Abstract
p73, the structural and functional homologue of p53, exists as two major forms: the transactivation-proficient, proapoptotic TAp73 or the transactivation-deficient, antiapoptotic DNp73. Expectedly, expression of both these major forms has to be coordinated precisely to achieve the desired cellular outcome. Genotoxic insults resulting in cell death lead to the stabilization of TAp73, mainly through posttranslational modifications, and the concomitant degradation of DNp73, through poorly understood mechanisms. We have therefore investigated the possible mechanisms of stress-induced DNp73 degradation and show here that c-Jun, the AP-1 family member activated by stress signals and involved in stabilizing TAp73, promotes DNp73 degradation. Genotoxic stress-mediated DNp73 degradation was found to occur in a c-Jun–dependent manner through a ubiquitin-independent but proteasome-dependent mechanism. Absence or down-regulation of c-Jun expression abrogated the reduction of DNp73 levels upon stress insults, whereas overexpression of c-Jun led to its degradation. c-Jun controlled DNp73 degradation through the nonclassical, polyamine-induced antizyme (Az) pathway by regulating the latter’s processing during stress response. Consistently, expression of c-Jun or Az, or addition of polyamines, promoted DNp73 degradation, whereas silencing Az expression or inhibiting Az activity in cells exposed to stress reduced c-Jun–dependent DNp73 degradation. Moreover, Az was able to bind to DNp73. These data together demonstrate the existence of a c-Jun–dependent mechanism regulating the abundance of the antiapoptotic DNp73 in response to genotoxic stress.
Keywords: DNp73, proteasome-dependent, ubiquitination, polyamines
Full-length TAp73 and the amino-terminally truncated Delta-N(DN)p73 are the two major forms of p73 that have been described (1). Expectedly, TAp73 possesses similar tumor-suppressive properties as p53 (2), whereas the DNp73 form lacking the transactivation domain acts as a dominant-negative inhibitor of both TAp73 and p53, and hence, can exhibit antiapoptotic properties (3, 4). Several studies have highlighted an obligatory role of endogenous TAp73 in regulating apoptosis in response to DNA damage (2, 5, 6). Conversely, DNp73 expression is elevated in many human cancers, and its overexpression has been shown to inhibit apoptosis (4, 7, 8). Consistently, exposure to multiple genotoxic signals leads to elevation of TAp73 and concomitantly to the reduction of DNp73 levels, therefore allowing apoptosis to ensue (5, 6, 9). Thus, coordinated regulation of the abundance of these two proteins appears to be critical in influencing the outcome of cellular response.
Posttranslational modifications regulate the levels of TAp73 and DNp73 through several factors, including Yes-associated protein (YAP), Itch, c-Abl, p38, promyelocytic leukemia protein (PML), etc., which have been shown to stabilize or destabilize both TAp73 and DNp73, due to their high homology (10). For example, c-Abl has been shown to phosphorylate both proteins, leading to their stabilization (11, 12). Conversely, factors such as the E3-ligase Itch were shown to equally destabilize TAp73 and DNp73 (13). However, although Itch was able to degrade both forms of p73 under normal conditions via the ubiquitin-proteasome pathway, reduction of its abundance upon DNA damage did not lead to the elevation of DNp73, in contrast to TAp73 (13), indicating that the levels of DNp73 and TAp73 are regulated by different mechanisms upon stress stimulation. These findings in turn suggest that other hitherto unidentified stress-regulated factors may be involved in keeping DNp73 levels low to allow for an effective apoptotic program.
Multiple mechanisms including ubiquitination, sumoylation, and neddylation have been implicated in regulating p73 degradation, primarily using TAp73 as a substrate (10). Besides, TAp73 and DNp73 have also been shown to be regulated by nonclassical pathways such as calpain cleavage (14), and by the NQO1 (15), UFD2a (16), and cyclin G–dependent (17), proteasome-mediated pathways. It is noteworthy that NQO1 and cyclin G–dependent p73 stability regulation is independent of the classical ubiquitin-proteasome–dependent pathway. These data therefore suggest that p73 may be regulated through multiple pathways.
Antizyme (Az) is an evolutionarily conserved protein that regulates cellular metabolism through its involvement in the degradation of substrates in a ubiquitin-independent manner (18). Az expression is regulated in a unique fashion during translation of the Az mRNA by polyamines, which induce a programmed +1 frameshift during translation, resulting in the expression of the functional full-length Az protein (19). The processed Az protein binds to and accelerates the degradation of ornithine decarboxylase (ODC), the rate-limiting enzyme in the polyamine biosynthesis pathway (20), and other factors such as Smad1, cyclinD1, and Aurora-A proteins (21–23), via the proteasome, and has also been shown to regulate proliferation and cell death (24).
