Abstract
To examine the fundamental mechanisms governing neural differentiation, we analyzed the transcriptome changes that occur during the differentiation of hESCs into the neural lineage. Undifferentiated hESCs as well as cells at three stages of early neural differentiation—N1 (early initiation), N2 (neural progenitor), and N3 (early glial-like)—were analyzed using a combination of single read, paired-end read, and long read RNA sequencing. The results revealed enormous complexity in gene transcription and splicing dynamics during neural cell differentiation. We found previously unannotated transcripts and spliced isoforms specific for each stage of differentiation. Interestingly, splicing isoform diversity is highest in undifferentiated hESCs and decreases upon differentiation, a phenomenon we call isoform specialization. During neural differentiation, we observed differential expression of many types of genes, including those involved in key signaling pathways, and a large number of extracellular receptors exhibit stage-specific regulation. These results provide a valuable resource for studying neural differentiation and reveal insights into the mechanisms underlying in vitro neural differentiation of hESCs, such as neural fate specification, neural progenitor cell identity maintenance, and the transition from a predominantly neuronal state into one with increased gliogenic potential.
Keywords: RNA-Seq, splicing isoforms, unannotated transcripts, neuron, glial
Neural commitment and subsequent differentiation is a complex process. Although the complexity of RNAs expressed in neural tissues is very high (1, 2), a comprehensive analysis of the genes and RNA isoforms that are expressed during the different stages of neural cell differentiation is largely lacking. Such information is expected to be important for understanding mechanisms of neural cell differentiation and ultimately providing therapeutic solutions for neural degenerative diseases, such as Parkinson's and Alzheimer's disease.
Our current knowledge of the mechanisms involved in neural cell formation is derived mostly from studying neurogenesis in the developing embryos of animal models (3, 4). However, neurogenesis in animals is a complex process involving many different cell types that differentiate asynchronously. This heterogeneity, along with the relatively small number of cells that can be readily obtained, makes the analysis of the temporal differentiation of individual cell types extremely difficult. One solution is to analyze hESCs during in vitro differentiation to different stages of neural development, which can be performed using a relatively large numbers of cells (5–9). Analysis of the transcriptome in these cells is expected to provide insights into the mechanisms and pathways involved in early cell fate specification, such as the acquisition of neurogenic potential and the transition to gliogenic potential, which may ultimately be extremely useful for pharmacologic screening and neurodegenerative disease therapies.
Many high-throughput methods have been used previously to study global transcription (10–14). The recent development of massively parallel sequencing of short reads derived from mRNA (RNA-Seq) makes it possible to globally map transcribed regions and quantitatively analyze RNA isoforms at an unprecedented level of sensitivity and accuracy (15–22). Although the use of short reads enables detection of transcribed regions and spliced adjacent exons, it has limitations. In particular, the relationship of nonadjacent exons and multiple exons within the same transcript cannot be deduced.
In this study we combined the strengths of several massively parallel sequencing technologies, including short Illumina single and paired-end reads (sequence reads from both ends of cDNA fragments; 35-bp reads) and longer Roche 454 FLX and Titanium sequencing reads (250–450-bp reads) to discern transcript structure and analyze transcriptome complexity at an unprecedented level (21, 23, 24). We applied these technologies to the analysis of early stages of neural differentiation of hESCs. Our results revealed an extraordinary degree of stage-specific transcription and splicing. From more than 150 million uniquely mapped sequence reads, we found thousands of unannotated transcriptionally active regions (TARs) and unannotated isoforms. Some unannotated TARs and splice isoforms are transcribed only at particular stages, implying functional roles in specific steps of neural differentiation. Moreover, we describe a phenomenon we call isoform specialization, whereby splicing isoform diversity is the highest in undifferentiated hESCs and decreases in cells undergoing neural differentiation. Finally, the characterization of dynamic changes of gene transcription levels has provided important insights into the in vitro neural differentiation of hESCs with regard to neural specification, neural progenitor identity maintenance, and the transition from a predominantly neuronal nature to one with increased gliogenic potential.
Results
RNA-Seq at Specific Stages of Neural Differentiation of hESCs.
We characterized changes in the transcriptome profiles during early human neural differentiation using H1 hESC cultures. Two differentiation strategies were used (Fig. 1, Materials and Methods, and SI Text). In approach A, the hESC H1 line was differentiated and cultured using feeder-free chemically defined adherent cell culture system through three stages: N1, an initiation stage; N2, a neural progenitor cell (NPC) stage that produces only neurons upon further differentiation; and N3, which produces both neurons and glial cells (Fig. 1 A and B) (7, 8, 25). In approach B, neural progenitors (N2-B) were generated from undifferentiated H1 hESCs via embryoid body-like neurosphere formation (SI Text) (9). In each case, we used standard protocols involving bone morphogenic protein signaling antagonists (Noggin) and basic fibroblast growth factor (bFGF) (Materials and Methods).
