Abstract
Germline mutations in the tumor suppressor gene PTEN (phosphatase and tensin homology deleted on chromosome 10) cause Cowden and Bannayan–Riley–Ruvalcaba (BRR) syndromes, two dominantly inherited disorders characterized by mental retardation, multiple hamartomas, and variable cancer risk. Here, we modeled three sentinel mutant alleles of PTEN identified in patients with Cowden syndrome and show that the nonsense Pten∆4–5 and missense PtenC124R and PtenG129E alleles lacking lipid phosphatase activity cause similar developmental abnormalities but distinct tumor spectra with varying severity and age of onset. Allele-specific differences may be accounted for by loss of function for Pten∆4–5, hypomorphic function for PtenC124R, and gain of function for PtenG129E. These data demonstrate that the variable tumor phenotypes observed in patients with Cowden and BRR syndromes can be attributed to specific mutations in PTEN that alter protein function through distinct mechanisms.
Keywords: cancer genetics, Cowden syndrome
The tumor suppressor gene PTEN (phosphatase and tensin homology deleted on chromosome ten) encodes a product with both protein and lipid phosphatase activity (1–3). The lipid phosphatase activity negatively regulates phosphoinositide-3 kinase (PI-3K) and the downstream Akt and mammalian target of rapamycin pathway components (4). Consistent with a requirement for lipid phosphatase activity in tumor suppression, the mutant PTENG129E allele, originally identified in patients with Cowden syndrome, selectively lacks lipid phosphatase activity (5). However, PTEN also has protein phosphatase activity, and although controversial, there is a body of evidence suggesting that PTEN's protein phosphatase activity may also contribute to tumor suppression (6). The Cowden syndrome PTENC124R allele, which encodes a protein product lacking lipid and protein phosphatase activity (7), provides genetic support for an involvement of dual PTEN phosphatase activities in tumor suppression. In addition, recent data suggest a role for nuclear PTEN in genomic stability that may be independent of its lipid phosphatase and PI-3K signaling activities (8). Despite intensive efforts in the past decade to understand the biochemical functions of PTEN, it remains unclear which of its many functions endow this gene with tumor suppressor status and whether the different functions contribute to tumor suppression selectively in an organ-specific and/or signaling pathway-specific manner.
Individuals with Cowden syndrome show a wide variation in disease manifestation, including cancer predisposition (9), that is thought to be driven by a combination of genotype as well as polymorphic heterogeneity in the population (10). The reason why polymorphic heterogeneity has been invoked is because identical mutations result in disparate phenotypes, ranging from mild developmental disorders in Cowden syndrome to very severe disorders in Bannayan–Riley–Ruvalcaba (BRR) syndrome (9). Here, we used homologous recombination in mice to model three different mutant alleles of PTEN originally identified in Cowden syndrome. We found that each mutant allele displayed distinct tumor phenotypes in organs targeted by Cowden syndrome. These functional differences are not manifested during embryonic development and do not correspond strictly to the level of Akt activation. Rather, the variable phenotypes can be attributed to functions beyond PI3K-Akt activation that include gain of function for the PtenG129E allele and loss of function for the Pten∆4–5 and PtenC124R alleles. Together, these results demonstrate that specific germline mutations have a strong influence in the variable predisposition to cancer of patients who have Cowden syndrome.
Results
Abnormal Embryonic Development in Pten∆4–5, PtenG129E, and PtenC124R Knockin Mice.
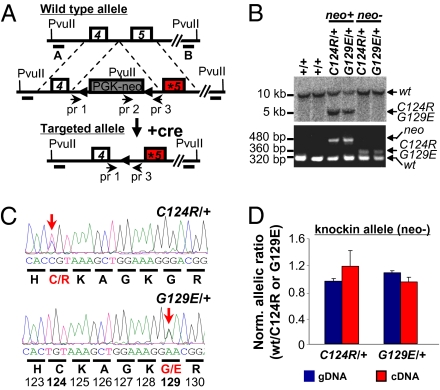
To explore the physiological consequence of inactivating distinct arms of PTEN's tumor suppressor arsenal, we generated, characterized, and compared mouse models that have a frameshift (Pten∆4–5) (11) leading to the premature stop and missense mutations in Pten (PtenG129E or PtenC124R) that can be found in patients with Cowden syndrome (12) (Fig.1A). Targeting of the three Pten mutant alleles was verified by Southern blot analysis, PCR, and direct sequencing of genomic DNA (11) (Fig.1 B and C and Fig. S1A, Left). Allelic expression imbalance analysis showed that the two missense mutant alleles lacking the neomycin (neo) cassette (neo−) were expressed at a similar level as the WT allele (Fig.1D), whereas the same mutant alleles containing the neo cassette (neo+) were expressed, as expected, at lower levels (Fig.S1A, Right).
