abrogated binding of NKp65 tetramers further corroborating the specificity of the NKp65-KACL interaction and validating OMA1 as a suitable blocking reagent for functional studies (Fig. 3D). Pretreatment with OMA1 also completely blocked binding of NKp65 tetramers to U937 cells endogenously expressing KACL (Fig. 3E). Subsequently, we determined the kinetics of KACL-NKp65 interaction by surface plasmon resonance (SPR). Soluble rNKp65 bound immobilized KACL with a fast association rate (kon, 5.8 × 105 M−1s−1) and dissociated relatively slowly (koff, 6.4 × 10−3s−1), yielding a fairly high calculated affinity (KD,calc of approximately 11 nM at 25 °C; Fig. 3F) that was confirmed in steady-state analyses (KD of approximately 13 nM at 25 °C; Fig. 3G). Hence, these results demonstrate that NKp65 is a high affinity receptor for KACL, and further establish and extend the principle of genetically linked C-type lectin-like receptor–ligand pairs recruited from the NKRP1 and CLEC2 subfamilies, respectively.
Fig. 3.
Identification of NKp65 as a high-affinity receptor for KACL. (A) KACL interacts with NKp65. KACL tetramers bind COS-7 transfected with a bicistronic NKp65-EGFP construct (Right), but not control transfectants (Left). (B) KACL tetramers bind imNKp65 (shaded), but not imNKp80 (solid), imNKR-P1A (dashed), or uncoated microspheres (dotted). (C) NKp65 specifically binds KACL. NKp65 tetramers (shaded) bind to imKACL (Left) but not to imAICL (Center) or imLLT1 (Right). NKp80 (solid) and NKR-P1A tetramers (dotted) bind imAICL and imLLT1, respectively, but not imKACL. (D) Binding of NKp65 tetramers to imKACL was blocked by preincubation of imKACL with OMA1 (solid) but not with an isotype control (shaded). Background staining of uncoated microspheres is dotted. (E) NKp65 binds KACL on U937. NKp65 tetramer binding to U937 is blocked by preincubation of U937 with OMA1 F(ab)2 (solid) but not with control F(ab)2 (shaded). Direct staining by OMA1 F(ab)2 (stippled) or control F(ab)2 (dotted) is also shown. (F) Kinetics of the binding of rNKp65 (8, 16, 31, 62, and 125 nM) to imKACL measured by SPR at 25 °C. Association rate is 5.8 × 105 M−1s−1, dissociation rate is 6.4 × 10−3s−1, and calculated KD is 11 nM. (G) Affinity of NKp65 for KACL was determined by injection of rNKp65 (63, 125, 250, 500, and 1,000 nM) over imKACL at 25 °C; KD is 13 nM. Average interval for the best fit curve (Right) is indicated.
NKp65 Engagement Triggers Cytotoxicity and Cytokine Secretion.
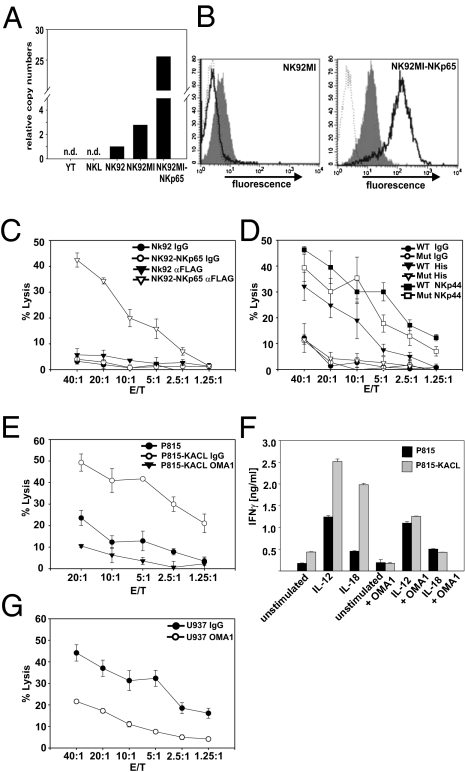
Next, we continued with the functional characterization of NKp65. The hemITAM present in the cytoplasmic domain of NKp65 was indicative of an activating receptor. A stimulatory function was also noted for the putative KACL receptor on NK92MI cells (Fig. 1F). Hence, we analyzed NK92MI cells for NKp65 expression. Both NK92MI and parental NK92 cells contained significant amounts of NKp65 transcripts, whereas no NKp65 transcripts were detected for YT and NKL cells (Fig. 4A). NK92MI cells also were markedly stained by KACL tetramers (Fig. 4B), strongly suggesting that NKp65 