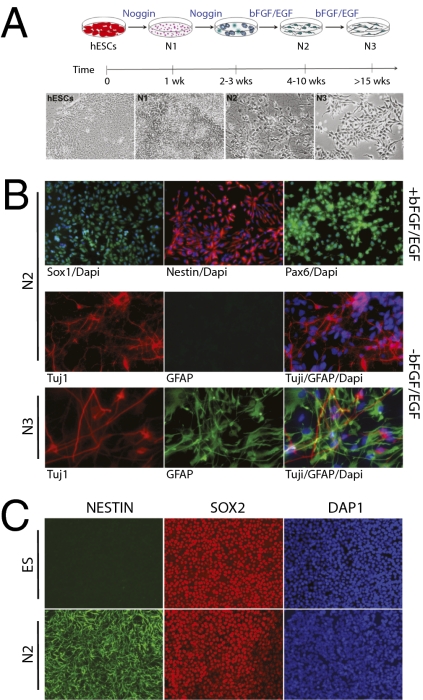
Fig. 1.
Characterization of neural differentiation cell cultures. (A) Schematic representation of the neural differentiation procedure and four stages by approach A: hESCs, N1, N2, and N3. (B) Immunofluorescence labeling of genes specifically expressed in N2 and N3 cells (H1 hESCs) prepared by approach A with or without growth factors. SOX1, NESTIN, and PAX6 are expressed in N2 cells with growth factors bFGF/EGF. TUJ1 is expressed in N2 cells without growth factors; GFAP is not expressed. GFAP is expressed in N3 cells after growth factor withdrawal, whereas TUJ1 expression is still visible. Blue: nuclei are stained by DAPI. (C) Immunostaining characterization of cell cultures (H1 hESCs) by approach B. SOX2 is expressed in hESCs but not NESTIN. Both NESTIN and SOX2 are expressed in N2 cells. Higher-resolution photos are in SI Text.
According to qualitative and quantitative analyses, the differentiation protocols were highly reproducible. The derived cell populations from each preparation were characterized by both immunoassays and FACS analysis for a large variety of markers to ensure that the cell cultures were highly homogeneous at the various stages (Fig. 1 and SI Text). (i) Undifferentiated hESCs (present in both approaches A and B) expressed all hESC surface antigens (e.g., TRA-1-60/81 and SSEA4) as well as transcription factors OCT4 and SOX2 (Fig. 1C and SI Text). (ii) N1 initiation stage cells (present only in approach A) lost TRA-1-60/81 expression but were still positive for SSEA4, although at a lower level, and began to express SSEA1. They also expressed OCT4 at a low level (SI Text). (iii) N2 NPCs generated by approach A lost OCT4 as well as SSEA4 expression (SI Text) but expressed NESTIN, PAX6, and SOX1 (Fig. 1B). The cells showed a typical morphology of NPCs: bipolar with small soma (Fig. 1A and SI Text). Glial fibrillary acidic protein (GFAP) was not expressed (Fig. 1B and SI Text). Upon withdrawal of growth factors, the N2 cells predominantly differentiated into neurons rather than glial cells, as indicated by the expression of neuronal marker TUJ1 (Fig. 1B). Similarly, >95% of N2 neural precursors generated by approach B expressed an array of neural markers (neuroepithelial marker PAX6, neural stem cell markers NESTIN, and SOX2, as well as neuronal stem/precursor markers MUSASHI) (Fig. 1C and SI Text). The N2 cell populations were negative for pluripotent hESC markers such as OCT4 (<0.01%; SI Text). (iv) N3 stage cells (only present in approach A) exhibited distinct morphology (Fig. 1A and SI Text), and GFAP was expressed in these cells (Fig. 1B). After bFGF/EGF withdrawal, more glial cells than neurons were generated (Fig. 1B). These events we observed in vitro are reminiscent of in vivo neurodevelopment, in which neurogenesis occurs before the onset of gliogenesis (25, 26). This phenomenon is also observed in the H7 hESC line (SI Text).
Complex Gene Structure Revealed Through the Integration of Short, Long, and Paired-End Sequences.
