Abstract
Nuclear lamins are components of the nuclear lamina, a structural scaffolding for the cell nucleus. Defects in lamins A and C cause an array of human diseases, including muscular dystrophy, lipodystrophy, and progeria, but no diseases have been linked to the loss of lamins B1 or B2. To explore the functional relevance of lamin B2, we generated lamin B2-deficient mice and found that they have severe brain abnormalities resembling lissencephaly, with abnormal layering of neurons in the cerebral cortex and cerebellum. This neuronal layering abnormality is due to defective neuronal migration, a process that is dependent on the organized movement of the nucleus within the cell. These studies establish an essential function for lamin B2 in neuronal migration and brain development.
Keywords: brain, lissencephaly, neuronal migration, nuclear envelope, nuclear lamina
The nuclear lamina is an intermediate filament meshwork lying beneath the inner nuclear membrane that provides a structural scaffolding for the nucleus (1). The lamina is also important for other processes, including gene transcription, chromatin organization, nuclear pore distribution, nuclear envelope assembly, and tethering of the nucleus to the cytoskeleton (1, 2). The main components of the nuclear lamina are nuclear lamins, a class of intermediate filament proteins that is generally divided into two groups, A-type (lamins A and C) and B-type (lamins B1 and B2) (3, 4). Lamins A and C are produced from LMNA by alternative splicing, whereas lamins B1 and B2 are encoded by distinct genes, LMNB1 and LMNB2, respectively. Lamins B1 and B2 are expressed in all cells and throughout development, whereas lamins A and C are expressed in differentiated cells, beginning at midgestation (3).
Interest in the nuclear lamins has intensified with the discovery that over a dozen human diseases, including muscular dystrophy, cardiomyopathy, lipodystrophy, and progeria, are caused by mutations in LMNA (5–7). To date, more than 340 missense, nonsense, frameshift, and splicing mutations have been identified (5). In contrast, no human diseases have been linked to these types of mutations in LMNB1 and LMNB2, and, so far, the only clear-cut association between B-type lamins and disease has been the finding of LMNB1 gene duplications in autosomal-dominant leukodystrophy (8).
The paucity of “lamin B diseases” is probably not due to complete redundancy of lamins B1 and B2, as Lmnb1-deficient mice are small during embryonic development and die soon after birth with defects in lungs and bones (9). Also, Lmnb1-deficient fibroblasts display misshapen cell nuclei, aneuploidy, and early senescence (9). To further examine the functional importance of the B-type lamins, we generated Lmnb2-deficient mice.
Results
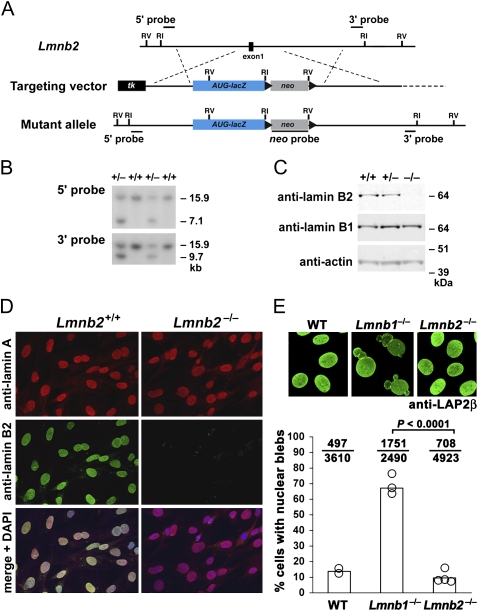
We used gene targeting to insert a lacZ reporter into exon 1 of Lmnb2 (Fig. 1 A and B), abolishing the production of lamin B2 transcripts. As expected, homozygous Lmnb2 knockout (Lmnb2−/−) fibroblasts lacked lamin B2 (Fig. 1 C and D). The production of the lamin B3 transcript, a testis-specific transcript that uses an alternative first exon (located within intron 4 of Lmnb2), was not affected by our knockout mutation (Fig. S1). Lmnb2−/− fibroblasts did not have misshapen cell nuclei (Fig. 1E), unlike Lmnb1−/− fibroblasts (9). Also, primary Lmnb2−/− fibroblasts had a normal number of chromosomes (40.2 ± 0.5, n = 5 independent cell lines versus 39.2 ± 0.6 in WT cells, n = 2 independent cell lines). In parallel experiments, freshly isolated Lmnb1−/− fibroblasts exhibited polyploidy (57.8 ± 3.4 chromosomes), as previously reported (9). We also examined levels of several nuclear envelope proteins and their intracellular localization in both WT and mutant fibroblasts, but we did not uncover any abnormalities (Fig. S2).
Fig. 1.
Inactivation of mouse Lmnb2. (A) Gene-targeting strategy to replace the coding sequences of Lmnb2 exon 1 with a lacZ cassette, beginning at the translation initiation site (ATG). The positions of relevant EcoRI and EcoRV sites and the 5′, 3′, and neo probes for Southern blot analysis are shown. (B) Southern blots identifying gene-targeting events in mouse embryonic stem cells. Genomic DNA was digested with EcoRV. (C) Western blots of mouse embryonic fibroblast extracts with antibodies against lamin B1 and lamin B2. Actin was used as a loading control. (D) Immunofluorescence microscopy of Lmnb2+/+ and Lmnb2−/− embryonic fibroblasts with antibodies against lamin A and lamin B2. (Bottom) Merged images with DAPI staining for nuclear DNA. (E) Lmnb2−/− fibroblasts did not have a higher frequency of cells with nuclear blebs than WT cells. (Upper) Confocal images of Lmnb2+/+, Lmnb1−/−, and Lmnb2−/− fibroblasts stained with a monoclonal antibody against LAP2β, a protein of the inner nuclear membrane. (Lower) graph showing the percentage of cells with nuclear blebs. Each open circle represents an independent cell line (two WT, three Lmnb1−/−, and four Lmnb2−/− cell lines). The ratio above each bar indicates the number of cells with nuclear blebs over the total number of cells evaluated.