expression on NK92MI cells is accountable for the KACL-specific lysis of P815-KACL cells. However, in the absence of a suitable NKp65-specific mAb, this could not be unequivocally demonstrated. Therefore, to directly assess functional activity of NKp65, we transduced NK92MI with tagged NKp65 (NK92MI-NKp65). NKp65 was brightly expressed on transduced cells (Fig. 4B), and in a redirected cytotoxicity assay, NK92MI-NKp65 lysed P815 loaded with tag-specific Ab, but not control Ig-loaded cells (Fig. 4C), demonstrating that NKp65 triggers NK cytotoxicity. To evaluate the relevance of the aminoterminal hemITAM, tyrosine 7 was mutated to phenylalanine and the resulting mutant (NKp65Y7F) introduced into NK92MI. Although mutant NKp65Y7F was expressed at similar levels as WT NKp65, and NK92MI-NKp65Y7F responded to NKp44 engagement by redirected lysis, no redirected cytolysis was observed when NKp65Y7F was engaged (Fig. 4D). Thus, tyrosine 7 is crucially involved in NKp65-mediated cytotoxicity. Next, we investigated NKp65-mediated cytotoxicity and cytokine secretion toward KACL-expressing cells. P815-KACL elicited a strong cytolytic response of NK92MI-NKp65 cells and cytotoxicity was abrogated by pretreatment with OMA1 (Fig. 4E). Moreover, P815-KACL stimulated IFN-γ secretion by NK92MI-NKp65 cells in a KACL-dependent manner, which became particularly apparent in the presence of IL-12 or IL-18 (Fig. 4F). Finally, we also showed that cytolysis of U937 by NK92MI-NKp65 cells is partially KACL-dependent (Fig. 4G). Together these data demonstrate that NKp65 is a functional activating receptor stimulating NK cytotoxicity and cytokine secretion upon engagement of its ligand KACL.
Fig. 4.
NKp65 triggers cytolytic activity and stimulates cytokine secretion. (A) NK92 and NK92MI, but not YT or NKL, express NKp65. Real-time PCR analysis of NKp65 transcripts (n.d., not detectable). (B) NKp65 expression on NK92MI. NK92MI transduced with tagged NKp65 or untransduced cells were stained for NKp65 with anti-FLAG mAb (solid) or KACL tetramers (shaded) or isotype control (dotted). (C) NKp65 triggers redirected cytolysis. Redirected lysis assay using P815 as targets and NK92MI-NKp65 or mock transductants as effectors either in the presence of anti-FLAG mAb M2 or isotype control. (D) Tyrosine 7 of NKp65 is critically involved in NKp65-mediated cytotoxicity. Redirected lysis assay using P815 as targets and NK92MI-NKp65 (WT) or NK92MI-NKp65Y7F (Mut) as effectors in the presence of anti-NKp44 mAb, anti–histidine-tag mAb or isotype control. (E) P815-KACL, but not P815-neo, are lysed by NK92MI-NKp65. Lysis of P815-KACL was inhibited to background levels in the presence of OMA1, but not in presence of an isotype control. (F) KACL stimulates IFN-γ secretion by NK92MI-NKp65. NK92MI-NKp65 were cocultured with P815-neo or P815-KACL in presence or absence of OMA1, IL-12, or IL-18, and subsequently, IFN-γ secretion was measured by ELISA. (G) KACL-dependent lysis of U937. Cytolysis of U937 in presence of OMA1 or isotype control by NK92MI-NKp65.
KACL on Keratinocytes Stimulates NKp65-Mediated Degranulation.