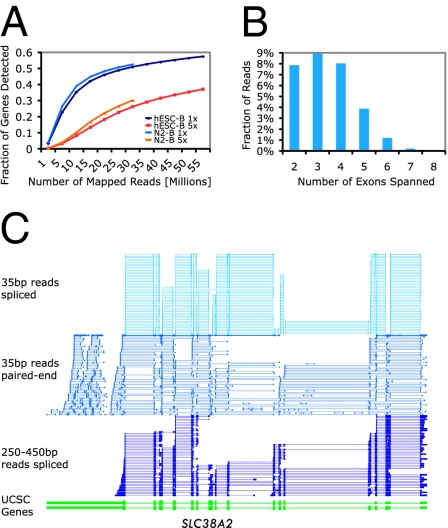
To characterize the transcription of cells at the specific differentiation stages, we generated a combination of 35-bp single reads, 35-bp paired-end reads, and 250–450-bp long reads. The paired-end reads were from cDNA fragments of different lengths, ≈300 bp, 300–600 bp, and 600–1,000 bp. A total of 140, 15, and 1.5 million uniquely mapped single, paired-end, and long reads, respectively (summarized in Table S1), were generated from two to three biologic replicates from each of the differentiation stages (Spearman correlations: 0.94–0.97). The fraction of genes detected at 1-fold average coverage approached saturation for the sequencing of hESC-B and N2-B cells (Fig. 2A).
Fig. 2.
Overview of transcript characterization by RNA-Seq. (A) Fraction of genes detected as a function of read depth. (B) Number of exons spanned by 450-bp reads. (C) Transcript complexity revealed by integrating short, long, and pair-end sequence information. Only the spliced reads for single-end and long reads are shown. For paired-end reads the same RNA fragments are shown as two vertical bars connected by a line.
We achieved extensive sequence depth primarily with the 35-bp single reads; the paired-end and 250–450-bp reads provided longer-range exon connectivity information and aided in defining complex splice isoforms. Longer reads, particularly 450-bp reads, can link up to eight exons (see Fig. 2B for distribution). Fig. 2C illustrates the structure of a 16-exon gene that was constructed using a combination of the sequencing technologies.
Identification of Unannotated Transcribed Regions and Their Connectivity Using Paired-End Reads.
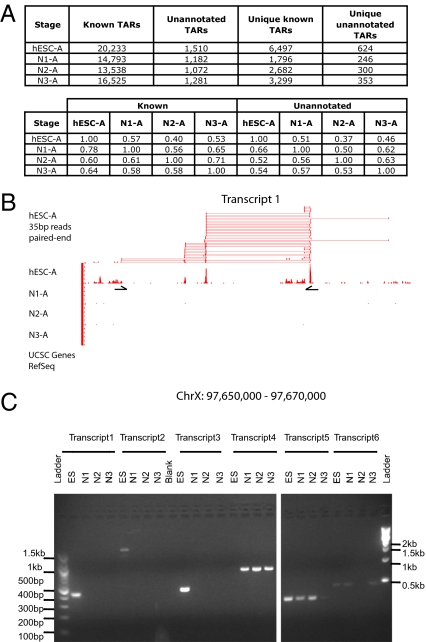
Consistent with our previous studies (10, 14, 27), thousands of unannotated TARs were identified. Specifically, if a TAR overlapped with University of California, Santa Cruz (UCSC) gene annotation it was categorized as “known,” and if there was no overlap it was classified as “unannotated.” Ninety percent of unannotated TARs were validated by RT-PCR from a random sample of 40 TARs identified from the different stages (SI Text). We also intersected TARs discovered by our RNA-Seq approach with previously published TARs expressed in the liver that were identified by tiling microarrays (10). Our study found a large number of hESC RNA that were not identified by the tiling microarray study (SI Text), consistent with previous observations that RNA-Seq has a higher sensitivity and dynamic range (19). Interestingly, we also found that a large number of unannotated TARs (35–65%) were specific for each stage; 624, 246, 300, and 353 unique unannotated TARs were found for hESC, N1, N2, and N3 stage cells, respectively (Fig. 3A, Top), raising the possibility that these might have stage-specific functions. Sequences and signal track files can be found in the Gene Expression Omnibus (GEO; accession number GSE20301).
Fig. 3.
Stage-specific unannotated TARs and their connectivity by paired-end reads. (A) (Upper) The number of unannotated, known, and unique TARs found at each stage. TARs that overlap with a UCSC gene are classified as “known.” Those that have no overlap with a UCSC gene annotation are called “unannotated.” TARs that do not overlap with TARs in other differentiation stages are called “unique” to that particular stage. (Lower) Fractions shared between stages for known TARs and unannotated TARs, respectively. (B) The transcript structure of an unannotated transcript (Transcript 1) that is uniquely transcribed in hESCs is reconstructed using paired-end reads. (C) RT-PCR validation of unannotated transcripts identified by groups of paired-end reads. Transcript 1 and its RT-PCR primer set are shown in B. Transcripts 1–3 are specifically expressed in hESCs, transcript 4 in N1–N3 cells, transcript 5 in ES-N2 cells, and transcript 6 in ES, N1, and N3 cells.