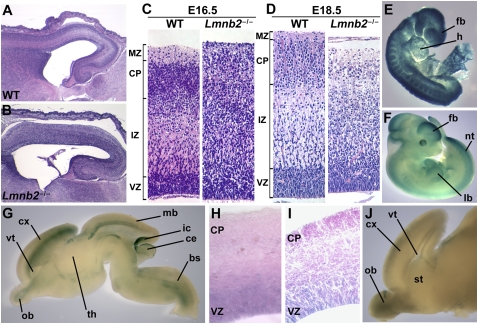
Heterozygotes (Lmnb2+/−) were phenotypically normal. Intercrosses of Lmnb2+/− mice yielded Lmnb2−/− embryos at the expected Mendelian frequency [22.3% of 112 embryos collected between embryonic day 11 (E11) and E19.5]. At birth, Lmnb2−/− pups were normal in size and appearance, but they died within 15–60 min. Histopathological studies of E16.5–E19.5 Lmnb2−/− embryos did not uncover abnormalities in any organ system, except for the brain. The brain was nearly normal in size (reduced by only 5–10%). However, the forebrain exhibited a dramatic neuronal layering defect at E16.5 (Fig. 2 A and B), with an accumulation of cells in the intermediate zone (Fig. 2C). Similar findings were apparent in E18.5 embryos (Fig. 2D and Fig. S3A) and newborns. At E18.5, the ventricular zone was virtually normal in size and with no noticeable accumulation of neurons; however, fewer neurons reached the cortical plate (Fig. S3 C and D).
Fig. 2.
Abnormal patterning of the cerebral cortex in the setting of Lmnb2 deficiency. (A–D) Lmnb2 deficiency alters neuronal layering in the cerebral cortex. H&E staining of paraffin-embedded sections of cerebral cortex from E16.5 WT (A) and Lmnb2−/− (B) embryos. (C) Higher-magnification views of sections from the same E16.5 WT and Lmnb2−/− embryos. Cortical layers are indicated on the Left, from Top to Bottom: MZ, marginal zone; CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone. Neuronal progenitors proliferate in the VZ; postmitotic cells leave the VZ and migrate along glial fibers to the CP. (D) Sections from E18.5 embryos showing similar defects. (E and F) Whole-mount β-galactosidase staining of Lmnb2+/− embryos at E8.5 (E) and E11.5 (F). Staining was ubiquitous at E8.5; at E11.5, staining was prominent in the forebrain (fb), midbrain, hindbrain, limb buds (lb), tailbud, somites, neural tube (nt), and retina. heart (h). (G) Whole-mount staining of an E16.5 Lmnb2+/− brain cut sagittally; β-galactosidase expression is found in the cortex (cx), olfactory bulb (ob), midbrain (mb), brainstem (bs), inferior colliculus (ic) and superficial layer of the cerebellum (ce); ventricle (vt); hypothalamus (th). (H) 40-μm section of the cortex of an E16.5 Lmnb2+/− embryo and (I) 20-μm section of the cortex of a newborn Lmnb2+/− pup, after β-galactosidase staining, revealing Lmnb2 expression in the VZ. (J) β-Galactosidase staining of the brain of a newborn Lmnb2+/− mouse, revealing Lmnb2 expression in the ob, cx, and vt.
At E13.5, before cortical neuronal migration gets underway, the neocortex of Lmnb2−/− embryos appeared similar to that of WT embryos, and the marginal zone was initially present at E16.5, as indicated by the expression of Reelin, a marker for Cajal-Retzius cells (10) (Fig. S4 A and B). However, Reelin expression was undetectable at E18.5 (Figs. S3B and S4C). Early stages of cortical plate formation were evident in Lmnb2−/− brains at E15.5 (with evidence of a marginal zone and a subplate) (Fig. S5 A and B). Thus, early development of the cortex appeared normal, and morphological defects appeared only when the second wave of neurons had begun their migration into the cortical plate (Fig. 2 and Fig. S5). These abnormalities are reminiscent of phenotypes in mice with defects in cortical neuron migration (11, 12).
At E8.5, β-galactosidase staining of Lmnb2+/− embryos revealed that Lmnb2 is expressed ubiquitously (Fig. 2E). At E11.5, β-galactosidase staining was prominent in the brain, limb buds, neural tube, and somites (Fig. 2F). In E16.5 embryos and newborns, staining was detected in the ventricular zone and upper layers of the cortex, the olfactory bulb, and cerebellum (Fig. 2 G–J). The in situ hybridization database GenePaint (www.genepaint.org) (13) also documents strong Lmnb2 expression in the ventricular zone at E14.5. The presence of lamin B2 in the cortical neurons of adult rats had been previously documented by immunohistochemistry (14). Here, we report robust immunostaining for lamin B2 in the cortex of E17.5 WT embryos (Fig. S6 A and B). A similar staining pattern was observed with antibodies against lamin B1 (Fig. S6B). As expected, lamin B2 was absent in the cortex of E17.5 Lmnb2−/− embryos (Fig. S6C).