We have previously shown that c-Jun, a member of the AP-1 family of transcription factors, is able to stabilize TAp73 but not DNp73 (25). c-Jun is a critical regulator of both cellular survival and death, with its levels being elevated upon exposure to a multitude of stress signals as well as growth factors (26). Because exposure to genotoxic stress signals lead to the degradation of DNp73, we investigated if c-Jun could negatively regulate DNp73 stability. The results presented here demonstrate that c-Jun is a crucial mediator of stress-induced DNp73 degradation, through the regulation of Az, which targets DNp73 for ubiquitin-independent degradation via the proteasome.
Results
Stress-Induced DNp73 Degradation Occurs via the Proteasome but Is Independent of Ubiquitination.
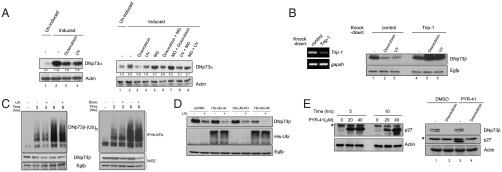
Reduction of DNp73 levels upon exposure to multiple stress signals was first confirmed using the p53 null SAOS-2 cell line in which DNp73α was inducibly expressed in a doxycycline-dependent manner (Fig. 1A, Left) (13) because endogenous DNp73 is almost undetectable by available antibodies. Expectedly, UV irradiation or treatment with the anticancer cytotoxic drug doxorubicin resulted in a reduction of DNp73α steady-state levels (Fig. 1A, Left, compare lanes 3 and 4 to lane 2, quantification shown below the blot). Treatment with the proteasome inhibitor MG132, either 3 h prior to (lanes 7 and 9, Fig. 1A, Right) or 3 h after (lanes 6 and 8, Fig. 1A, Right) UV and doxorubicin treatment decreased the reduction of DNp73 levels, suggesting that the degradation occurs via the proteasome. To further confirm the role of proteasome, we silenced the expression of Trip1/S8/hSug1, a critical 19S proteasomal component (27) (Fig. 1B, Left). This specifically abrogated the reduction of DNp73β after UV or doxorubicin treatment (Fig. 1B, Right, compare lanes 5 and 6 to 2 and 3). Moreover, half-lives of both DNp73α and DNp73β were reduced after doxorubicin treatment (Fig. S1), confirming that genotoxic stress-mediated DNp73 degradation occurs via the proteasome through altered protein turnover.
Fig. 1.
DNp73 is regulated by a proteasome-dependent but ubiquitin-independent mechanism upon exposure to genotoxic stress. (A) Extracts from SAOS-2-DNp73α cells induced for 24 h were treated with the indicated agents for 6 h (Left), or in the presence of MG132 (Right) and were used for immunodetection. Actin levels indicate loading control. Values below the blots indicate fold change in expression relative to lane 1 (= 1.0). (B) H1299 cells were transfected with indicated siRNA 24 h before transfection of DNp73β and Egfp (for transfection efficiency) and were treated 24 h later as described above (Right). Knock-down efficiency is shown by RT-PCR analysis (Left). (C) DNp73β and 6×His-ubiquitin plasmids were transfected into H1299 cells 24 h before UV (Left) or doxorubicin (Doxo.) (Right) treatment in the presence of MG132. Lysates purified through a Ni-NTA-agarose column (Top) and whole cell extracts (WCE) were also used for immunodetection (Bottom). (D) DNp73β and Egfp plasmids were cotransfected with various 6×His-ubiquitin forms into H1299 cells, which were UV irradiated 24 h later and analyzed. (E) H1299 cells were treated with PYR-41 (Left) or transfected with DNp73β 24 h before PYR-41 treatment for 4 h prior to doxorubicin treatment for a further 6 h (Right). Asterisk (*) indicates nonspecific band seen with the anti-p27 antibody. All experiments were performed at least three times independently.
We next evaluated if ubiquitination is involved in this degradation process. In vivo ubiquitination assays in the presence of MG132 revealed that UV or doxorubicin treatment did not lead to the addition of transfected ubiquitin molecules onto DNp73β up until 6 h, by which time the steady-state levels had reduced (Fig. 1C). Similar results were obtained in experiments without MG132 with transfected ubiquitin (Fig. S2A), or in the presence of MG132 with endogenous ubiquitin (Fig. S2B). Consistently, UV irradiation of cells cotransfected with DNp73β and ubiquitin mutants that prevent the formation of all types of polyubiquitin chains [all lysine mutants (KO)] or Lysine-48 type chains that are required to mediate ubiquitin-dependent proteasomal degradation of substrates (K48R/Ubi-48) (28) did not abrogate the reduction of DNp73 levels (Fig. 1D), which suggested that the degradation process was indeed independent of ubiquitination. The ability of Ubi-KO mutant to inhibit polyubiquitination of DNp73 was confirmed by in vivo ubiquitination assays (Fig. S3). Additionally, coexpression of DNp73 with wild-type ubiquitin or Ubi-KO did not affect the decrease in half-life of DNp73 upon doxorubicin treatment (Fig. S4). To further confirm this conclusion, we utilized the chemical inhibitor of the E1 ubiquitin-activating enzyme, 4[4-(5-nitro-furan-2-ylmethylene)-3,5-dioxo-pyrazolidin-1-yl]-benzoic acid ethyl ester (PYR-41) (29). PYR-41 treatment caused an increase in p27 levels, which is normally degraded by ubiquitination (30) (Fig. 1E, Left), confirming the utility of this reagent. Doxorubicin treatment of cells transfected with DNp73β and further treated with PYR-41 did not prevent the degradation of DNp73 (Fig. 1E, Right, compare lanes 2 and 4), indicating that ubiquitination is not essential for the reduction DNp73. The data therefore demonstrate that DNp73 levels are reduced upon stress treatment in a ubiquitin-independent but proteasome-dependent manner.