As expected, the majority of the paired-end reads fell within the same known exons. However, a small fraction of paired-end reads linked unannotated TARs to either UCSC-known annotated genes (0.35%, 0.46%, 1.03%, and 0.58% for hESC, N1, N2, and N3, respectively) or to other unannotated TARs (0.36%, 0.38%, 1.50%, and 0.89% for hESC, N1, N2, and N3, respectively). Although the percentage of “linking” reads was low, their large number allowed for unambiguous connection of TARs, and unannotated spliced gene structures could be identified by an overlapping group of paired-end reads. Fig. 3B shows an unannotated transcript with at least five exons that was uniquely transcribed in hESCs and accurately constructed using a group of overlapping paired-end reads. This transcript and expression pattern was validated by RT-PCR (Fig. 3C). Twelve such multiexonic unannotated transcripts were further examined using RT-PCR; 11 were validated and 6 were verified to be stage specific (Fig. 3C).
Alternative Splicing During Early Neural Differentiation of hESCs.
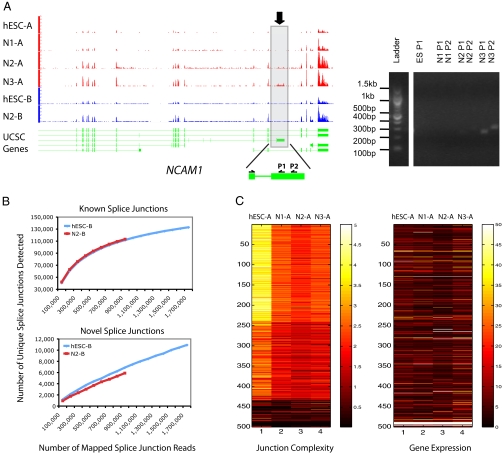
In addition to cell type–specific gene expression, such as for OCT4 and GFAP, we observed many interesting differentiation-stage-specific alternative splicing isoforms. For instance, an isoform of neural cell adhesion molecule 1 (NCAM1) was prevalent at the N3 stage, low level at the N2 stage, but not detectable at the N1 and hESC stages (Fig. 4A). In addition, an isoform of serine/threonine kinase 2 (SLK) was specifically transcribed in hESCs (SI Text), consistent with an independent observation by Gage and colleagues (28). Although the number of known splice junctions detected in our study was near saturation (Fig. 4B, Top), the number of unannotated splice junctions continued to increase with read depth, indicating that there are many more unannotated isoforms yet to be discovered (Fig. 4B, Bottom).
Fig. 4.
Splicing analysis. (A) One of the transcript isoforms of neural cell adhesion molecule 1 (NCAM1) (marked by a rectangle and arrow) is primarily expressed in N3 and very weakly at N2 but not at N1 and hESC stages. The y axis of the RNA-Seq signal tracks represents the read density normalized by the number of mapped reads per million for each cell type. Two sets of RT-PCR primers were designed on the alternative exon and the adjacent constant exon, which generated two products of slightly different sizes. The DNA ladder and the RT-PCR products were from the same gel. (B) (Upper) The number of known splice junctions detected nears saturation. (Lower) The number of unannotated splice junctions does not saturate at this read depth. (C) Splicing diversity is the highest in hESCs and decreases when cells commit to neural differentiation. The top 500 highly expressed genes shown here were clustered by splice junction diversity (k-means clustering, k = 3) (see SI Text for details). The splice junction diversity value was defined as the number of unique splice junctions detected in the composite gene model given all of the mapped splice junction reads; thus the junction diversity values were normalized for the number of annotated splice junctions in the composite gene model and the number of mapped reads per million. Splice junction diversity is independent of transcript abundance for this set of genes.
Of particularly high interest is how splice isoform diversity changes as a function of cell differentiation, which has not been examined previously. We therefore quantified the number of unique splice junctions per composite gene model for each differentiation stage (see SI Text for details). To analyze the splice junction diversity, the 500 most highly transcribed genes were selected on the basis of the sum of their transcription values in the four stages. These abundant transcripts were chosen because they provide large numbers of reads and allow for significant splicing differences to be identified. Our analysis revealed higher isoform diversity in hESCs compared with the neural stages (the median of the junction values for hESC, N1, N2, and N3 are 3.1, 2.2, 1.9, and 2.1, respectively). Interestingly, within the chosen set, this observation is independent of transcript abundance (Fig. 4C). These data suggest that isoform diversity simplifies during differentiation, a process we have named isoform specialization.