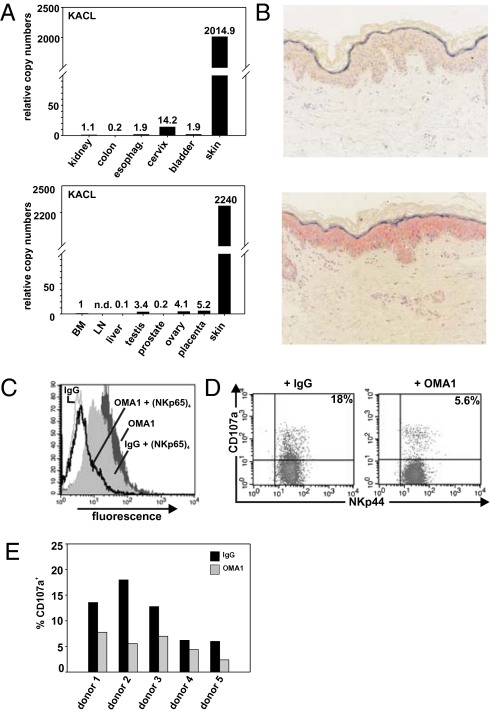
KACL transcripts are abundantly present in human skin, but barely detectable in other tissues (Fig. 5A and ref. 15). To further pinpoint KACL expression in skin, we performed in situ hybridization of nondiseased human skin samples and observed a fairly homogenous distribution of KACL transcripts throughout the epidermis indicating KACL expression by keratinocytes (Fig. 5B). Subsequently, we addressed surface expression of KACL on keratinocytes isolated from nondiseased surgical skin samples. Using OMA1, we detected marked KACL expression on freshly isolated keratinocytes (Fig. 5C). KACL expression by keratinocytes was further corroborated by the specific and distinct binding of NKp65 tetramers that was completely blockable by pretreatment with OMA1 (Fig. 5C). Finally, we assessed whether KACL on keratinocytes can be functionally recognized by the NKp65 receptor. Coculture with freshly isolated keratinocytes from various donors triggered a marked degranulation of NK92MI-NKp65 cells that was partially blocked by addition of OMA1 (Fig. 5 D and E). Thus, the specific expression of KACL by keratinocytes allows for a dedicated functional recognition of keratinocytes via the activating receptor NKp65.
Fig. 5.
KACL on keratinocytes stimulates NKp65-mediated degranulation. (A) Predominance of KACL transcripts in human skin. Relative levels of KACL transcripts in human tissues were determined by real-time PCR. (B) KACL is expressed in the epidermis of human skin. In situ hybridization of human skin with a sense KACL probe (control, Upper) and an antisense KACL probe (Lower). (C) Keratinocytes express surface KACL. Freshly isolated human keratinocytes were stained with OMA1 F(ab)2 or control F(ab)2, or with NKp65 tetramers after preincubation with control F(ab)2 or OMA1 F(ab)2. (D and E) KACL-mediated activation of NK92MI-NKp65 by keratinocytes. Degranulating (CD107a+) NK92MI-NKp65 (NKp44+ cells) after coculture with freshly isolated keratinocytes in presence of OMA1 or isotype control. (D) One representative dot plot analysis and (E) summary of five independent experiments using keratinocytes of different donors.
Discussion
We describe here the activating immunoreceptor NKp65, the high-affinity interaction of NKp65 with the recently reported orphan CTLR KACL/CLEC2A, a highly restricted expression of KACL by human keratinocytes, and functional consequences of NKp65–KACL interaction. NKp65 and KACL both are non–disulfide-linked homodimeric CTLRs and are encoded by genes that are separated by only a few thousand base pairs in the human NKC. This report further establishes and extends the principle of genetically linked C-type lectin-like receptor–ligand pairs in humans that was originally reported in a seminal study by Yokoyama and colleagues for certain CTLRs encoded in the mouse NKC (9). This latter study prompted the characterization of an obviously corresponding receptor/ligand pair in humans (i.e., NKR-P1A/LLT1) (11, 14), and aided in the identification of AICL as a genetically linked ligand for the human activating NK receptor NKp80 (13). Based on this trait of genetic linkage and the relatedness of the respective CTLD sequences in mouse and man (Fig. 2B), we would like to suggest to group these CTLR together into distinct subfamilies (i.e., NKRP1 and CLEC2 families).
As our current study provides further documentation that humans, like mice, possess several of these genetically coupled receptor/ligand pairs of CTLRs, it evokes several questions. First, why are genes of receptors and ligands tightly coupled, although there is no apparent polymorphism? In contrast to other NKC-encoded CTLR such as NKG2D, CD94, or Dectin-1, there are no clear homologies between certain NKRP1 or CLEC2 genes of mouse and man, apart from CD69. But do some of these genes encode for CTLR serving a similar function across species, i.e., are there functional homologues? Are there even more functional members of these CTLR subfamilies to be discovered that may have escaped description because of a highly restricted expression pattern? Does CD69, which, like KACL, AICL, and LLT1, also is clearly a member of the CLEC2 subfamily, also engage a yet unrecognized receptor encoded in this subregion of the NKC? What is the immunological function of these multiple receptor/ligands pairs? Are these receptor–ligand pairs functionally redundant or do they exert some specialized function that has not yet been appreciated?