To determine if cortical neuron migration was perturbed in Lmnb2−/− embryos, we performed neuronal birthdating experiments by injecting BrdU. When embryos are pulse-labeled with BrdU, progenitors within the ventricular zone incorporate BrdU into their DNA. Newly born neurons do not reenter the cell cycle; therefore, their BrdU content remains constant as they migrate into the cortical plate. Meanwhile, progenitors continue to divide, diluting their BrdU content, and neurons born several days after the BrdU pulse contain less BrdU. To examine neuronal migration in Lmnb2-deficient mice, we injected BrdU at E13.5 and collected embryos at E18.5 and E19.5. In WT embryos, the neurons that stained most intensely for BrdU were found within the lower portion of the cortical plate (Fig. 3A, Fig. S7 A and B). Neurons born at later times normally migrate higher into the cortical plate; consequently, more superficial layers in WT brains stain less intensely for BrdU (Fig. 3A). In Lmnb2−/− embryos, the most intense BrdU staining was observed in superficial layers of the cortical plate (Fig. 3A, Fig. S7 A and B), suggesting a defect in the ability of newer neurons to migrate past older BrdU-positive neurons. Costaining with antibodies against BrdU and Ctip2 [a marker for cortical layers V and VI (15)] supported this interpretation (Fig. 3B). An injection of BrdU in WT mice at E15.5 labeled neurons in more superficial layers of the cortex. In contrast, the neurons that were labeled at E15.5 in Lmnb2−/− embryos were found in lower layers of the cortex, implying that those cells were defective in their ability to migrate into more superficial layers of the cortex (Fig. S7C).
Fig. 3.
Lmnb2 deficiency causes defective neuronal migration in the brain. (A) Birthdating experiment demonstrating defective neuronal migration in the cortex of Lmnb2−/− embryos. Pregnant females were injected with BrdU at E13.5; brains were collected at E18.5, sectioned, and stained with DAPI and a rat monoclonal antibody against BrdU. Neurons born at E13.5, the time of the BrdU injection, stain very intensely for BrdU and were present within the lower levels of the cortical plate in WT mice. Neurons born later (therefore containing less BrdU) migrate to more superficial layers of the cortex. In Lmnb2−/− embryos, intense BrdU staining is noted in the most superficial layers of the cortical plate, consistent with an inability of the neurons born later to migrate past older BrdU-positive neurons. Arrow indicates the orientation of neuronal migration. (B) Sections from the same experiment stained with a sheep polyclonal antibody against BrdU (red) and a rat monoclonal against Ctip2, a marker for cortical layers V and VI (green). In WT embryos, neurons with the strongest BrdU staining were found in deep layers of the cortical plate together with Ctip2 staining. In contrast, in the Lmnb2−/− embryos, BrdU-positive and Ctip2-positive cells were both found in the more superficial part of the cortex. (C) Immunostaining of cortical sections of E19.5 WT and Lmnb2−/− embryos with antibodies against Ctip2 (green) and NeuN (red). In WT brains, a large number of mature neurons positive for NeuN were detected above layers V and VI (positive for Ctip2), whereas in Lmnb2−/− brains, many NeuN-positive cells were found below Ctip2-positive cells. (D) Immunostaining of cortical sections of E19.5 WT and Lmnb2−/− embryos with antibodies specific for Ctip2 (green) and FoxP1, a marker for layers III–V (red). In Lmnb2−/− embryos, FoxP1-positive neurons accumulated in lower levels of the cortex. (E) Immunostaining of cortical sections of E19.5 WT and Lmnb2−/− embryos with antibodies against Cux1, a marker for layers II–IV (green) and FoxP1 (red). In WT brains, Cux1-positive neurons were mainly located above the cells that stain for FoxP1, whereas in Lmnb2−/− brains, many Cux1-positive cells were located below FoxP1-positive cells. (F) Immunostaining of cortical sections of E19.5 WT and Lmnb2−/− embryos with antibodies specific for FoxP2, a marker for layer VI (green), and for Ctip2 (red). In WT embryos, Ctip2 staining was detected in the upper part of the FoxP2 territory, corresponding to layer V, whereas in Lmnb2−/− embryos the two cell populations were intermixed.
A defect in neuronal migration was further supported by immunohistochemical studies at E19.5 with cortical layer–specific markers (Fig. 3 C–F). In WT cortex, the newer neurons, positive for NeuN, migrated past older Ctip2-positive neurons and into more superficial layers of the cortex (Fig. 3C). In Lmnb2−/− embryos, most NeuN-positive neurons tended to accumulate in lower levels of the cortex, below the Ctip2-positive cells (Fig. 3C). Similar findings were observed in sections stained for FoxP1, a marker for superficial layers of the cortical plate (layers III–V) (Fig. 3D) (16). In Lmnb2−/− embryos, FoxP1-positive neurons accumulated in lower levels of the cortical plate and did not reach their normal position (Fig. 3D). Similar abnormalities of cortical layering were observed with other pairs of layer specific markers: Cux1 (layers II–IV) and FoxP1 (Fig. 3E), and FoxP2 (layer VI) and Ctip2 (Fig. 3F). In each case, neuronal populations were intermixed in the Lmnb2−/− brains.