Distinct Classes of Genes Are Differentially Transcribed During Neural Differentiation.
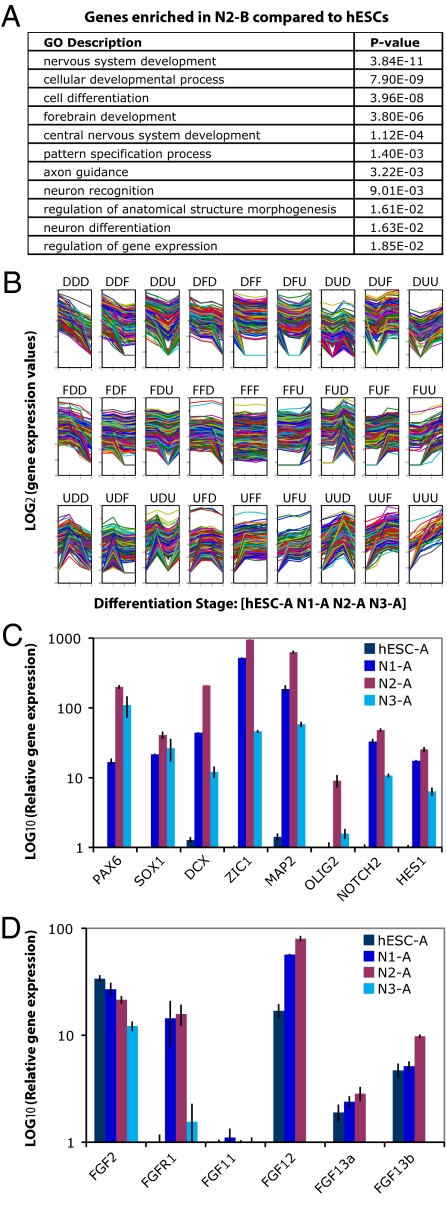
We next examined the types of genes that exhibited differences in transcription using Gene Ontology analysis. We found that genes involved in nervous system development, neuron differentiation, brain development, regulation of gene expression, and pattern specification were significantly overrepresented among up-regulated genes in N2-B cells compared with hESCs (Fig. 5A).
Fig. 5.
Dynamics of gene expression during neural differentiation. (A) The enriched Gene Ontology (GO) categories among up-regulated genes (>2-fold) in N2 cells compared with hESCs. (B) Quantification of dynamic transcriptome changes during neural differentiation process [x axis: differentiation stages hESC, N1, N2, and N3; y axis: log2(gene expression values by RNA-Seq)]. Twenty-seven patterns were identified from clustering the expression changes of the UCSC gene annotation set. U, up-regulation; D, down-regulation; F, flat (<2-fold change). (C) qPCR validation of the genes that show the highest expression at N2. y axis: log2(relative gene expression level for each stage normalized using HPRT). (D) qPCR validation of FGF family gene expression (Note: qPCRs were performed for two isoforms of FGF13.) Expression of FGF12 and FGF13a was not detected in N3 by qPCR.
Genes were clustered according to the dynamic transcriptome changes between the four stages (ES→N1, N1→N2, and N2→N3) (Fig. 5B). We found that a group of genes containing SOX1, SOX2, PAX6, MAP2, DCX, ZIC1, NOTCH2, HES1, and OLIG2 had the highest transcript levels at the N2 stage and validated the relative transcript levels by quantitative PCR (qPCR) (Fig. 5C). H7 hESCs showed similar gene expression patterns by qPCR (SI Text). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that the neuroactive ligand–receptor interaction pathway was enriched among genes that were up-regulated at the N1 and/or N2 stage but down-regulated at the N3 stage [these genes are marked by an asterisk (*) in SI Text]. A wide variety of receptor genes were transcribed in N1 and N2 cells, suggesting that these cells may be capable of differentiating into glutamatergic, GABAergic, dopaminergic, cholinergic, adrenergic, and serotoninergic neuronal subtypes. Although the overall proliferation rate between N2 and N3 cells in cultures with bFGF/EGF supplement is similar, the receptors were diminished at the N3 stage. This is consistent with the fact that the N3 stage cells expressed glial markers and had a propensity to differentiate into glial cells after bFGF/EGF withdrawal (7).
A coordinated interplay among signaling pathways, such as Wnt and FGF is critical for neural specification (3). Components of the Wnt pathway previously implicated in noncanonical Wnt signaling [FZD5 (frizzled homolog 5, Drosophila), WNT5A, and WNT5B] were found to be down-regulated upon neural differentiation. This is consistent with Wnt pathway function in maintaining the undifferentiated state of stem cells. Interestingly, a group of FGF family genes exhibited increased expression at the N2 stage and then decreased at the N3 stage. This group included FGF11, FGF13, and FGF14, which do not bind to FGF receptor (Fig. 5D); their roles during neural differentiation are not well understood.