Fig. 1.
Generation, targeting, and verification of point mutations of Pten alleles. (A) Endogenous WT Pten allele (Top), the two targeted missense mutations in exon 5 (*) of Pten allele containing the selectable phosphoglycerate kinase promoter (PGK)-neo cassette (flanked by LoxP sites, triangles; Middle), and the two targeted mutant Pten knockin alleles (PtenC124R and PtenG129E) lacking the PGK-neo cassette (after mating with EIIA-cre-expressing mice; Bottom). A and B, DNA probes used for Southern blot analysis; pr, primers used for PCR genotyping. (B) Southern blot analysis (Upper) and genotyping PCR (Lower) of tail DNA with the indicated genotypes. Genomic DNA was digested with PvuII and probed with probe A, and expected band sizes are indicated for each allele. PCR amplification using primer pairs 1–3 and 2–3 (primer information provided in Table S3) yielded specific fragment size for different alleles as indicated. (C) Sequence analysis of tail DNA isolated from PtenC124R/+ and PtenG129E/+ mice. Chromatograms demonstrating the successful targeting of the Pten locus and translated amino acids are shown below the codons. Red letters and bold numbers denote the two targeted amino acids (C124R and G129E). Arrows point to targeted nucleotides. (D) Allelic expression imbalance analysis of allele-specific expression. The graph shows the proportion of mRNA expressed from the WT allele over the indicated mutant allele in lungs of PtenC124R/+ and PtenG129E/+ mice. neo−, mice lacking the PGK-neo cassette. Genomic DNA (gDNA) was used as an internal control.
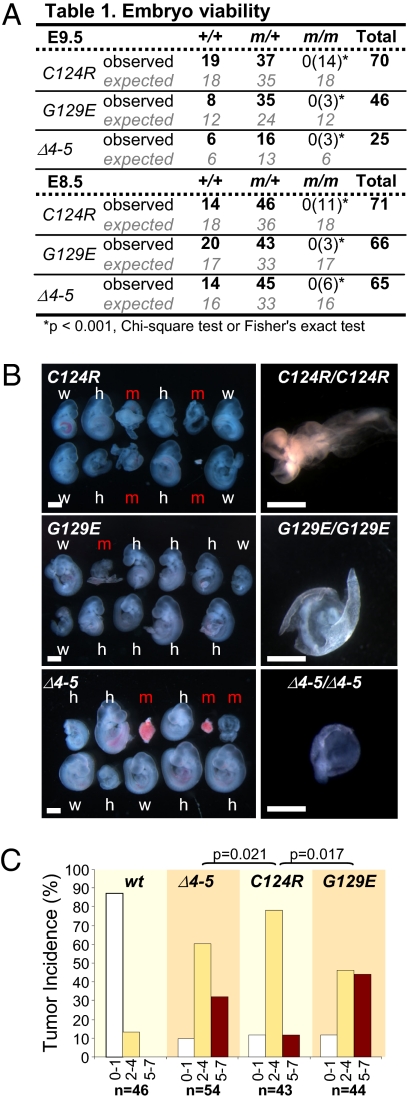
We first evaluated the effect of Pten∆4–5, PtenG129E, and PtenC124R on embryonic development by examining offspring derived from intercrosses between heterozygous knockin mice (Fig. 2A). The intercrosses yielded no live homozygous progeny at birth for any of the three mutant alleles; however, mutant embryos were recovered at embryo day (E) 8.5 and E9.5, albeit at a lower than expected frequency. Development of the few mutant embryos that were obtained was severely compromised, with defects in anterior-posterior patterning, failure of axial rotation, and absence of overt tissue differentiation leading to resorption by E9.5 (Fig. 2 A and B and Fig. S1B). Heterozygous Pten∆4–5/+, PtenG129E/+, and PtenC124R/+ embryos were morphologically indistinguishable from WT littermates (Fig. 2 A and B and Fig. S1C). Thus, overall, there were no obvious differences among homozygous knockin embryos of the three Pten mutant genotypes.
Fig. 2.