c-Jun Is Required for Reduction in DNp73 Levels After Exposure to Genotoxic Stress.
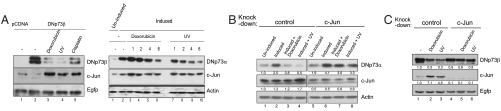
Because it appeared that DNp73 degradation may occur via a novel mechanism (independent of ubiquitination), we explored alternate possibilities. c-Jun, a key regulator of stress-mediated death, has been shown to regulate the abundance of TAp73 (25). We therefore investigated if c-Jun would have any role in the reduction of DNp73 levels. Treatment of cells with doxorubicin, UV, or cisplatin induced the expression of c-Jun, concurrent with reduction in DNp73 abundance (Fig. 2A, Left, compare lane 2 with lanes 3–5). Time-course analysis indicated that induction of c-Jun, which occurs at about an hour after exposure to doxorubicin or UV, preceded the reduction in DNp73 levels occurring around 4 and 2 h, respectively (Fig. 2A, Right, compare lane 2 with lanes 5 and 6 and 8–10).
Fig. 2.
Reduction of DNp73 levels after genotoxic stress occurs in a c-Jun–dependent manner. (A) H1299 cells transfected with DNp73β and Egfp plasmids were treated with the indicated agents for 6 h and analyzed by immunoblotting (Left). A time-course analysis was performed similarly using SAOS-2-DNp73α inducible cells (Right). (B) SAOS-2-DNp73α inducible cells were transfected with siRNA 24 h prior to induction of DNp73α expression for 24 h, followed by doxorubicin or UV treatment for 6 h. (C) H1299 cells transfected with indicated shRNA and DNp73β/Egfp plasmids were subjected to UV or doxorubicin treatment 24 h later for 6 h and analyzed.
c-Jun expression was therefore silenced to determine if its increase would be causal to reduction of DNp73, in two cellular systems. Silencing c-Jun expression in DNp73α-inducible cells prior to stress treatment abrogated the reduction of DNp73α levels after UV or doxorubicin treatment (Fig. 2B, compare lanes 3 and 4 to 7 and 8). Secondly, silencing c-Jun expression in cells transfected with DNp73β and subsequently treated with UV or doxorubicin alleviated the reduction of DNp73β levels (Fig. 2C, compare lanes 2 and 3 to 5 and 6), suggesting that c-Jun is required for DNp73 degradation.
We next evaluated the role of the E3 ligase Itch in DNp73 degradation upon stress stimulation. Silencing Itch expression did not affect DNp73 degradation upon doxorubicin treatment (Fig. S5A). To further confirm this, we assessed the stability of DNp73β in mouse embryonic fibroblasts (MEFs) proficient or deficient in Itch or c-Jun, by generating cells stably expressing DNp73β. UV irradiation led to a reduction of DNp73 in both wild-type and in itch-/- cells, but this was not the case in c-jun-/- cells (Fig. S5B), implying that stress-induced DNp73 degradation occurs primarily through c-Jun. Moreover, absence of c-Jun did not affect Itch levels, therefore suggesting that c-Jun–mediated regulation of DNp73 levels could occur independent of Itch. Together, the data demonstrate that c-Jun is required for the reduction of DNp73 levels upon exposure to genotoxic signals.
c-Jun Expression Results in Degradation of DNp73.