With regard to the last issue, this study reporting a tissue-specific expression of KACL introduces another aspect into this discussion, raising the possibility that at least some members of the CLEC2 subfamily (together with their putative receptors) may fulfill a dedicated function in tissue-specific immunity. From our data presented in this study it is evident that NKp65/KACL does not simply represent “just another” receptor–ligand pair involved in NK cell activation. Rather, KACL is clearly distinct by its skin-specific expression from other CLEC2 family members that are broadly expressed by hematopoietic cells. Further, both NKp65 and KACL are non–disulfide-linked homodimers and thereby differ from other members of the human NKRP1 and CLEC2 subfamilies including NKp80, NKR-P1A, AICL, LLT1, as well as CD69, all being disulfide-linked homodimers (19, 20, 23, 24). It remains to be addressed whether this trait is related to a particular function or a skin-associated function of this receptor–ligand pair. Noteworthy is also the strong affinity of the NKp65–KACL interaction (KD of approximately 0.01 μM at 25 °C) which is considerably higher (approximately 400-fold) than the NKp80–AICL interaction (KD of approximately 4.1 μM at 25 °C), the only CTLR–CTLR interaction for which affinity data are available (13). Further, to the best of our knowledge, NKp65 is the first NK-associated receptor that contains a so-called hemITAM previously reported for myeloid-specific receptors (21, 22). Tyrosine 7 of this hemITAM is essential for NKp65-stimulated cytotoxicity and it will be addressed in upcoming studies whether NKp65 signals through Syk kinase as reported for other hemITAM-bearing receptors (22). Interestingly, tyrosine 7 is also shared by NKp80 and NKR-P1A, likewise suggesting a crucial involvement of tyrosine 7 in signaling of these latter receptors.
At present, our knowledge on the physiological expression of NKp65 is rather limited. We isolated the full-length NKp65 cDNA from primary NK cells, but in contrast to NK92MI cells, human peripheral blood NK cells contain only low levels of NKp65 transcripts, suggesting that NKp65 is barely expressed on blood NK cells (Fig. S4). Accordingly, we did not obtain evidence for NKp65 surface expression on freshly isolated or IL-2/IL-12–stimulated peripheral blood NK cells by staining with KACL tetramers (Fig. S4). Taking into consideration that KACL expression is restricted to skin, in contrast to other CLEC2 family members, one may speculate that expression of NKp65 likewise is restricted to a subset of skin-associated lymphocytes. For example, a subset of CD4 memory T cells has recently been reported which is prone to home to the skin, produces IL-22, and is found in lesions of psoriatic skin (25, 26). It will be of immediate interest to address expression and function of NKp65 on these T cells and on skin-associated NK cells (27) when NKp65-specific mAb become available. At any rate, our findings demonstrate that NKp65 is an activating receptor capable of mediating the dedicated immunorecognition of keratinocytes via engaging the genetically linked CTLR KACL.
In summary, we find KACL specifically expressed on human keratinocytes and detected by a previously unknown genetically linked activating CTLR capable of stimulating cytotoxicity and cytokine secretion via a hemITAM. Considering the particular immunobiology of human skin, it will be of considerable interest to address functional implications of this unique NKC-encoded receptor–ligand pair in skin-affecting diseases such as psoriasis and graft-versus-host disease, as well as in wound healing.
Materials and Methods
Cells.
Surgical samples of human skin were obtained from the Dermatology Department of the University of Tübingen with approval of the local ethics committee. After removing the cutaneous fat layer, skin pieces were incubated for 16 h at 37 °C in trypsin solution. Subsequently, the epidermal layer was separated from the dermis and resuspended keratinocytes filtered through a cell strainer before used in experiments. Human NK cell lines NKL, YT, and NK92MI (ATCC) and freshly isolated human NK cells were used in NK effector assays. NK92MI cells were transduced with retroviruses generated by transfection of phoenix-ampho cells (Nolan Laboratory, Stanford, CA) with the vector pMXsIP kindly provided by Toshio Kitamura (Tokyo, Japan) and containing cDNA of NKp65 or NKp65Y7F with carboxy-terminal FLAG- and six-histidine tags. Transduced cells were selected in puromycin-containing medium.
Recombinant Proteins and Antibodies.