Pten∆4–5, PtenC124R, and PtenG129E mutations cause early embryonic lethality and exhibit allele-specific tumor syndromes. (A) Offspring from Pten (PtenC124R, PtenG129E, or Pten∆4–5) heterozygous intercrosses were examined. The number of observed and expected E8.5 and E9.5 embryos is indicated. Fisher's exact or χ2 tests were used to compare differences between observed and expected homozygous embryos. The number of dead embryos is shown in parentheses. m, mutant allele (PtenC124R, PtenG129E, or Pten∆4–5). (B) (Left) Stereomicroscopic images of E9.5 WT (w), heterozygous (h), and homozygous (m) Pten-mutant littermate embryos. (Right) Higher magnification images of severely affected homozygous mutant embryos. (Scale bar: 1 mm.) (C) Organ distribution of tumor lesions in animals harboring the indicated Pten alleles. To simplify the graphical representation of data, mice were grouped based on the number of organs (0–1, white bars; 2–4, yellow bars; 5–7, red bars) afflicted with tumors. All uncategorized data were analyzed by Poisson regression methods, and significant differences are shown; all Pten mutant-WT allele comparisons were significant (P < 0.0001; not indicated).
Organ-Selective Cancer Predisposition in Pten∆4–5/+, PtenG129E/+, and PtenC124R/+ Mice.
The tumor spectrum in animals harboring one copy of each of the three mutant Pten alleles was determined by analyzing groups of >40 adult male and female mice. Similar to previous studies that analyzed heterozygous mice harboring a Pten knockout allele (13–17), mice carrying one copy of each of the three Pten mutant alleles exhibited neoplasms in multiple organs that are characteristically involved in patients who have Cowden syndrome, confirming these are causal mutations that drive this syndrome. Interestingly, pathological examination of 9-month-old mice revealed differences in the frequency and severity of the individual proliferative lesions found in the three genetic groups (Table S1). The intergroup differences were amplified when the propensity of an individual animal to develop tumors at two or more organ sites was considered (Fig. 2C). A significant bias for lesions also occurred in specific organ pairs for each of the three genetic cohorts. Mammary gland-lymph node (P < 0.001), uterus-lymph node (P = 0.032), thyroid-lymph node (P = 0.003), and prostate-stomach (P = 0.003) lesions were frequently present in Pten∆4–5/+ mice; mammary gland-lymph node (P = 0.050) lesions were present in PtenG129E/+ mice; and mammary gland-intestine (P = 0.007), mammary gland-adrenal (P = 0.049), and prostate-stomach (P = 0.002) lesions were present in PtenC124R/+ mice (Table S2). Consistent with observations made in human patients with Cowden syndrome, there was a gender bias in the organ-specific manifestation of lesions, with lymph node lesions more frequently found in female mice (for Pten∆4–5, P < 0.001) and adrenal and stomach lesions more commonly found in male mice (for Pten∆4–5, P = 0.008 and P = 0.012, respectively; for PtenC124R, P = 0.034) (Fig. S2). These findings challenge the current view of polymorphic heterogeneity in the population as the principal reason for the variable penetrance observed in patients with Cowden syndrome. We suggest that this variability may instead be attributed, at least in part, to allele-specific effects of PTEN mutations.
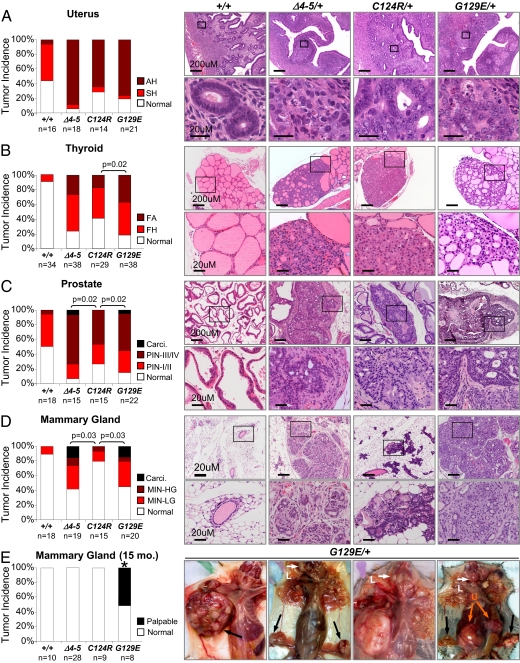