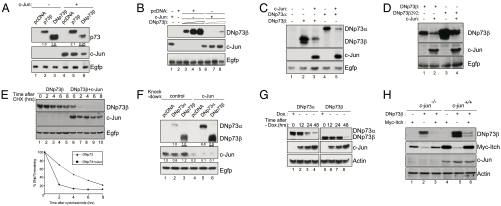
Whether expression of c-Jun could directly lead to DNp73 degradation was next analyzed by transiently coexpressing c-Jun with either TAp73β or DNp73β. As previously shown, c-Jun expression increased the steady-state levels of TAp73β (Fig. 3A, compare lanes 2 and 5) (25). By contrast, c-Jun expression caused a marked decrease in DNp73β levels (Fig. 3A, compare lanes 3 and 6), even with increasing amounts of DNp73β, suggesting that c-Jun may be a potent negative regulator of DNp73 abundance (Fig. 3B). Moreover, both the levels of DNp73α and DNp73β were reduced in the presence of c-Jun (Fig. 3C, compare lanes 3 and 5 to 2 and 4). Furthermore, the levels of R292H-DNp73β—a DNA-binding domain mutant—was also reduced in the presence of c-Jun (Fig. 3D), indicating that c-Jun could regulate DNp73 levels independent of the carboxyl-terminal isoforms and their DNA-binding ability. In addition, treatment of cells transfected with DNp73β alone or together with c-Jun with the protein synthesis inhibitor cycloheximide revealed that the highly stable DNp73β’s half-life was reduced to less than 2 h in the presence of c-Jun (Fig. 3E), indicating that c-Jun indeed regulated the stability of DNp73β.
Fig. 3.
c-Jun promotes DNp73 degradation. (A–D) The indicated plasmids were coexpressed in H1299 cells, and the steady-state levels of the proteins were determined 24 h later. Ratio of DNp73 and c-Jun was kept at 1∶3. In B, the following amounts of DNp73β was used: 0.5 μg, 1 μg, and 2 μg. The underlined values are for DNp73. (E) H1299 cells transfected with DNp73β alone or with c-Jun were treated with 50 μg/mL cycloheximide (CHX) for half-life determination. Graph shows percentage of remaining DNp73. (F) The indicated p73 plasmids were coexpressed with siRNA prior to determination of DNp73 levels. (G) DNp73α or DNp73β plasmids were transfected into the c-Jun-inducible cell line [by removal of doxycycline (Dox.)] for determination of DNp73 levels. (H) Wild-type and c-jun-/- were transfected with DNp73β and Myc-Itch plasmids and analyzed 24 h later.
To further verify the role of c-Jun in regulating DNp73 abundance, we utilized several other cellular systems. In the first case, silencing endogenous c-Jun resulted in elevated expression of transfected DNp73α and DNp73β (Fig. 3F, compare lanes 2 and 3 to 5 and 6). Secondly, transfection of DNp73α or DNp73β and subsequent induction of c-Jun expression by removal of doxycycline in a c-Jun inducible cell line (31) resulted in reduction of DNp73 levels over time (Fig. 3G).
Finally, we evaluated if c-Jun was acting independent of Itch in the degradation of DNp73. Coexpression of Itch with DNp73β led to a dramatic reduction of DNp73β levels in wild-type MEFs, as previously reported (Fig. 3H, compare lanes 5 and 6) (13). Similar results were obtained with c-jun-/- MEFs (Fig. 3H, compare lanes 2 and 3), indicating that DNp73β degradation by Itch occurs independent of c-Jun and that Itch and c-Jun probably regulate DNp73β abundance via independent pathways. Altogether, the results demonstrate that c-Jun expression is capable of destabilizing DNp73 in several cellular systems.
Az1 Negatively Regulates DNp73 Abundance.
As we were unable to detect reciprocal binding between c-Jun and p73 (25), we explored various other possibilities through which c-Jun could regulate p73 levels. Az belongs to a class of proteins that is involved in ubiquitin-independent protein degradation via the proteasome (18). We therefore coexpressed DNp73β with the cDNA encoding for processed Az1, an established member of the Az family (23), which resulted in a reduction in the steady-state levels of DNp73β (Fig. 4A, compare lanes 3 and 4), similar to the effect on Aurora-A (Fig. 4A, compare lanes 5 and 6), another protein whose abundance is negatively regulated by Az1 (23). Similar results were obtained with both DNp73α and DNp73β (Fig. S6). Hence, we evaluated the role of Az1 in stress-mediated or c-Jun-mediated DNp73 degradation by silencing the expression of Az1 (Fig. 4B and C). Whereas DNp73 levels were reduced dramatically upon doxorubicin treatment in control siRNA-treated cells, this was partially inhibited in Az1 siRNA-treated cells, although c-Jun levels were induced in both cases (Fig. 4B), suggesting that Az1 may act downstream of c-Jun to regulate DNp73 abundance. This was confirmed by silencing Az1 expression that prevented c-Jun–mediated DNp73 degradation (Fig. 4C). To further establish the role of Az1 in stress-mediated and c-Jun–dependent DNp73 degradation, we utilized the natural inhibitor of Az1, AZI (32). Coexpression of AZI and DNp73 prior to doxorubicin treatment decreased the reduction in DNp73 levels (Fig. 4D, compare lanes 3 to 4 and 5 to 6). Similarly, coexpression of AZI with c-Jun and DNp73 prevented the reduction of DNp73 levels by c-Jun (Figure 4E), suggesting that Az1 is indeed required for c-Jun–mediated DNp73 degradation. In addition, silencing the expression of c-Jun did not prevent Az1-mediated DNp73 degradation (Fig. 4F), indicating that c-Jun acted upstream of Az1 in regulating DNp73 degradation.