Discussion
Our study revealed a high level of transcriptome complexity and dynamics during early neural differentiation of hESCs. Alternative splicing has been suggested as a driving force for the evolution of higher eukaryotic phenotypic complexity (29), and it has been shown recently that ≈94% of human genes undergo alternative splicing (30). Previous studies have shown that massively parallel sequencing technology provides the ability to monitor spliced isoforms at the level of individual splice junctions (15, 30). Our work incorporates the use of paired-end sequencing and demonstrates that this approach in conjunction with long reads can elucidate multiple exon connections and thereby reconstruct transcribed regions.
Importantly, our study demonstrated that greater splice junction diversity is present in hESCs relative to cells undergoing neural differentiation (Fig. 4 B and C). We suggest that this high transcript diversity contributes to the pluripotency of hESCs. Upon differentiation, more specialized transcripts are used, a process that we call isoform specialization. A previous study (31) reported elevated global transcription level in murine ESCs compared with NPCs; increased transcription in ESCs would support a larger number of isoforms.
RNA-Seq also enabled the detection of unknown RNAs (unannotated TARs) (Fig. 3A). A larger fraction of these unannotated TARs were transcribed in a stage-specific fashion compared with the annotated mRNAs (Fig. 3A). It is possible that at least some of the unannotated TARs may play an important role in specifying cell differentiation. The largest number of unannotated TARs was observed in hESCs, consistent with our conclusion that the transcriptome complexity is the greatest in pluripotent hESCs.
One important advantage of massively parallel sequencing is that it is more quantitative than other methods (19). RNA-Seq can accurately measure gene transcription changes at a genome-wide scale, even for low-abundance transcription factors. Thus, these data are a valuable reference data set for researchers studying this process.
Our results provide important insights into the hESCs neural differentiation in vitro. Specifically, we have revealed aspects of neural specification, neural progenitor identity maintenance, and the transition from neurogenesis to gliogenesis. Our data suggest the temporal order of transcription of the key transcription factors SOX1 and PAX6 during human neural specification. SOX1 is a member of the SOXB1 transcription factor family that plays important roles in neuroectodermal lineage commitment and maintenance (32, 33). PAX6 is a highly conserved transcription factor essential for central nervous system development (34). The temporal order of their transcription and their roles in human neuroectodermal specification are not fully understood. In mice SOX1 was found to be the earliest transcribed neural marker, preceding PAX6. PAX6 is first transcribed in radial glial cells during the differentiation of mouse ESCs (35), and it has been reported to be involved in the progression of neuroectoderm toward radial glia (36). However, in our experiments using hESCs, PAX6 mRNAs appeared before SOX1 mRNA, consistent with the immunostaining observations of Gerrard et al. (7). Thus, PAX6 may have an earlier role in neural lineage choice in human ESCs than in mouse ESCs.
The transcription of a wide variety of receptor genes at the N1 and N2 stages indicates that if the proper differentiation conditions are applied, these cells could potentially differentiate into glutamatergic, GABAergic, dopaminergic, cholinergic, adrenergic, and serotoninergic neuronal subtypes. Two possibilities can explain why these neuroactive ligand–receptors are not retained in N3 cultures. First, the receptors may be lost in N3 cells owing to cell death and/or less proliferation of proneuronal cells; the proneuronal cells would then be gradually replaced by the proglial cells. However, this cannot explain the complete absence of GFAP when neuronal differentiation is induced at an earlier stage. The second possibility is that a series of gene repression and activation events lead to the transition of the cells from a proneuronal nature to a proglial nature. Our finding that FGF family genes, including nonFGF-receptor-binding FGF11, FGF13, and FGF14, increase at the N2 stage and decrease at the N3 stage (Fig. 5D) raises the possibility that modifying their levels may help to maintain hESC-derived neural cells at the neuronal stage.
Overall, our approach can serve as a template for the investigation of dynamic temporal or spatial transcriptome changes during various developmental processes. Future improvements of sequencing technologies, including longer reads, higher throughput, and reduced cost will aid in the definition of transcriptomes and alternative splicing in specific temporal and spatial contexts.
Materials and Methods
hESC Culture and Neural Differentiation Using Two Approaches.
Approach A.