Based on these initial observations, we performed a thorough histopathological analysis of neoplastic lesions in organ sites frequently involved in Cowden syndrome, including the uterus, thyroid, mammary gland (2, 18–20) and the prostate (21). Detailed criteria for grading lesions are presented in Fig. S3. In the uterus, proliferative lesions of the endometrium (simple hyperplasia and atypical hyperplasia) were encountered in Pten∆4–5/+, PtenC124R/+, and PtenG129E/+ female mice with equal frequency (Fig. 3A), similar to what has been reported in patients with Cowden syndrome. Advanced thyroid follicular lesions, however, were more frequently observed in PtenG129E/+ mice compared with Pten∆4–5/+ or PtenC124R/+ mice (Fig. 3B and Fig. S4A). In the prostate, both Pten∆4–5/+ and PtenG129E/+ mice were more likely to have lesions of higher histological grade, which included prostatic adenocarcinoma, than PtenC124R/+ mice (Fig. 3C). In the mammary gland, advanced lesions such as mammary intraepithelial neoplasia (MIN), invasive carcinoma, and adenosquamous carcinoma were observed in both Pten∆4–5/+ and PtenG129E/+ female mice but rarely in PtenC124R/+ female mice (Fig. 3D; P = 0.03). Strikingly, only lesions in PtenG129E/+ female mice (n = 8) advanced to form large palpable tumors with unique adenosquamous morphology and a prominent stromal component (Fig. 3E and Fig. S4B), which are classic characteristics of breast carcinoma seen in individuals with Cowden syndrome (22). Despite small sample sizes in our database comprising >400 PTEN mutation-positive individuals, all three PTENG129E-affected women had breast cancer, whereas two PTENC124R-affected individuals only had fibrocystic disease of the breast. Moreover, we also observed high-grade MIN in PtenG129E/+ male mice but not in males of other genotypes (Fig. S4C), consistent with an increased incidence of breast cancer observed in male patients with Cowden syndrome with this particular PTEN mutation (5, 23). The development of more severe lesions in select organs of PtenG129E/+ mice when compared with Pten∆4–5/+ mice suggests that the PtenG129E allele, in addition to lacking lipid phosphatase activity, may have gained protumorigenic functions. Together, these findings demonstrate the critical role of specific PTEN mutant alleles in driving the broad clinical manifestations of Cowden syndrome that often present in an organ-selective fashion and with variable penetrance and expressivity.
Fig. 3.
Allele-specific and organ-specific tumor development in Pten∆4–5/+, PtenC124R/+, and PtenG129E/+ mice. Histopathological grades of lesions were compared between WT, Pten∆4–5/+, PtenC124R/+, and PtenG129E/+ 9-month-old mice in the uterus (A), thyroid (B), prostate (C), and mammary gland (D). Note the significant difference in the severity of lesions among the various animals with mutant Pten alleles. (Right) Histology with the highest grade lesions found in each genetic group. (Lower) Magnified view of the boxed region in the upper panels. Detailed histopathological criteria used to grade the lesions are included in Fig. S3. AH, atypical hyperplasia; SH, simple hyperplasia; PIN-I/II, prostatic intraepithelial neoplasia grade I or II; PIN-III/IV, prostatic intraepithelial neoplasia grade III or IV; FA, follicular adenoma; FH, follicular hyperplasia; Carci., carcinoma; MIN, mammary intraepithelial neoplasia; LG, low grade; HG, high grade; Normal, no gross or microscopic tumor. Comparisons between genetic groups in A–D were analyzed by χ2 or Fisher's exact tests and adjusted by Holm's method. (E) Large palpable tumor masses in 12–15-month-old PtenG129E/+ female mice. Black and orange arrows point to mammary gland and uterine (U) tumors, respectively. White arrows point to enlarged lymph nodes (L). Normal, no gross mammary gland tumors; Palpable, palpable mammary gland tumors. Comparisons between genetic groups in E were analyzed by a binomial exact test. All comparisons between PtenG129E/+ and other genetic groups were found to be significant (*P < 0.01).
Organ-Specific Pten Inactivation During Tumor Initiation and Progression.