Fig. 4.
Az1 negatively regulates DNp73 abundance. (A) DNp73β or Aurora-A levels were determined after transfection of the respective plasmids with the processed HA-Az1 and Egfp plasmids in H1299 cells. (B and C) DNp73β and siRNA were transfected into H1299 cells, which were either treated with doxorubicin (B) or cotransfected with c-Jun plasmid (C) prior to determination of DNp73 levels. Silencing efficiency is shown at the bottom of each blot by RT-PCR analysis. (D) Indicated plasmids were transfected into H1299 cells, which were treated with doxorubicin 24 h later, and used for analysis. (E) Effect of c-Jun on DNp73β expression in the presence of the AZI was determined by cotransfecting the indicated plasmids in H1299 cells. (F and G) H1299 cells were transfected with indicated plasmids in the presence of shRNA (F) or treated with 20 μM of PYR-41/DMSO for a further 16 h (G) prior to harvest. Values below the blots indicate fold change in expression relative to lane 1 (= 1.0). (H) H1299 cells were transfected with indicated plasmids, and cell lysates were used for coimmunoprecipitation (I.P.) using anti-FLAG conjugated beads. Interacting proteins were detected using indicated antibodies.
To confirm the ubiquitin-independence of this process, we coexpressed Az1 with DNp73 in the presence of PYR-41, which resulted in reduction of DNp73 levels, similar to control DMSO-treated cells, indicating that Az1-mediated DNp73 degradation indeed occurred independent of ubiquitination (Fig. 4G compare lanes 2 and 3 to 5 and 6).
We evaluated if DNp73 and Az1 were able to interact. Coexpression of DNp73 and Az1 followed by immunoprecipitation by either anti-FLAG-Az1 or anti-FLAG-DNp73 led to the pull-down of DNp73 or Az1, respectively (Fig. 4H, Left and Right panels), confirming that Az1 and DNp73 can indeed interact in vivo. Taken together, these data show that stress-induced, c-Jun–mediated regulation of DNp73 abundance occurs through Az1.
Az1 Processing upon Stress Stimulation Is Dependent on c-Jun.
How c-Jun degraded DNp73 via Az1 upon stress stimulation was next investigated. Coexpression of TAM67, a transactivation-domain deficient c-Jun mutant, with DNp73 did not lead to reduction of the latter’s steady-state levels (Fig. 5A), suggesting that the transactivation function of c-Jun may be required for its ability to degrade DNp73. Thus, we analyzed Az1 mRNA levels upon UV irradiation or doxorubicin treatment, which showed no changes even though c-jun levels were induced (Fig. 5B), indicating that Az1 is probably not a direct target of c-Jun in this process. Because Az1 expression is known to be regulated in a polyamine-dependent manner during translation through a programmed +1 frameshift, resulting in the expression of the functional full-length protein (Fig. 5C) (19), we examined if the processed Az1 [Az1(p)] was inducible by genotoxic stresses using an antibody that specifically recognizes Az1(p) (33). Putrescine, a polyamine family member, was added to cells separately as a positive control for induction of Az1(p) (Fig. 5D). Similar to putrescine addition, doxorubicin treatment or UV irradiation also resulted in the appearance of Az1(p) in wild-type cells (Fig. 5E, Left and Right panels, respectively). However, this was not the case in c-jun-/- cells, indicating that c-Jun is required for Az1(p) production after exposure to doxorubicin or UV. We also did not observe any changes in the levels of Az1 mRNA in c-Jun deficient cells (Fig. S7), confirming the regulation of Az1 by c-Jun is transcription-independent.
Fig. 5.
Stress induces the expression of processed Az in a c-Jun–dependent manner. (A) Either FLAG-c-Jun or FLAG-TAM67 was cotransfected with DNp73β and Egfp plasmids in H1299 cells and used for analysis. (B) mRNA from doxorubicin or UV-treated H1299 cells were collected for semiquantitative RT-PCR (Top blots) and real-time PCR (Bottom) analyses. Results show means of triplicate experiments. (C) Schematic shows effects of polyamines on Az1 frameshifting (+1). (D) Wild-type MEFs were treated with putrescine to detect processed Az1 [Az1(p)] induction. (E) Doxorubicin- (Left) and UV-treated (Right) wild-type and c-jun-/- MEFs were used for immunoblot analysis. (F and G) H1299 cells transfected with DNp73β and Egfp plasmids were treated with various concentration of putrescine for 16 h and used for immunoblot analysis (F). Similarly, cells were transfected together with shRNA constructs and treated with 10 mM putrescine 24 h later for a further 16 h prior to analysis (G). (H) Schematic illustrates the proposed mechanism by which genotoxic stresses can induce the degradation of DNp73. c-Jun expression is induced by genotoxic insults (UV or doxorubicin), which in turn is required for the +1 frameshift during the translation of Az1, resulting in the expression of the functional processed Az protein [Az1(p)]. Az1(p) induces the degradation of DNp73 via the proteasome, independent of ubiquitination (Left). In the absence of c-Jun, Az1(p) is not induced upon genotoxic stress, leading to failure of DNp73 degradation (Right).