H1 hESCs were cultured in Matrigel-coated plates in mouse embryonic fibroblast conditioned medium supplemented with 8 ng/mL bFGF as previously described (37). Cells were propagated at a 1:3 ratio by treatment with 200 U/mL collagenase IV and mechanical dissection. Neural differentiation was carried out as previously described (7). Briefly, hESCs were split with EDTA at 1:5 ratios into culture dishes coated with poly-L-lysine/laminin (Sigma-Aldrich) and cultured in N2B27 medium supplemented with 100 ng/mL mouse recombinant Noggin (R&D Systems). At this stage, cells were defined as passage 1 (P1), and N1 cells were collected at Day 11 of the differentiation. Cells of P1 and P2 were split by collagenase into small clumps, similar to hESC culture, and continuously cultured in N2B27 medium with Noggin. From P3, cells were plated at the density of 5 × 104 cells/cm2 after disassociation by TrypLE express (Invitrogen) into single-cell suspension, and cultured in N2B27 medium with the addition of 20 ng/mL bFGF and EGF. Cells can be maintained under this culture condition for a long time with a stable proliferative capacity. N2 cells were collected at P9 and N3 at P22. To induce postmitotic cell types, bFGF and EGF were withdrawn, and neural progenitors were continually cultured in N2B27 for 7 days. Cells from the different stages were analyzed for homogeneity by flow cytometry analysis. Cells from N2 and N3 were stained with anti-SOX1 (Abcam), SOX2 (Abcam), and MUSASHI (Chemicon) antibodies; 10–20,000 cells were analyzed (see SI Text for details).
Approach B.
All experiments involving hESCs were approved by the Yale Embryonic Stem Cell Oversight Committee. hESC line H1 (WA01, WiCell) was maintained in undifferentiated state by culturing on Matrigel-coated plates (BD) in feeder-free and serum free, component-defined conditions. Briefly, the cells were cultured in DMEM/F12 medium (Invitrogen) supplemented with 1% MEM-nonessential amino acids (Invitrogen), 1 mM L-glutamine, 1% penicillin-streptomycin, 50 ng/mL bFGF (FGF-2) (Millipore), 1× N2 supplements, and 1× B27 supplements (Invitrogen) (38), with daily media change. H1 cells were passaged every 4–6 days by dissociation with 1 mg/mL collagenase IV (Invitrogen). The hESCs used were between passages 30 and 70 with normal karyotype and expressed conventional hESC markers. hESCs were differentiated by neural sphere formation with some modifications of previously published protocols (9). Cells were fixed and analyzed by standard fluorescent immunocytochemical techniques. See SI Text for details.
RNA Sequencing.
Construction of Solexa sequencing library.
mRNA samples were extracted and double-polyA purified from cell cultures using Oligotex Direct mRNA Kits followed by Oligotex mRNA Kits, according to the manufacturer's instructions (Qiagen). mRNA (500 ng) was used in each sequencing library. mRNA was fragmented using 10× Fragmentation Buffer (Ambion), and double-stranded cDNA was synthesized using SuperScript II (Invitrogen) RT and random primers. DNA sequencing followed the instructions of the mRNA-Sequencing Sample Prep Kit (Illumina) as previously described (16).
454 sequencing library preparation.
mRNA was prepared as described above. mRNA samples (200–500 ng) were heat fragmented. Single-stranded cDNA library was synthesized, and adapters were ligated (SI Text) and sequenced using the emPCR II Kit (Amplicon A) and on the 454 Genome Sequencer FLX instrument according to the manufacturer's instructions. GS FLX Titanium cDNA libraries were prepared and sequenced at the 454 Life Sciences Sequencing Centre.
RT-PCR and real-time quantitative RT-PCR.
RT-PCR was carried out as described previously (12, 14). RNA without RT treatment was used as negative control. See SI Text for primers and details.
Bioinformatic analyses.
Please see SI Text.
Supplementary Material
Acknowledgments
We thank S. Marjani for critical reading and editing of the manuscript; H. Monahan, A. Urrutikoetxea-Uriguen, G. Zhong, K. Nelson, and P. Zumbo for technical assistance; and W. Zhong, J. Rozowsky, J. Du, and T. Gianoulis for discussions. J.Q.W., A.S, S.W., M.G. and M.S. were supported by grants from the National Institutes of Health and the State of Connecticut. P.N. and W.C. were partially supported by a grant from the Institute on Governance Trust (IOG).
Footnotes
The authors declare no conflict of interest.
Data deposition: Data were submitted to Gene Expression Omnibus (GEO; accession number GSE20301).
This article contains supporting information online at www.pnas.org/cgi/content/full/0914114107/DCSupplemental.