Although neuron layering was dramatically perturbed in Lmnb2−/− mice, the size of the brain was minimally reduced and the overall number of cells in the cortex did not appear to be altered. However, the number of cells positive for the mitotic antigen Ki-67 in the ventricular zone at E15.5 was somewhat lower in Lmnb2−/− brains than in WT brains (Fig. S8). Other organs, aside from the brain, were normal in size and free of histopathological abnormalities, providing no support for a widespread defect in cell division or migration. Interestingly, the growth of Lmnb2−/− fibroblasts was normal, and the mobility of Lmnb2−/− embryonic fibroblasts was normal in a “wound-healing” assay (Fig. S9).
The development of the cerebellum also depends on neuronal migration with granule cells moving inward past the Purkinje cells (17, 18). In Lmnb2−/− embryos, the morphology of the cerebellum was profoundly abnormal, with a complete absence of foliation that normally results from granule cell migration (Fig. 4 A and B). A discrete Purkinje cell layer was absent in Lmnb2−/− cerebellum, as judged by routine histology and immunohistochemical staining for calbindin, a Purkinje cell marker (Fig. 4B). Given these findings, one would predict that Lmnb2 expression would be evident in the cerebellum. Indeed, β-galactosidase staining revealed that Lmnb2 is expressed in the superficial layer of E16.5 cerebellum, and also in the vicinity of the Purkinje cell layer of newborn mice (Fig. 4C). Immunostaining of E17.5 embryos revealed that lamins B1 and B2 are present in all cells of the cerebellum, but staining was higher in an inner band of cells, corresponding to the Purkinje cell layer (Fig. 4D). In addition, we examined the hippocampus in WT and Lmnb2−/− mice (Fig. S10). Not surprisingly, the layering of the hippocampus was perturbed in Lmnb2−/− embryos. Although the CA1 and CA3 layers of the hippocampus appeared fairly normal in mutant embryos at E18.5, the layers were less compact at E19.5 and P0, compared with WT mice.
Fig. 4.
Abnormal cerebellar morphology in Lmnb2−/− mice. (A) Cross-sections of cerebellum in newborn WT and Lmnb2−/− mice. The cerebellum of Lmnb2−/− embryos was smooth and smaller in size; asterisks show fissures in the WT cerebellum (absent in the Lmnb2−/− cerebellum). (B) Cerebellar sections of E19.5 WT and Lmnb2−/− embryos stained with H&E (Top) and with an antibody against calbindin (Bottom). Calbindin stains the layer of Purkinje cells (arrow in WT section; layer is also visible in the H&E-stained section). Note the reduced thickness of the external granule layer in Lmnb2−/− cerebellum (arrowhead). Cp, choroid plexus. (C) Cross-section of the cerebellum from an E16.5 Lmnb2+/− embryo and a newborn Lmnb2+/− mouse (P0) after whole-mount staining for β-galactosidase. Lmnb2 expression is noted in the superficial layer of the cerebellum, the external granule layer (arrowhead) and in the vicinity of the Purkinje cell layer (arrow). (D) Immunostaining of a cerebellar section of an E17.5 Lmnb2+/− embryo with antibodies against lamin B1 (green) and lamin B2 (red). Nuclear DNA was stained with DAPI. Lamins B1 and B2 are detected in all cells, with a stronger signal in the Purkinje layer (arrow).
Discussion
Human geneticists have made remarkable progress in uncovering diseases associated with defects in lamins A and C, and additional studies with cultured cells and mouse models are beginning to define the functions of these proteins in health and disease (5–7). Meanwhile, the functional relevance of the other nuclear lamins, and in particular lamin B2, has remained obscure. In the current study, we shed light on this issue. We report that mice lacking lamin B2 are entirely normal in size at birth but die shortly thereafter. Detectable pathology is confined to the brain, and the most striking finding is a defect in the layering of neurons in the cerebral cortex. The cortical abnormality was striking at E16.5 and remained obvious in newborn mice. BrdU birthdating experiments and immunohistochemical studies revealed that the neuronal layering defect was due to defective migration of neurons from the ventricular zone to the cortical plate.
Lmnb2−/− mice had a smaller cerebellum with a complete absence of foliation, and the hippocampus was also abnormal. Defective migration of cortical neurons is a hallmark of lissencephaly, a human brain disease characterized by a smooth brain devoid of folds (19, 20), and some forms of lissencephaly are also accompanied by cerebellar hypoplasia or dysplasia (11). A deficiency in Reelin also causes severe abnormalities in the cortex, cerebellum, and hippocampus (21), and we did observe markedly reduced levels of Reelin expression at later stages of development in Lmnb2−/− embryos. Although we cannot exclude a significant role for Reelin in the Lmnb2 phenotype, we do not believe that altered Reelin expression is the entire story. First, Lmnb2−/− mice exhibit a perinatal lethal phenotype, whereas Reelin-deficient mice do not (21). Second, the subplate was separated from the marginal zone in Lmnb2−/− mice at E15.5—a process known to be blocked in the setting of Reelin deficiency (21).