As polyamine addition induces Az1(p) expression, we hypothesized that the polyamine pathway may be involved in DNp73 degradation. This was tested by treating cells transfected with DNp73 with putrescine, which led to the reduction of DNp73 levels, as well as the other known target of the Az pathway, cyclinD1 (Fig. 5F). This degradation could be inhibited by the proteasomal inhibitor MG132 (Fig. S8). Moreover, silencing c-Jun expression upon putrescine addition did not prevent DNp73β degradation (Fig. 5G). Consistently, putrescine was able to induce Az1(p) expression in c-jun-/- cells (Fig. S9), indicating that c-Jun acts upstream of the polyamine pathway. These data together suggest that c-Jun and the polyamine pathway are involved in the control of Az1(p) processing, thereby regulating the abundance of DNp73 upon exposure to genotoxic stresses.
Discussion
A unique feature of p73 is that both of its distinct forms are differently regulated by stress signals. With tumor-suppressive properties, TAp73 is often upregulated upon exposure to cellular insults, contributing to cell death (5, 6). By contrast, DNp73 levels are rapidly reduced upon exposure to stress signals, expectedly, to allow the manifestation of apoptosis (9, 13), indicating that selective regulation of the different p73 forms is crucial in maintaining the homeostatic balance between life and death of a cell that is undergoing genotoxic insults. Thus far, no other regulators of p73 have been identified that perform antagonistic functions in regulating the abundance of these different p73 proteins. The data presented here therefore provide an example of reciprocal regulation of the tumor suppressor and its inhibitor by a common factor, in this case, c-Jun. Importantly, the mechanism utilized by c-Jun in the degradation of DNp73 occurs via a nonclassical ubiquitin-independent mechanism that is often associated with the regulation of polyamine biosynthesis. This highlights the existence of distinct pathways that have probably evolved for the reciprocal regulation of the abundance of these functionally different but highly homologous proteins.
We have previously demonstrated that c-Jun is required for the stabilization of TAp73 (25), probably through the transactivation of YAP, which is a positive regulator of TAp73 stability (34, 35). YAP has been shown to displace Itch by competitive binding to TAp73, and hence, could be responsible for c-Jun-mediated TAp73 stability (34). However, coexpression of c-Jun with DNp73 did not lead to the latter’s stabilization, but to the contrary, led to its degradation (this report), although the Itch/YAP binding site is common to both TAp73 and DNp73. Thus, both Itch and YAP may not contribute to c-Jun–mediated DNp73 degradation upon stress activation, a notion that is supported by the fact that DNp73 was degraded in itch-/- cells, but not in c-jun-/- upon exposure to stress (13 and this report).
DNp73 was found to be degraded in a ubiquitin-independent but proteasome-dependent manner upon stress activation. We therefore explored other possibilities and found that the Az/polyamine pathway was crucial for DNp73 degradation. Overexpression of processed Az1, or the physiological activation of the Az pathway by addition of polyamines, led to DNp73 degradation. Conversely, blocking Az1 expression by siRNA or with the natural inhibitor AZI abrogated stress-induced and c-Jun–mediated DNp73 degradation. As with all its substrates targeted for degradation, Az1 was found to be able to bind to DNp73 and to cause the degradation of DNp73 in the absence of E1 ubiquitin–activating enzyme activity, suggesting that DNp73 abundance is indeed regulated by Az1. Consistently, stress insults such as doxorubicin or UV were found to induce the expression of the processed Az1 product only in wild-type cells, but not in c-jun-/- cells. The results from the TAM67 experiments together with those in which c-Jun’s expression was silenced/absent during putrescine addition suggest that c-Jun probably transactivates some targets and operates upstream of the polyamine biosynthesis pathway to regulate DNp73 stability. Preliminary analysis of other members of the Az pathway such as spermine synthase, spermidine synthase, or ODC did not reveal any significant differences between cells proficient or deficient for c-Jun, or had no effect on c-Jun–dependent DNp73 degradation. Thus, the identity of the regulator of Az1 processing that is induced by c-Jun remains to be elucidated. Nonetheless, these data together establish that DNp73 can indeed be subjected to degradation by the nonclassical Az pathway upon stress stimulation, in a c-Jun–dependent manner.