The complete NKp65 cDNA was isolated from IL-2/IL-12–stimulated human NK cells by a 5′RACE kit (Invitrogen) according to the manufacturer's instruction (accession no. GQ398770). Soluble recombinant rKACL (Ile-46 through Leu185) and rNKp65 (Ser63 through Val207) were purified from supernatants of 293T stably transfected with the respective cDNA containing BirA, c-myc, and six-histidine tags as previously reported for rNKp80, rNKR-P1A, rAICL, and rLLT1 (13). For gel filtration analysis, rCTLR were loaded onto a Superdex 200 gel filtration column at a flow rate of 0.5 mL/min. Ectodomains were biotinylated using BirA ligase and, before use, either immobilized on streptavidin-coated microspheres (Bangs Laboratories) or tetramerized using PE-labeled streptavidin (Molecular Probes) (13). KACL-specific mAb OMA1 was generated by standard hybridoma technology (28). In brief, BALB/c mice were immunized with P815-KACL and resulting hybridoma screened for specific reactivity against COS-7 expressing a KACL-pIRES-DsRed construct. F(ab’)2 fragments were generated by pepsin digestion (Pierce).
SPR.
Using a BIAcore X apparatus (BIAcore) KACL ectodomains were immobilized to CM5 chips by amine coupling. In kinetic analyses (flow rate, 50 μL/min), RU from the control flow cell (imAICL) were subtracted from response units (RU) of the KACL-derivatized surface (black traces) with overlaid gray traces representing fitting of a 1:1 Langmuir model to the association and dissociation phases. In steady-state analyzes (15 μL/min), RU from the KACL-derivatized surface were corrected by RU from the LLT1-derivatized control cell. Raw data were analyzed and illustrated using BIAevaluation software (BIAcore).
Cytotoxicity, Degranulation, and Cytokine Analyses.
Direct cytotoxicity of NK cells as well as redirected lysis of P815I was assessed in a standard 51Cr release assay (28). In redirected lysis assays, P815 were loaded with 5 μg/mL antihistidine (Qiagen), 2 μg/mL anti-NKp44 (R&D Systems), or 5 μg/mL anti-FLAG. In blocking experiments, antibodies (OMA1, isotype) were used at a final concentration of 10 μg/mL Degranulating NK92MI-NKp65 were quantified by analysis of surface CD107a after overnight coculture with freshly isolated keratinocytes in presence of mAb OMA1 or an isotype control (10 μg/mL). Golgi-Stop (BD Biosciences) was added for the last 4 h. IFN-γ secreted by NK92MI-NKp65 after 16-h coculture with P815 transfectants was quantified using a sandwich ELISA (BD Biosciences). Assays were performed in presence or absence of blocking anti-KACL mAb OMA1 (10 μg/mL), IL-12 (1 ng/mL), or IL-18 (10 ng/mL), respectively. All samples were done in triplicates.
In Situ Hybridization.
In situ hybridization was performed on sections of paraffin-embedded skin with digoxigenin-labeled RNA probes generated by in vitro transcription of KACL full-length cDNA in pBluescript using DIG RNA labeling kit (Roche) and T7 (sense) and T3 polymerases (antisense), respectively. Tissues were surgical samples of nondiseased human skin. For more details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Birgit Sauer, Wiebke Ruschmeier, and Beate Pömmerl for excellent technical assistance. This work was supported by Grants SFB 685/A1 and STE 828/5-1 from the Deutsche Forschungsgemeinschaft (to A.S.).
Footnotes
*This Direct Submission article had a prearranged editor.
Conflict of interest statement: A.S. and J.S. have filed a patent application on NKp65. None of the other authors declare a conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. GQ398770).
This article contains supporting information online at www.pnas.org/cgi/content/full/0913108107/DCSupplemental.