The neuronal migration defect in Lmnb2−/− mice was initially surprising to us because diseases caused by lamin A and lamin C defects are confined largely to mesenchymal tissues (skeletal muscle, heart, adipose tissue, connective tissue, and bone). In hindsight, however, the involvement of a nuclear lamin in neuronal migration makes sense, for the simple reason that cortical neuron migration is utterly dependent on the ability of the neurons to move their own nucleus (12, 22). Migration of neurons from the ventricular zone to the cortical plate is a saltatory process that involves the coupled translocation of the centrosome and the nucleus (22). The initial step is a forward movement of the centrosome toward the leading process of the neuron. Next, cytoplasmic motors pull the cell nucleus along microtubules toward the centrosome. These cytoplasmic motors have been noted to associate with the nuclear envelope (23). After the nucleus is translocated forward, the trailing process of the cell body is remodeled, resulting in net forward movement of the soma. Repeated cycles of this process—extension of the centrosome followed by nucleokinesis—make it possible for cortical neurons to migrate over large distances (22).
The cytoplasmic molecules involved in nuclear movement and neuronal migration have been studied intensively. For example, LIS1, NDE1, and NDEL1 have been shown to regulate the dynein motors required for nuclear translocation along microtubules (24, 25), and knockouts or knockdowns of these proteins result in impaired neuronal migration (12). In humans, LIS1 mutations are the most common cause of defective neuronal migration and lissencephaly (19, 20). Although the cytoplasmic molecules underlying nuclear movement and neuronal migration have come into focus, the nuclear proteins required for this process have remained mysterious. Quite remarkably, very little attention has been paid to which proteins within the nucleus are being “tugged on” by cytoplasmic motors. The current study sheds light on this issue, strongly suggesting that nuclear translocation within migrating neurons depends on the nuclear lamina. This discovery was foreshadowed by Drosophila studies; Patterson et al. (26) showed that lamin B is required for the migration of photoreceptor nuclei during eye formation in Drosophila and suggested that these findings might be relevant to the pathogenesis of diseases caused by LMNA mutations (e.g., muscular dystrophy and cardiomyopathy).
Lamins are nuclear proteins, but are known to be mechanically linked to the cytoskeleton via nuclear-envelope-spanning complexes of SUN- and KASH-domain proteins (27–29). Because Klarsicht, a KASH protein, interacts with the Drosophila B-type lamin and is required for nuclear movement in photoreceptor cells (26), it seems possible that neuronal migration in mammals could also involve SUN- and KASH-domain proteins. In the future, defining molecular partners for lamin B2 could uncover other nuclear proteins involved in neuronal migration, just as screens for LIS1-interacting proteins uncovered additional cytoplasmic players in nuclear migration (12).
Other studies have linked the nuclear lamins to elements of the cytoskeleton (30). For example, Lammerding and coworkers (31) showed that the nuclei of cells spin in the absence of lamin B1, strongly suggesting that lamin B1 helps to anchor the nucleus to the cytoskeleton. Although the current studies focused entirely on Lmnb2-deficient mice, the involvement of lamin B1 in nuclear–cytoskeletal interactions raises the question of whether lamin B1 might also be important for neuronal migration in the brain. This issue needs to be investigated.
The discovery of neuronal migration defects in the setting of Lmnb2 deficiency could explain the absence of reported associations between LMNB2 mutations and the types of human diseases commonly associated with LMNA mutations (e.g., muscular dystrophy, cardiomyopathy). LIS1 mutations cause devastating developmental brain abnormalities, and we suspect that consequences of LMNB2 deficiency will be similar. Over the next few years, we predict that LMNB2 mutations will be identified in children with severe developmental brain disorders. If this prediction is borne out, the spectrum of diseases associated with the nuclear envelope would be significantly expanded (5–7).
Materials and Methods
Generation of Lmnb2-Deficient Mice.
Arms for a sequence-replacement gene-targeting vector were amplified from genomic DNA of E14Tg2A ES cells derived from 129/Ola mice (32) and cloned into the TopoI vector (Invitrogen). The 5′ arm (containing sequences upstream of the ATG in exon 1) was amplified with primers 5′–TGG CGG CCG CGA ATT CGT GCA GGG CAG TGT GAA TCA GAT AGG G–3′ and 5′–GAG CTA GCA TGG CGG AGG TGG CGG CGG CGG GAG AAT CAC CTC–3′; the 3′ arm was amplified with primers 5′–ATA AGA ATG CGG CCG CGA ATT CGT ACG TGG GTC GGC ACC GGG GAC GC–3′ and 5′–ATA GTT TAG CGG CCG CGG GTC TGC TCC AGG AAT GCA ATG CGG GG–3′. The 5′ arm was excised with NotI and NheI and cloned upstream of a promoterless lacZ cassette (to insert lacZ at the site of the Lmnb2 ATG). An EcoRI fragment spanning the entire 5′ arm and lacZ was subcloned into pKSloxPNT (33) (between the PGK-tk and the PGK-neo cassettes). The 3′ arm was inserted into the NotI site downstream from the neo cassette. The vector was linearized with XhoI and electroporated into E14Tg2A ES cells. After selection with G418 (125 μg/mL; Gibco, Invitrogen) and ganciclovir (2 μM; Sigma), ES cell colonies were picked and screened for recombination with Southern blots on EcoRV digests with 5′ and 3′ flanking probes. The 5′ probe was amplified from E14Tg2A genomic DNA with primers 5′-GAA CTT CCA GGA TTG AGA GTC TAG G-3′ and 5′-CCA GAC TTC CCC AGG CCT TAC TCA C-3′; the 3′ probe was amplified with 5′-GGG AAT AAG AGC CTC ACC CAC TCC AG-3′ and 5′-GCC CAT GAG CTG CAA CAC TGC CCA ACC-3′. Screening of 230 ES cell clones identified 13 targeted clones. After verifying that the targeted clones were euploid and lacked random integration events, two independent ES cell lines were injected into C57BL/6 blastocysts. Male chimeras from both clones were mated with C57BL/6 females and transmitted the targeted mutation to their offspring. No discernible phenotypic differences were observed in mice derived from the two different ES cell clones. All mice were analyzed on a mixed genetic background (C57BL/6 and 129/Ola). No phenotypic differences were observed between Lmnb2+/+ and Lmnb2+/− mice. All animal protocols used in the present study were reviewed and approved by the Animal Research Committee at UCLA.