Az1 has been shown to be a potent tumor-suppressor gene, expression of which can inhibit cellular growth and tumorigenesis (24). It also targets several positive regulators of cellular proliferation, such as Smad1, cyclinD1, and Aurora-A (21–23). Consistent with this is our data that has identified DNp73, which has antiapoptotic properties, as yet another target degraded by Az1. Therefore, one could envisage that Az1 processing and activation would lead to DNp73 degradation, freeing the proapoptotic TAp73 protein from inhibitory effects of DNp73, leading to an appropriate stress response.
In summary, the data presented here provide insights into a pathway that connects stress signaling to the regulation of the abundance of DNp73. Stress signals activate c-Jun, setting in motion a cascade of events regulating the polyamine biosynthesis pathway, culminating in the processing of Az1 (Fig. 5H, Left panel). Az1 then causes the degradation of DNp73 independent of ubiquitination. Absence of c-Jun or inhibition of the Az pathway abrogates this phenomenon (Fig. 5H, Right panel). It is noteworthy that this pathway probably works under stress stimulation only, and DNp73 can be degraded by other classical ubiquitin-mediated pathways under other conditions. Looking ahead, the findings presented here suggest that DNp73-overexpressing cancers may be particularly more sensitive to chemotherapeutic regimens using genotoxic drugs, especially in combination with polymaine analogues that have also been shown to inhibit cell growth and induce apoptosis in breast cancer cells (36).
Materials and Methods
Cells and Reagents.
Cells were treated as follows: doxorubicin (5 μM), cisplatin (20 μM), MG132 (5 μM/16 h or 20 μM/3 h), putrescine (10 mM/16 h), PYR-41 (20–40 μM/3–6 h), or were UV irradiated (70 J/m2). Cycloheximide (50 μg/mL) was added 24 h posttransfection for half-life determination. Cell lines details are provided in SI Text.
Plasmids, siRNAs, Transfection, and RNA Analyses.
Cells were transfected using Lipofectamine Plus–Reagent. Plasmids used and sequences for siRNAs/shRNAs used are provided in SI Text. Semiquantitative RT-PCR was performed as described (25), and real-time quantitative PCR was carried out using SYBR Green Master Mix (Applied Biosystems); details are provided in SI Text.
Immunoblot Analysis, Coimmunoprecipitation and in Vivo Ubiquitination Assays.
Immunolot analysis was performed as described (25). For coimmunoprecipitation, Anti-FLAG® M2-Agarose beads (Sigma) were used for pull-down before immunoblotting. For in vivo ubiquitination assays, cells were treated without or with MG132 before harvesting and pulled-down with Nickel-NTA-Agarose beads (Qiagen), prior to immunoblot analysis. Details are provided in SI Text.
Supplementary Material
Acknowledgments.
We thank K.S. lab members for critical reading of the manuscript, and are grateful to Dr. John Mitchell (Northern Illinois University) for providing the polyclonal anti-Az antibody and advice on the use of it. I.D. was partially supported by a Singapore Millennium Foundation scholarship. We thank the National Medical Research Council of Singapore, Biomedical Research Council of Singapore, and the Singhealth Research Foundation for their generous funding and support to K.S.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906782107/DCSupplemental.
References
- 1.Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat Rev Cancer. 2002;2:605–15. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 2.Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature. 1997;389:191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 3.Nakagawa T, et al. Autoinhibitory regulation of p73 by Delta Np73 to modulate cell survival and death through a p73-specific target element within the Delta Np73 promoter. Mol Cell Biol. 2002;22:2575–2585. doi: 10.1128/MCB.22.8.2575-2585.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaika AI, et al. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J Exp Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Irwin MS, et al. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–10. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 6.Lin KW, Nam SY, Toh WH, Dulloo I, Sabapathy K. Multiple stress signals induce p73beta accumulation. Neoplasia. 2004;6:546–57. doi: 10.1593/neo.04205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishimoto O, et al. Possible oncogenic potential of DeltaNp73: A newly identified isoform of human p73. Cancer Res. 2002;62:636–641. [PubMed] [Google Scholar]
- 8.Belloni L, et al. DNp73alpha protects myogenic cells from apoptosis. Oncogene. 2006;25:3606–12. doi: 10.1038/sj.onc.1209321. [DOI] [PubMed] [Google Scholar]
- 9.Maisse C, Munarriz E, Barcaroli D, Melino G, De Laurenzi V. DNA damage induces the rapid and selective degradation of the DeltaNp73 isoform, allowing apoptosis to occur. Cell Death Differ. 2004;11:685–687. doi: 10.1038/sj.cdd.4401376. [DOI] [PubMed] [Google Scholar]