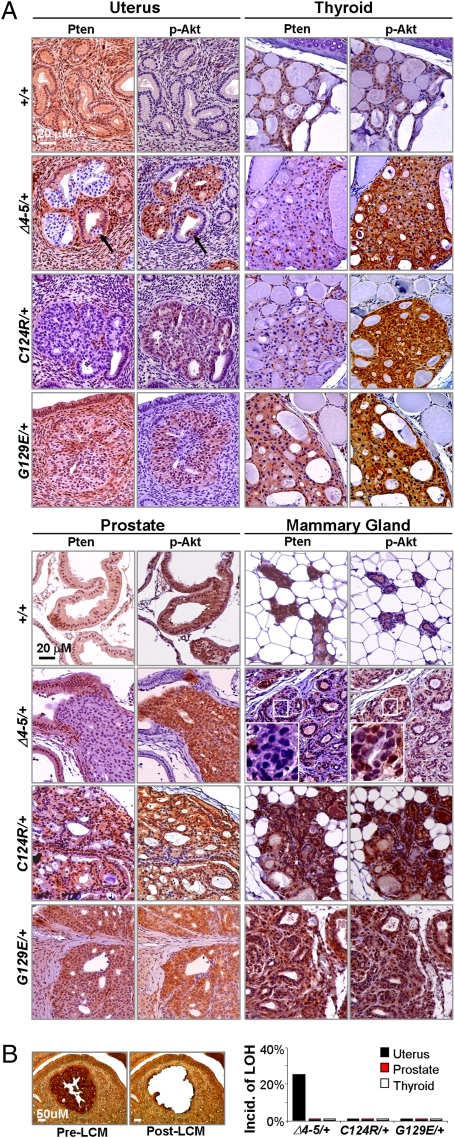
Molecular analysis of embryos and tumors with each of the three different Pten mutant genotypes revealed intriguing tissue-specific mechanisms of how Pten mutant alleles differentially contribute to tumor development and progression. As might have been expected, there was focal loss of WT Pten protein expression in Pten∆4–5/+ epithelial lesions of the endometrium, thyroid, and prostate (Fig. 4A). In the endometrial lesions analyzed, there was loss of heterozygosity (LOH) in 25% of cases (Fig. 4B). In the remaining endometrial lesions, as well as in the limited number of thyroid and prostate Pten∆v4–5/+ tumors analyzed, LOH was not observed (Fig. 4B), consistent with the low percentage of LOH (11%) observed in Cowden syndrome-related nonmalignant neoplasias (24). These results suggest that the frequency of LOH in neoplastic lesions of all three mutant heterozygotes was low and that loss of expression from the WT Pten allele in the majority of Pten∆4–5/+ tumors was mediated by a posttranscriptional mechanism.
Fig. 4.
Expression of Pten and p-Akt in proliferative lesions of various organs. (A) Tissue sections from the uterus and thyroid (Upper) or prostate and mammary gland (Lower) of mice with the indicated genotypes were stained with Pten- and p-Akt-specific antibodies. Complete loss of Pten occurred in Pten∆4–5/+ lesions of the uterus, thyroid, and prostate, whereas decreased levels of Pten were observed in PtenC124R/+ lesions of the uterus and thyroid. Pten protein expression was lost in a mosaic fashion in advanced lesions of Pten∆4–5/+ mice but persisted in PtenC124R/+ MIN and PtenG129E/+ mammary gland carcinomas. (Right) Magnified images of the boxed region. (B) LOH analysis of WT Pten allele detected by PCR from laser capture microdissected (LCM) lesions of the uterus, prostate, and thyroid of 9-month-old heterozygotes. (Left) Representative images showing pre- and post-LCM tissue of “quick” p-Akt immunostained sections. (Right) Incidence of LOH in lesions from the indicated organs. For LOH in the uterus, 25 Pten∆4–5/+, 5 PtenC124R/+, and 5 PtenG129E/+ mice were examined. LOH in other organs was sampled from 5 mice per genotype group.
Interestingly, Pten protein in lesions of the uterus, thyroid, and prostate was frequently absent in hyperplastic clonal areas containing as few as three to five cells (Fig. 4A and Fig. S5 A–C), suggesting that loss of Pten expression in these organs was an early or initiating event during tumor development. In mammary preneoplastic lesions, however, Pten∆4–5/+ female mice retained normal amounts of Pten protein, and its loss, which was manifested in a mosaic pattern, was only detected in advanced adenocarcinomas (Fig. 4A and Fig. S5D). When we examined epithelial cells from three adenocarcinomas in Pten∆4–5/+ and PtenG129E/+ female mice, no LOH was detected in any of these samples. From these data, we conclude that the level of WT Pten protein that accumulates in Pten∆4–5/+ mammary glands is haploinsufficent to suppress tumor initiation, and that loss of expression from the WT allele is a late event that may be required for malignant progression of mammary cancers. Together, these observations support a tissue-selective requirement for loss of Pten protein expression.
Allele-Specific Mechanism of Pten-Mediated Tumor Suppression.
Inactivation of tumor suppression by the two missense mutations (PtenG129E and PtenC124R) is thought to result from the ablation of enzymatic phosphatase activity (1, 25, 26). It was therefore surprising to find that lesions in the uterus, thyroid, and prostate of PtenC124R/+ mice had little (if any) detectable Pten protein (Fig. 4A). This was specific to this cohort of mice, because Pten protein in the corresponding lesions of PtenG129E/+ mice was clearly present. Interestingly, the levels of Pten protein in preneoplastic mammary gland lesions in PtenC124R/+ mice were uniformly high, suggesting that, as in Pten∆4–5/+ mice, tumor initiation in mammary glands did not require LOH (note that carcinoma was never detected in this genetic group) (Fig. 4A). These results suggest that the two missense mutations (PtenG129E and PtenC124R) contribute to tumorigenesis differently, with the PtenC124R allele producing a protein product that is particularly labile.