Mice were genotyped by PCR. The WT allele was identified by amplifying a 350-bp product with forward primer 5′-CGG GTT TTA CTG GAA AGC TG-3′ and reverse primer 5′-CGG AGC AGC AAC CTA TCA TT-3′; the mutant allele was identified by amplifying a 550-bp product with the same forward primer and reverse primer 5′-GAC AGT ATC GGC CTC AGG AA-3′ (located within the lacZ cassette).
Studies with Lmnb2−/− Fibroblasts.
Mouse embryonic fibroblasts from E13.5 sibling embryos were isolated and cultured as described previously (34). A wound-healing assay was performed on WT and Lmnb2−/− fibroblasts (n = 3 lines/genotype) at passage 4. A monolayer of confluent fibroblasts was scraped with a pipette tip and washed with PBS before adding fresh culture medium. Photographs of the scraped cells were recorded immediately after the wound (t = 0) and 80, 190, 280, and 390 min later. The ability of cells to migrate into the gap was quantified by measuring the distance between the two migration fronts at six independent positions on the plate; data were plotted as a percentage of the wound that had closed.
RNA and Protein Analysis.
Transcript levels were measured by qPCR in ES cells and in testis of adult male mice. Total RNA were extracted with TRI reagent (Sigma), treated with DNase I (Ambion, Applied Biosystems), and reverse-transcribed with a mixture of random primers, oligo(dT), and SuperScript III (Invitrogen). qPCR reactions were performed in triplicates with 50 ng cDNA, 200 nM of oligonucleotides, and 10 μl SYBR Green PCR Master Mix (Qiagen) on a 7900 Fast Real-Time PCR System (Applied Biosystems). Expression levels were calculated with the comparative cycle threshold method and normalized to β2-microglobulin. Primers located in exons 9 and 10 were used to detect all transcripts of Lmnb2 (5′-GAG GAC ATT GCC TAC AAG TTC AC-3′ and 5′-TTC CAC ACA AGG GTT GAT G-3′); primers in exons 1 and 2 were used to measure lamin B2 transcripts (5′-GCT GGA GCT GGA GAA TGA TAG-3′ and 5′-GTC AGC CAG CTC TGA CTC GTA-3′); primers in exons 1b and 5 were used to measure lamin B3 transcripts (5′-GAC TTG GAA CAA CCA CCT CAG-3′ and 5′-TGT CCA CCT CCA CCA AGC-3′). The primers for β2-microglobulin were 5′-TGC TTG TCT CAC TGA CC-3′ and 5′-TAT GTT CGG CTT CCC ATT CT-3′.
Proteins from embryonic fibroblast cell extracts were separated on NuPAGE 4–12% Bis-Tris gels (Invitrogen) and transferred to nitrocellulose membrane (Bio-Rad). Primary antibodies were used at the dilutions indicated in Table S1. IR-Dye 800CW- and 700DX-conjugated secondary antibodies (Rockland) were used at 1:8,000 and detected with an Odyssey Infrared Imaging System (Li-Cor Biosciences).
Immunofluorescence Microscopy.
Mouse embryonic fibroblasts grown on coverslips were washed in PBS containing 1 mM Ca2+ and 1 mM Mg2+ (PBS/Ca/Mg) and fixed in methanol for 10 min. After permeabilizing cells with 0.2% Triton for 5 min and blocking for 1 h in PBS/Ca/Mg containing 10% FBS and 0.2% BSA, cells were incubated for 1 h with primary antibodies (Table S1). Alexa Fluor 488-conjugated goat anti-mouse IgG (Molecular Probes, Invitrogen) and Cy3-conjugated donkey anti-rabbit IgG (Jackson ImmunoResearch) secondary antibodies were diluted 1:400. DNA was stained with 2 μg/mL DAPI and coverslips mounted with Prolong Gold antifade reagent (both Invitrogen). Images were taken on a Zeiss Axiovert 200 M microscope with a 40× or a 63× oil objective with an AxioCam MRm and ApoTome, and on a Leica TCS-SP MP laser-scanning confocal microscope with a 100× objective. The number of cells with misshapen nuclei (nuclear blebs) was assessed in a blinded fashion by immunofluorescence microscopy (35).
β-Galactosidase Staining on Tissues.