References
- 1.Nelson SB, Sugino K, Hempel CM. The problem of neuronal cell types: A physiological genomics approach. Trends Neurosci. 2006;29:339–345. doi: 10.1016/j.tins.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Watakabe A, Komatsu Y, Nawa H, Yamamori T. Gene expression profiling of primate neocortex: Molecular neuroanatomy of cortical areas. Genes Brain Behav. 2006;5(Suppl 1):38–43. doi: 10.1111/j.1601-183X.2006.00193.x. [DOI] [PubMed] [Google Scholar]
- 3.Suter DM, Krause KH. Neural commitment of embryonic stem cells: Molecules, pathways and potential for cell therapy. J Pathol. 2008;215:355–368. doi: 10.1002/path.2380. [DOI] [PubMed] [Google Scholar]
- 4.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: Lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- 6.Reubinoff BE, et al. Neural progenitors from human embryonic stem cells. Nat Biotechnol. 2001;19:1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- 7.Gerrard L, Rodgers L, Cui W. Differentiation of human embryonic stem cells to neural lineages in adherent culture by blocking bone morphogenetic protein signaling. Stem Cells. 2005;23:1234–1241. doi: 10.1634/stemcells.2005-0110. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz PH, Brick DJ, Stover AE, Loring JF, Müller FJ. Differentiation of neural lineage cells from human pluripotent stem cells. Methods. 2008;45:142–158. doi: 10.1016/j.ymeth.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen MA, Itsykson P, Reubinoff BE. Neural differentiation of human ES cells. Curr Protoc Cell Biol. 2007 doi: 10.1002/0471143030.cb2307s36. Chapter 23:Unit 23.7. [DOI] [PubMed] [Google Scholar]
- 10.Bertone P, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- 11.Carninci P, et al. FANTOM Consortium; RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 12.Wu JQ, et al. Large-scale RT-PCR recovery of full-length cDNA clones. Biotechniques. 2004;36:690–696. doi: 10.2144/04364DD03. 698–700. [DOI] [PubMed] [Google Scholar]
- 13.Temple G, et al. MGC Project Team The completion of the Mammalian Gene Collection (MGC) Genome Res. 2009;19:2324–2333. doi: 10.1101/gr.095976.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu JQ, et al. Systematic analysis of transcribed loci in ENCODE regions using RACE sequencing reveals extensive transcription in the human genome. Genome Biol. 2008;9:R3. doi: 10.1186/gb-2008-9-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 16.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 17.Cloonan N, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 18.Nagalakshmi U, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilhelm BT, et al. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 21.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 22.Sultan M, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 23.Emrich SJ, Barbazuk WB, Li L, Schnable PS. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugarbaker DJ, et al. Transcriptome sequencing of malignant pleural mesothelioma tumors. Proc Natl Acad Sci USA. 2008;105:3521–3526. doi: 10.1073/pnas.0712399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elkabetz Y, Studer L. Human ESC-derived neural rosettes and neural stem cell progression. Cold Spring Harb Symp Quant Biol. 2008;73:377–387. doi: 10.1101/sqb.2008.73.052. [DOI] [PubMed] [Google Scholar]
- 26.Temple S. The development of neural stem cells. Nature. 2001;414:112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 27.Rozowsky J, et al. Novel transcribed regions in the human genome. Cold Spring Harb Symp Quant Biol. 2006;71:111–116. doi: 10.1101/sqb.2006.71.054. [DOI] [PubMed] [Google Scholar]
- 28.Yeo GW, et al. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLOS Comput Biol. 2007;3:1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 30.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Efroni S, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kan L, et al. Sox1 acts through multiple independent pathways to promote neurogenesis. Dev Biol. 2004;269:580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Zhao S, Nichols J, Smith AG, Li M. SoxB transcription factors specify neuroectodermal lineage choice in ES cells. Mol Cell Neurosci. 2004;27:332–342. doi: 10.1016/j.mcn.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Osumi N, Shinohara H, Numayama-Tsuruta K, Maekawa M. Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells. 2008;26:1663–1672. doi: 10.1634/stemcells.2007-0884. [DOI] [PubMed] [Google Scholar]
- 35.Mori T, Buffo A, Götz M. The novel roles of glial cells revisited: The contribution of radial glia and astrocytes to neurogenesis. Curr Top Dev Biol. 2005;69:67–99. doi: 10.1016/S0070-2153(05)69004-7. [DOI] [PubMed] [Google Scholar]
- 36.Suter DM, Tirefort D, Julien S, Krause KH. A Sox1 to Pax6 switch drives neuroectoderm to radial glia progression during differentiation of mouse embryonic stem cells. Stem Cells. 2008;27:49–58. doi: 10.1634/stemcells.2008-0319. [DOI] [PubMed] [Google Scholar]
- 37.Xu C, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 38.Yao S, et al. Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc Natl Acad Sci USA. 2006;103:6907–6912. doi: 10.1073/pnas.0602280103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.