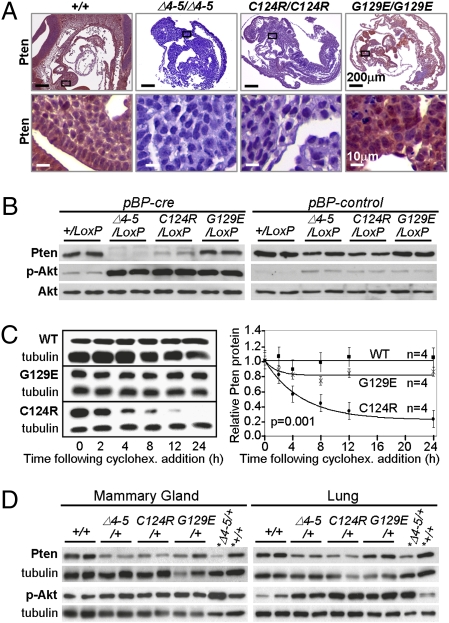
The unexpectedly low levels of PtenC124R protein in tumors prompted us to reexamine its expression in normal tissues. We used immunohistochemistry (IHC) to compare Pten protein levels in E9.5 Pten+/+, Pten∆4–5/∆4–5, PtenG129E/G129E, and PtenC124R/C124R embryos. Remarkably, PtenC124R protein was almost undetectable in most tissues of homozygous embryos (Fig. 5A), even though its mRNA levels were not altered. In contrast, there were normal amounts of Pten protein expressed from the PtenG129E allele. To determine whether the absence of PtenC124R protein could be attributable to reduced protein stability, we generated and compared mouse embryonic fibroblasts (MEFs) containing a conditional PtenLoxP null allele (with LoxP sites flanking exons 4–5) and either a WT allele or each of the three mutant knockin alleles (Pten+/LoxP, Pten∆4–5/LoxP, PtenG129E/LoxP, and PtenC124R/LoxP; Fig. S6A). Retroviral transduction of cre recombinase (cre) in the MEF cultures resulted in efficient deletion of PtenLoxP (Fig. S6B), generating cells that express Pten solely from the remaining WT allele or from each of the three knockin alleles (Fig. S6B). Western blot analysis confirmed normal steady-state levels of Pten protein in cre-Pten+/LoxP and cre-PtenG129E/LoxP MEFs (compared with control-Pten∆4–5/LoxP MEFs), undetectable Pten in cre-Pten∆4–5/LoxP MEFs, and decreased Pten in cre-PtenC124R/LoxP MEFs (Fig. 5B). Quantitative RT-PCR analysis showed equivalent amounts of Pten mRNA in MEFs of all genotypes (Fig. S6C). Pulse-chase experiments using cycloheximide demonstrated a significant reduction in the half-life of PtenC124R protein when compared with either WT or PtenG129E protein (P = 0.001; ∼3.7 vs. >24 h; Fig. 5C). From these results, we suggest that one mechanism by which the PtenC124R allele promotes Cowden syndrome and sporadic cancers is through the inherent instability of its protein product.
Fig. 5.
Decreased PtenC124R protein stability. (A) Pten IHC staining of homozygous embryos (E9.5) with the indicated genotypes. (Lower) High magnification of boxed areas in the upper panels. (B) Western blots of control- (pBP-control) or cre-retrovirus- (pBP-cre) infected MEFs of the indicated genotypes were probed with antibodies specific for Pten, p-Akt S473, and total Akt. Complete cre-mediated deletion of exons 4 and 5 was confirmed by PCR (Fig. S6B). (C) (Left) Western blots of cycloheximide- (cyclohex.) treated MEFs treated as in B and expressing WT Pten (WT), PtenC124R (C124R), and PtenG129E (G129E). (Right) Protein half-lives of WT Pten (WT), PtenC124R (C124R), and PtenG129E (G129E) were calculated as described in SI Text and plotted. n, number of independent experiments. ANOVA with a repeated-measures model was used to evaluate differences across the various time points and between genotypes. (D) Western blots of lysates derived from mammary glands and lungs of two 9-month-old female mice with the indicated genotypes probed with Pten- and p-Akt (S473)-specific antibodies as indicated. *Lysates derived from MEFs with the indicated genotypes were included in these blots. Tubulin protein levels were monitored as a loading control.