β-Galactosidase staining was performed on whole embryos and on dissected brains after removal of the meninges. Tissues were fixed for 5 min in 4% paraformaldehyde in PBS, washed twice for 10 min in ice-cold PBS, and then stained for 16–24 h at 37 °C in X-gal buffer [PBS, 20 mM potassium ferricyanate, 20 mM potassium ferrocyanate, 2 mM MgCl2, 0.2% Nonidet P-40, 0.1% sodium deoxycholate, and 0.8 mg/mL 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (Xgal)] (36). Staining specificity was established by parallel studies with WT tissues, which did not stain for β-galactosidase activity. After staining, tissues were washed in PBS and postfixed in 4% paraformaldehyde in PBS. Thick sections of brain tissues were cut manually with double-edge stainless steel blades; 40-μm thin sections were cut from stained tissues after embedding in O.C.T. (Optimal Cutting Temperature; Tissue-Tek, Sakura Finetek). Sections were counterstained with eosin, dehydrated and mounted in Permount (Fisher Scientific). Images were recorded with a Leica MZ6 dissecting microscope and a Leica DFC290 digital camera.
Histological and Immunochemical Staining of Mouse Tissues.
Paraffin-embedded sections of mouse embryos (5-μm thick) were stained with hematoxylin and eosin according to ref. 37. For immunostaining, sections were rehydrated and boiled for 10 min in 10 mM sodium citrate, pH 6; endogenous peroxidase activities were quenched by incubating with 0.3% H2O2 for 30 min. After permeabilization with 0.1% Tween-20 and blocking for 20 min in 2.5% horse serum (Vector Laboratories), sections were incubated overnight at 4 °C with primary antibodies diluted as indicated in Table S1. Mouse monoclonal antibodies were detected with Vectastain Elite ABC kits, and rabbit polyclonal antibodies were detected with ImmPRESS Reagent, (both Vector Laboratories) and DAB Substrate (Roche). Sections were counterstained with 0.5% methyl green (Sigma) in 0.1 M sodium acetate, dehydrated, and mounted with Permount (Fisher). Light microscopy pictures were taken on a Nikon Eclipse E600 microscope with a camera RT slider 2.3.0 and SPOT 4.1.1 (both Diagnostic Instruments).
For most of the immunochemical studies, mouse tissues were frozen in O.C.T.; 10-μm thick sections were fixed for 5 min with ice-cold acetone or with ice-cold methanol followed by five dips in acetone, and permeabilized with 0.1% Tween-20. Background staining for mouse antibodies was minimized with the Mouse-on-Mouse kit (Vector Laboratories). To detect BrdU, sections were pretreated with 1 N HCl for 10 min on ice, 2 N HCl for 10 min at room temperature followed by 10 min at 37 °C, and 0.1 M sodium borate (pH 8.5) for 12 min. Otherwise, sections were blocked with 2.5% horse serum for 1 h at room temperature and incubated overnight at 4 °C with primary antibodies at the dilutions indicated in Table S1. Alexa Fluor 488- and Alexa Fluor 568-conjugated secondary antibodies were used at a 1:200 dilution, and Alexa Fluor 555–conjugated streptavidin was diluted at 20 μg/mL (all from Molecular Probes, Invitrogen). After counterstaining with DAPI, sections were mounted with Prolong Gold antifade (Invitrogen), and images were recorded with an Axiovert 200M microscope using a 10× objective with an AxioCam MRm and an ApoTome (all from Zeiss).
Note Added in Proofs.
While the present manuscript was under revision, a report by Zhang et al. revealed that indeed both SUN1/2 and Nesprin 1/2 proteins are essential for neurogenesis and neuronal migration in mice (Neuron 64:173-187; October 29 2009).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AR050200, HL76839, HL86683, HL89781, and GM66152, March of Dimes Grant 6-FY2007-1012, and an Ellison Medical Foundation Senior Scholar Award.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908790107/DCSupplemental.
References
- 1.Stewart CL, Roux KJ, Burke B. Blurring the boundary: The nuclear envelope extends its reach. Science. 2007;318:1408–1412. doi: 10.1126/science.1142034. [DOI] [PubMed] [Google Scholar]
- 2.Zheng Y, Tsai MY. The mitotic spindle matrix: A fibro-membranous lamin connection. Cell Cycle. 2006;5:2345–2347. doi: 10.4161/cc.5.20.3365. [DOI] [PubMed] [Google Scholar]
- 3.Dechat T, et al. Nuclear lamins: Major factors in the structural organization and function of the nucleus and chromatin. Genes Dev. 2008;22:832–853. doi: 10.1101/gad.1652708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutchison CJ. Lamins: Building blocks or regulators of gene expression? Nat Rev Mol Cell Biol. 2002;3:848–858. doi: 10.1038/nrm950. [DOI] [PubMed] [Google Scholar]
- 5.Worman HJ, Fong LG, Muchir A, Young SG. Laminopathies and the long strange trip from basic cell biology to therapy. J Clin Invest. 2009;119:1825–1836. doi: 10.1172/JCI37679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson KL, Zastrow MS, Lee KK. Lamins and disease: Insights into nuclear infrastructure. Cell. 2001;104:647–650. [PubMed] [Google Scholar]