- 10.Oberst A, et al. Regulation of the p73 protein stability and degradation. Biochem Biophys Res Commun. 2005;331:707–712. doi: 10.1016/j.bbrc.2005.03.158. [DOI] [PubMed] [Google Scholar]
- 11.Gong J, et al. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–808. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 12.Tsai KK, Yuan ZM. c-Abl stabilizes p73 by a phosphorylation-augmented interaction. Cancer Res. 2003;63:3418–24. [PubMed] [Google Scholar]
- 13.Rossi M, et al. The ubiquitin-protein ligase Itch regulates p73 stability. EMBO J. 2005;24:836–848. doi: 10.1038/sj.emboj.7600444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munarriz E, et al. Calpain cleavage regulates the protein stability of p73. Biochem Biophys Res Commun. 2005;333:954–60. doi: 10.1016/j.bbrc.2005.05.188. [DOI] [PubMed] [Google Scholar]
- 15.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosoda M, et al. UFD2a mediates the proteasomal turnover of p73 without promoting p73 ubiquitination. Oncogene. 2005;24:7156–7169. doi: 10.1038/sj.onc.1208872. [DOI] [PubMed] [Google Scholar]
- 17.Ohtsuka T, Ryu H, Minamishima YA, Ryo A, Lee SW. Modulation of p53 and p73 levels by cyclin G: Implication of a negative feedback regulation. Oncogene. 2003;22:1678–1687. doi: 10.1038/sj.onc.1206306. [DOI] [PubMed] [Google Scholar]
- 18.Coffino P. Regulation of cellular polyamines by antizyme. Nat Rev Mol Cell Biol. 2001;2:188–194. doi: 10.1038/35056508. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov IP, Gesteland RF, Atkins JF. Antizyme expression: A subversion of triplet decoding, which is remarkably conserved by evolution, is a sensor for an autoregulatory circuit. Nucleic Acids Res. 2000;28:3185–3196. doi: 10.1093/nar/28.17.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2003;22:1488–1496. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y, et al. A novel link between the proteasome pathway and the signal transduction pathway of the bone morphogenetic proteins (BMPs) BMC Cell Biol. 2002;3:15. doi: 10.1186/1471-2121-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newman RM, et al. Antizyme targets cyclin D1 for degradation. A novel mechanism for cell growth repression. J Biol Chem. 2006;281(21):14529–14532. doi: 10.1074/jbc.M407349200. [DOI] [PubMed] [Google Scholar]
- 23.Lim SK, Gopalan G. Antizyme1 mediates AURKAIP1-dependent degradation of Aurora-A. Oncogene. 2007;26:6593–603. doi: 10.1038/sj.onc.1210482. [DOI] [PubMed] [Google Scholar]
- 24.Fong LY, Feith DJ, Pegg AE. Antizyme overexpression in transgenic mice reduces cell proliferation, increases apoptosis, and reduces N-nitrosomethylbenzylamine-induced forestomach carcinogenesis. Cancer Res. 2003;63:3945–3954. [PubMed] [Google Scholar]
- 25.Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K. c-Jun regulates the stability and activity of the p53 homologue, p73. J Biol Chem. 2004;279:44713–22. doi: 10.1074/jbc.M407672200. [DOI] [PubMed] [Google Scholar]
- 26.Eferl R, Wagner EF. AP-1: A double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 27.Yamada HY, Gorbsky GJ. Inhibition of TRIP1/S8/hSug1, a component of the human 19S proteasome, enhances mitotic apoptosis induced by spindle poisons. Mol Cancer Ther. 2006;5:29–38. doi: 10.1158/1535-7163.MCT-05-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–83. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, et al. Inhibitors of ubiquitin activating enzyme (E1), a new class of potential cancer therapeutics. Cancer Res. 2007;67:9472–81. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 30.Pagano M, et al. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–5. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 31.Hettinger K, et al. c-Jun promotes cellular survival by suppression of PTEN. Cell Death Differ. 2007;14:218–29. doi: 10.1038/sj.cdd.4401946. [DOI] [PubMed] [Google Scholar]
- 32.Mangold U. Antizyme inhibitor: Mysterious modulator of cell proliferation. Cell Mol Life Sci. 2006;63:2095–101. doi: 10.1007/s00018-005-5583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell JL, et al. Overproduction of stable ornithine decarboxylase and antizyme in the difluoromethylornithine-resistant cell line DH23b. Biochem J. 1996;317:811–6. doi: 10.1042/bj3170811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy D, Adamovich Y, Reuven N, Shaul Y. The Yes-associated protein 1 stabilizes p73 by preventing Itch-mediated ubiquitination of p73. Cell Death Differ. 2007;14:743–51. doi: 10.1038/sj.cdd.4402063. [DOI] [PubMed] [Google Scholar]
- 35.Danovi SA, et al. Yes-Associated Protein (YAP) is a critical mediator of c-Jun-dependent apoptosis. Cell Death Differ. 2008;15:217–9. doi: 10.1038/sj.cdd.4402226. [DOI] [PubMed] [Google Scholar]
- 36.Huang Y, et al. A novel polyamine analog inhibits growth and induces apoptosis in human breast cancer cells. Clin Cancer Res. 2003;9:2769–77. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.