The activation of Akt, through its PI-3K-mediated phosphorylation at either residue 308 or 473, is thought to be attributable to the inactivation of Pten (1). Indeed, the level of phosphorylated Akt (p-Akt) was increased in cre-Pten∆4–5/LoxP MEFs relative to either cre-Pten+/LoxP or control-Pten∆4–5/LoxP MEFs. Interestingly, p-Akt levels were similarly increased in MEFs expressing PtenG129E or PtenC124R protein (Fig. 5B). Moreover, IHC revealed that E9.5 Pten∆4–5/∆4–5, PtenG129E/G129E, and PtenC124R/C124R embryos had equivalent levels of p-Akt, indicating that signaling downstream of PI-3K in embryos of all three mutant genotypes was similarly engaged (Fig. S1B). Because Pten∆4–5/+, PtenG129E/+, and PtenC124R/+ mice had different tumor and gender biases characteristic of Cowden syndrome with distinct pathological findings and frequencies, we investigated the extent of Akt phosphorylation in tumor samples. Again, IHC of all tumor types and Western blot analysis of lysates derived from mammary glands, lungs, and MEFs failed to reveal any difference in the amounts of p-Akt between the three genotypes (Figs. 4A and 5 B and D). Therefore, the severity of cancer phenotypes attributed to PtenC124R, PtenG129E, and Pten∆4–5 alleles did not correspond to Akt activation (8, 27, 28). These data indicate that unique functions, beyond Akt activation, distinguish the three mutant alleles from one another in their ability to instigate cancer development and progression.
Discussion
Germline PTEN mutations cause a broad array of seemingly unrelated syndromes, which are collectively called the PTEN hamartoma tumor syndrome and include BRR and Cowden syndromes (9). Puzzlingly, certain identical mutations, such as R130× and R233×, can result in either Cowden or BRR syndrome (29), suggesting that polymorphic heterogeneity strongly influences disease outcome. Even individuals with Cowden syndrome have a wide variation in disease manifestation and in the penetrance of phenotypes, including cancer predisposition (2, 12, 30). Whether this variability is attributable to gene mutations that differentially alter the function of Pten protein directly or to polymorphic variation and/or mutations in genetic modifiers of PTEN has been a subject of controversy (16).
Mouse modeling of three important mutant alleles of PTEN identified in patients with Cowden syndrome demonstrates that nonsense (Pten∆4–5) and missense (PtenG129E and PtenC124R) mutations in Pten display distinct tumor phenotypes in organs targeted by Cowden syndrome. These functional differences are not manifested during embryonic development and do not appear to depend on differential PI-3K-Akt signaling. We have not formally ruled out, however, the possibility that the timing of Akt activation during tumor initiation/progression may differ between the three Pten knockin genetic groups. Nonetheless, it is interesting that phenotypic differences in human patients with Cowden syndrome also do not strictly correspond to the level of Akt activation. Rather, the striking differences in tumor spectrum and severity, with phenotypes in PtenG129E/+ > Pten∆4–5/+ >> PtenC124R/+ mice, may be partially accounted for by a gain of function in PtenG129E/+ individuals and by reduced protein stability in PtenC124R/+ individuals. The gain-of-function phenotypes manifested in PtenG129E/+ mice may be attributable to the remaining protein phosphatase activity of mutant protein or to interference of function by mutant protein in protein complexes. Although further experimentation will be required to define fully the mechanisms underlying our observations, the in vivo models we describe provide the physiological and genetic foundation necessary to unravel this puzzle. In summary, the results demonstrate that specific germline mutations disrupt PTEN function by distinct mechanisms that may explain the organ-selective variability associated with cancer predisposition in Cowden syndrome. These findings raise the possibility that specific somatic alterations, whether genetic or epigenetic, in PTEN will also have a differential impact on sporadic cancers that develop in patients who do not have Cowden syndrome. We believe that the genotype-phenotype relations revealed here may be used to guide more effective and personalized targeted therapies for cancer patients with PTEN mutations.
Methods
PtenC124R/+ and PtenG129E/+ knockin mice were generated using a standard homologous recombination approach. Isolation of primary MEFs from E13.5 embryos, retroviral infections, and cell culture conditions were done using standard methods. PtenLoxP deletion by cre was confirmed by genotyping of MEF DNA and Western blot analysis using PTEN antibody. Statistics analyses were performed for all the data. Detailed methods can be found in SI Text.