- 7.Burke B, Stewart CL. Life at the edge: The nuclear envelope and human disease. Nat Rev Mol Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- 8.Padiath QS, et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- 9.Vergnes L, Péterfy M, Bergo MO, Young SG, Reue K. Lamin B1 is required for mouse development and nuclear integrity. Proc Natl Acad Sci USA. 2004;101:10428–10433. doi: 10.1073/pnas.0401424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcántara S, et al. Regional and cellular patterns of reelin mRNA expression in the forebrain of the developing and adult mouse. J Neurosci. 1998;18:7779–7799. doi: 10.1523/JNEUROSCI.18-19-07779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta A, Tsai LH, Wynshaw-Boris A. Life is a journey: A genetic look at neocortical development. Nat Rev Genet. 2002;3:342–355. doi: 10.1038/nrg799. [DOI] [PubMed] [Google Scholar]
- 12.Wynshaw-Boris A. Lissencephaly and LIS1: Insights into the molecular mechanisms of neuronal migration and development. Clin Genet. 2007;72:296–304. doi: 10.1111/j.1399-0004.2007.00888.x. [DOI] [PubMed] [Google Scholar]
- 13.Visel A, Thaller C, Eichele G. GenePaint.org: An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32(Database issue):D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takamori Y, et al. Differential expression of nuclear lamin, the major component of nuclear lamina, during neurogenesis in two germinal regions of adult rat brain. Eur J Neurosci. 2007;25:1653–1662. doi: 10.1111/j.1460-9568.2007.05450.x. [DOI] [PubMed] [Google Scholar]
- 15.Arlotta P, et al. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. doi: 10.1016/j.neuron.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 16.Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- 17.Hatten ME. Central nervous system neuronal migration. Annu Rev Neurosci. 1999;22:511–539. doi: 10.1146/annurev.neuro.22.1.511. [DOI] [PubMed] [Google Scholar]
- 18.Sudarov A, Joyner AL. Cerebellum morphogenesis: The foliation pattern is orchestrated by multi-cellular anchoring centers. Neural Develop. 2007;2:26. doi: 10.1186/1749-8104-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayala R, Shu T, Tsai LH. Trekking across the brain: The journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 20.Gleeson JG, Walsh CA. Neuronal migration disorders: From genetic diseases to developmental mechanisms. Trends Neurosci. 2000;23:352–359. doi: 10.1016/s0166-2236(00)01607-6. [DOI] [PubMed] [Google Scholar]
- 21.Rice DS, Curran T. Role of the reelin signaling pathway in central nervous system development. Annu Rev Neurosci. 2001;24:1005–1039. doi: 10.1146/annurev.neuro.24.1.1005. [DOI] [PubMed] [Google Scholar]
- 22.Solecki DJ, Govek EE, Tomoda T, Hatten ME. Neuronal polarity in CNS development. Genes Dev. 2006;20:2639–2647. doi: 10.1101/gad.1462506. [DOI] [PubMed] [Google Scholar]
- 23.Salina D, et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 24.Wynshaw-Boris A, Gambello MJ. LIS1 and dynein motor function in neuronal migration and development. Genes Dev. 2001;15:639–651. doi: 10.1101/gad.886801. [DOI] [PubMed] [Google Scholar]
- 25.Vallee RB, Tsai JW. The cellular roles of the lissencephaly gene LIS1, and what they tell us about brain development. Genes Dev. 2006;20:1384–1393. doi: 10.1101/gad.1417206. [DOI] [PubMed] [Google Scholar]
- 26.Patterson K, et al. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15:600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- 28.Gieni RS, Hendzel MJ. Mechanotransduction from the ECM to the genome: Are the pieces now in place? J Cell Biochem. 2007;104:1964–1987. doi: 10.1002/jcb.21364. [DOI] [PubMed] [Google Scholar]
- 29.Fridkin A, Penkner A, Jantsch V, Gruenbaum Y. SUN-domain and KASH-domain proteins during development, meiosis and disease. Cell Mol Life Sci. 2009;66:1518–1533. doi: 10.1007/s00018-008-8713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burke B, Roux KJ. Nuclei take a position: Managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 31.Ji JY, et al. Cell nuclei spin in the absence of lamin B1. J Biol Chem. 2007;282:20015–20026. doi: 10.1074/jbc.M611094200. [DOI] [PubMed] [Google Scholar]
- 32.Nichols J, Evans EP, Smith AG. Establishment of germ-line-competent embryonic stem (ES) cells using differentiation inhibiting activity. Development. 1990;110:1341–1348. doi: 10.1242/dev.110.4.1341. [DOI] [PubMed] [Google Scholar]
- 33.Hanks M, Wurst W, Anson-Cartwright L, Auerbach AB, Joyner AL. Rescue of the En-1 mutant phenotype by replacement of En-1 with En-2. Science. 1995;269:679–682. doi: 10.1126/science.7624797. [DOI] [PubMed] [Google Scholar]
- 34.Kim E, et al. Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J Biol Chem. 1999;274:8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- 35.Yang SH, et al. Blocking protein farnesyltransferase improves nuclear blebbing in mouse fibroblasts with a targeted Hutchinson-Gilford progeria syndrome mutation. Proc Natl Acad Sci USA. 2005;102:10291–10296. doi: 10.1073/pnas.0504641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manipulating the Mouse Embryo: A Laboratory Manual. Cold Spring Harbor. NY: Cold Spring Harbor Laboratory Press; 2003. pp. 687–691. [Google Scholar]
- 37.Kuroda H, Fuentealba L, Ikeda A, Reversade B, De Robertis EM. Default neural induction: Neuralization of dissociated Xenopus cells is mediated by Ras/MAPK activation. Genes Dev. 2005;19:1022–1027. doi: 10.1101/gad.1306605. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.