References
- 1.Di Santo JP. Natural killer cells: diversity in search of a niche. Nat Immunol. 2008;9:473–475. doi: 10.1038/ni.f.201. [DOI] [PubMed] [Google Scholar]
- 2.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–510. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 3.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 4.Bauer S, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. [PubMed] [Google Scholar]
- 5.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 7.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 8.Yokoyama WM, Seaman WE. The Ly-49 and NKR-P1 gene families encoding lectin-like receptors on natural killer cells: the NK gene complex. Annu Rev Immunol. 1993;11:613–635. doi: 10.1146/annurev.iy.11.040193.003145. [DOI] [PubMed] [Google Scholar]
- 9.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 10.Carlyle JR, et al. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci USA. 2004;101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldemir H, et al. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol. 2005;175:7791–7795. doi: 10.4049/jimmunol.175.12.7791. [DOI] [PubMed] [Google Scholar]
- 12.Rosen DB, et al. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol. 2005;175:7796–7799. doi: 10.4049/jimmunol.175.12.7796. [DOI] [PubMed] [Google Scholar]
- 13.Welte S, Kuttruff S, Waldhauer I, Steinle A. Mutual activation of natural killer cells and monocytes mediated by NKp80-AICL interaction. Nat Immunol. 2006;7:1334–1342. doi: 10.1038/ni1402. [DOI] [PubMed] [Google Scholar]
- 14.Rosen DB, et al. Functional consequences of interactions between human NKR-P1A and its ligand LLT1 expressed on activated dendritic cells and B cells. J Immunol. 2008;180:6508–6517. doi: 10.4049/jimmunol.180.10.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spreu J, Kienle EC, Schrage B, Steinle A. CLEC2A: a novel, alternatively spliced and skin-associated member of the NKC-encoded AICL-CD69-LLT1 family. Immunogenetics. 2007;59:903–912. doi: 10.1007/s00251-007-0263-1. [DOI] [PubMed] [Google Scholar]
- 16.Boles KS, Barten R, Kumaresan PR, Trowsdale J, Mathew PA. Cloning of a new lectin-like receptor expressed on human NK cells. Immunogenetics. 1999;50:1–7. doi: 10.1007/s002510050679. [DOI] [PubMed] [Google Scholar]
- 17.Hamann J, Montgomery KT, Lau S, Kucherlapati R, van Lier RA. AICL: a new activation-induced antigen encoded by the human NK gene complex. Immunogenetics. 1997;45:295–300. doi: 10.1007/s002510050208. [DOI] [PubMed] [Google Scholar]
- 18.Huarte E, et al. PILAR is a novel modulator of human T-cell expansion. Blood. 2008;112:1259–1268. doi: 10.1182/blood-2007-12-130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathew PA, et al. The LLT1 receptor induces IFN-gamma production by human natural killer cells. Mol Immunol. 2004;40:1157–1163. doi: 10.1016/j.molimm.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 20.Llera AS, Viedma F, Sánchez-Madrid F, Tormo J. Crystal structure of the C-type lectin-like domain from the human hematopoietic cell receptor CD69. J Biol Chem. 2001;276:7312–7319. doi: 10.1074/jbc.M008573200. [DOI] [PubMed] [Google Scholar]
- 21.Fuller GL, et al. The C-type lectin receptors CLEC-2 and Dectin-1, but not DC-SIGN, signal via a novel YXXL-dependent signaling cascade. J Biol Chem. 2007;282:12397–12409. doi: 10.1074/jbc.M609558200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robinson MJ, Sancho D, Slack EC, LeibundGut-Landmann S, Reis e Sousa C. Myeloid C-type lectins in innate immunity. Nat Immunol. 2006;7:1258–1265. doi: 10.1038/ni1417. [DOI] [PubMed] [Google Scholar]
- 23.Lanier LL, Chang C, Phillips JH. Human NKR-P1A. A disulfide-linked homodimer of the C-type lectin superfamily expressed by a subset of NK and T lymphocytes. J Immunol. 1994;153:2417–2428. [PubMed] [Google Scholar]
- 24.Vitale M, et al. Identification of NKp80, a novel triggering molecule expressed by human NK cells. Eur J Immunol. 2001;31:233–242. doi: 10.1002/1521-4141(200101)31:1<233::AID-IMMU233>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 25.Duhen T, Geiger R, Jarrossay D, Lanzavecchia A, Sallusto F. Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat Immunol. 2009;10:857–863. doi: 10.1038/ni.1767. [DOI] [PubMed] [Google Scholar]
- 26.Trifari S, Kaplan CD, Tran EH, Crellin NK, Spits H. Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1 and T(H)2 cells. Nat Immunol. 2009;10:864–871. doi: 10.1038/ni.1770. [DOI] [PubMed] [Google Scholar]
- 27.Ebert LM, Meuter S, Moser B. Homing and function of human skin gammadelta T cells and NK cells: relevance for tumor surveillance. J Immunol. 2006;176:4331–4336. doi: 10.4049/jimmunol.176.7.4331. [DOI] [PubMed] [Google Scholar]
- 28.Welte SA, et al. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur J Immunol. 2003;33:194–203. doi: 10.1002/immu.200390022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.