Supplementary Material
Acknowledgments
The authors thank John C. Thompson for excellent technical assistance, the Ohio State University Comprehensive Cancer Center Nucleic Acids and Transgenics Shared Facilities for technical assistance, and Dr. Philip Popovich and Wenmin Lai for guidance in the laser capture microdissection technique. This work was funded by National Institutes of Health Grants R01CA85619, R01HD47470, and P01CA097189 (to G.L.) and R01CA053271 and P01CA097189 (to M.C.O.). H.W. is supported by an American Cancer Society Postdoctoral Fellowship. M.C.O received awards from both the Susan Komen Foundation and the Evelyn Simmers Foundation, and G.L. received an award from the Susan Komen Foundation and US Department of Defense. G.L. is the recipient of the Pew Charitable Trusts Scholar Award and the Leukemia and Lymphoma Society Scholar Award.
Footnotes
*This Direct Submission article had a prearranged editor.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912524107/DCSupplemental.
References
- 1.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 2.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 3.Myers MP, et al. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manning BD, Cantley LC. AKT/PKB signaling: Navigating downstream. Cell. 2007;129:1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liaw D, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 6.Leslie NR, Maccario H, Spinelli L, Davidson L. The significance of PTEN's protein phosphatase activity. Adv Enzyme Regul. 2009;49:190–196. doi: 10.1016/j.advenzreg.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 7.Nelen MR, et al. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 8.Shen WH, et al. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell. 2007;128:157–170. doi: 10.1016/j.cell.2006.11.042. [DOI] [PubMed] [Google Scholar]
- 9.Gustafson S, Zbuk KM, Scacheri C, Eng C. Cowden syndrome. Semin Oncol. 2007;34:428–434. doi: 10.1053/j.seminoncol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Pezzolesi MG, Platzer P, Waite KA, Eng C. Differential expression of PTEN-targeting microRNAs miR-19a and miR-21 in Cowden syndrome. Am J Hum Genet. 2008;82:1141–1149. doi: 10.1016/j.ajhg.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trimboli AJ, et al. Pten in stromal fibroblasts suppresses mammary epithelial tumours. Nature. 2009;461:1084–1091. doi: 10.1038/nature08486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marsh DJ, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-Ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 13.Suzuki A, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 14.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 15.Podsypanina K, et al. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman D, et al. Genetic background controls tumor development in PTEN-deficient mice. Cancer Res. 2006;66:6492–6496. doi: 10.1158/0008-5472.CAN-05-4143. [DOI] [PubMed] [Google Scholar]
- 17.Stambolic V, et al. High incidence of breast and endometrial neoplasia resembling human Cowden syndrome in pten+/− mice. Cancer Res. 2000;60:3605–3611. [PubMed] [Google Scholar]
- 18.Risinger JI, Hayes AK, Berchuck A, Barrett JC. PTEN/MMAC1 mutations in endometrial cancers. Cancer Res. 1997;57:4736–4738. [PubMed] [Google Scholar]
- 19.Dahia PL, et al. Somatic deletions and mutations in the Cowden disease gene, PTEN, in sporadic thyroid tumors. Cancer Res. 1997;57:4710–4713. [PubMed] [Google Scholar]
- 20.Steck PA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 21.Cairns P, et al. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 22.Schrager CA, Schneider D, Gruener AC, Tsou HC, Peacocke M. Clinical and pathological features of breast disease in Cowden's syndrome: An underrecognized syndrome with an increased risk of breast cancer. Hum Pathol. 1998;29:47–53. doi: 10.1016/s0046-8177(98)90389-6. [DOI] [PubMed] [Google Scholar]
- 23.Fackenthal JD, et al. Male breast cancer in Cowden syndrome patients with germline PTEN mutations. J Med Genet. 2001;38:159–164. doi: 10.1136/jmg.38.3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsh DJ, et al. Germline PTEN mutations in Cowden syndrome-like families. J Med Genet. 1998;35:881–885. doi: 10.1136/jmg.35.11.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers MP, et al. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramaswamy S, et al. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vivanco I, et al. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell. 2007;11:555–569. doi: 10.1016/j.ccr.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 28.Chang CJ, et al. PTEN nuclear localization is regulated by oxidative stress and mediates p53-dependent tumor suppression. Mol Cell Biol. 2008;28:3281–3289. doi: 10.1128/MCB.00310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eng C. PTEN: One gene, many syndromes. Hum Mutat. 2003;22:183–198. doi: 10.1002/humu.10257. [DOI] [PubMed] [Google Scholar]
- 30.Eng C, Peacocke M. PTEN and inherited hamartoma-cancer syndromes. Nat Genet. 1998;19:223. doi: 10.1038/897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.