Abstract
In vertebrate somitogenesis, the expression of segmentation clock genes oscillates and the oscillation is synchronized over nearby cells. Both experimental and theoretical studies have shown that the synchronization among cells is realized by intercellular interaction via Delta–Notch signaling. However, the following questions emerge: (i) During somitogenesis, dynamic rearrangement of relative cell positions is observed in the posterior presomitic mesoderm. Can a synchronized state be stably sustained under random cell movement? (ii) Experimental studies have reported that the synchronization of cells can be recovered in about 10 or fewer oscillation cycles after the complete loss of synchrony. However, such a quick recovery of synchronization is not possible according to previous theoretical models. In this paper, we first show by numerical modeling that synchronized oscillation can be sustained under random cell movement. We also find that for initial perturbation, the synchronization of cells is recovered much faster and it is for a wider range of reaction parameters than the case without cell movement. When the posterior presomitic mesoderm is rectangular, faster synchronization is achieved if cells exchange their locations more with neighbors located along the longer side of the domain. Finally, we discuss that the enhancement of synchronization by random cell movement occurs in several different models for the oscillation of segmentation clock genes.
Keywords: zebrafish, somitogenesis, Delta–Notch, mathematical models
In vertebrate development, somites bud off from the anterior end of the tissue not yet differentiated to somites, called the presomitic mesoderm (PSM), one by one moving posteriorly. The time interval between the formation of one somite and the next is almost constant during somitogenesis, and it is species-specific. In the PSM, there are segmentation clock genes with oscillating expression, and the timing of segmentation is considered to be controlled by the oscillatory expression of these genes because their period of oscillation is very close to the period of segmentation (1–5).
The oscillatory expression of the segmentation clock genes is known to be caused by the negative feedback regulation by their own products (6–8). Neighboring cells are in contact with each other (9, 10). In the PSM, oscillatory expressions are synchronized among neighboring cells. This synchronized oscillation is necessary for normal segmentation, and disruption of the synchronization results in a defective somite boundary (11–13).
Theoretical models of segmentation in vertebrates have been developed to explain the spatiotemporal periodicity of the segmentation process (14–18), the oscillatory expression of segmentation clock genes (19–24), and the wave-like gene expression observed in the anterior PSM (23, 25–29). Previous theoretical studies have also addressed mechanisms of synchronization of the segmentation clock between cells (13, 24, 25, 30). In zebrafish, the synchronization of the segmentation clock is realized by intercellular interaction via Delta–Notch signaling (11–13, 30, 31). Previous models have confirmed that this interaction successfully leads to the synchronization of oscillation among cells (24, 25, 30).
However, previous analyses did not answer the following questions: (i) In the actual segmentation process, dynamic rearrangement of relative cell positions due to cell proliferation and movement is observed in the PSM (12, 32, 33). Can the synchronization of the segmentation clock be achieved and stably sustained under such random cell movement? (ii) Cells can restore synchronization within about 10 or fewer oscillation cycles after the complete loss of synchrony caused by, for example, N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) treatment, which blocks Delta–Notch signaling within a cell (13). However, in previous models, recovery of the synchronization takes a long time (e.g., tens or hundreds of cycles) after a disturbance in the initial phase of oscillation. As reported by Tiedemann et al. (25), when the disturbance is somewhat large, different patches of cells are synchronized to different phases and these patches persist for a long time. This tendency has been observed in different models (see SI Text). What mechanisms can cause the quick recovery of synchronization after external perturbations?
In this paper, we first show by numerical modeling that a stable synchronized state can be sustained under random cell movement. In addition, we show that the synchronization is recovered faster after external perturbations and it is achieved for wider ranges of reaction parameters than is the case without cell movement. Moreover, there exists an optimal magnitude of anisotropy in the direction of cell movement that allows synchronization to be achieved most quickly for a given PSM geometry (e.g., its aspect ratio). Finally, we confirm that the enhancement of synchronization by random cell movement does not depend on the detailed modeling of molecular events.
Model
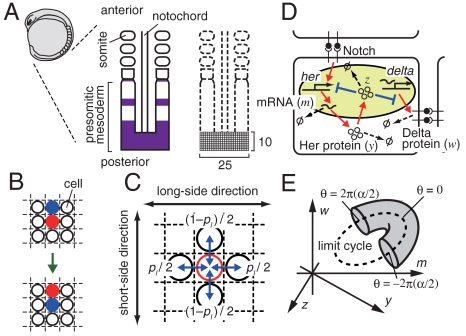
We represent the posterior part of the PSM as a two-dimensional lattice (Fig. 1A). Each cell in the lattice is identified by index j (j = 1,2,…,N). In our simulation, the lattice size was chosen as 25 × 10 (N = 250) unless otherwise indicated, because the tailbud region of the PSM is approximately 250 × 100 μm2 (12) and the diameter of each cell is approximately 10 μm.
Fig. 1.
Model scheme for the segmentation clock in vertebrate somitogenesis. (A) The presomitic mesoderm (PSM) and the two-dimensional lattice representing the posterior part of the PSM. The purple area indicates the expression of the her gene. (B) Exchange of location between two cells. (C) Each cell exchanges its location with one of its two neighbors in the long-side direction of the lattice with probability pl/2, and it exchanges its location with one of its two neighbors in the short-side direction of the lattice with probability (1 - pl)/2. (D) Negative feedback regulation of the her gene in a cell and intercellular interaction between neighboring cells via Delta–Notch signaling. Production, activation, and transport are represented by red arrows, whereas suppression is shown by blue lines ending with perpendicular bars. Decay of mRNA and proteins is denoted by arrows leading to symbol Ø. (E) Schematic representation of how an initial condition was prepared. The circle drawn with the broken line represents the limit cycle with all the N cells perfectly synchronized in the phase space of m, y, z, and w (Appendix). Initial states of cells are distributed in the shaded region that wraps a part of the limit cycle (see Model and Appendix for details).
We adopt a Neumann neighborhood—each cell has four nearest neighbors. To model random cell movement in the PSM, we let the cells in the lattice exchange their locations with one of their four neighbors at random times (Fig. 1B). For a system with N cells, the probability that an exchange of location between a pair in N cells occurs during a small time interval Δt is assumed to be λNΔt. We chose the value of coefficient λ such that each cell experiences an exchange of its location with one of its neighbors within 5–20 min on average, which is based on the fact that a cell in the posterior part of the chick PSM moves at 1.04 ± 0.33 μm/ min (32).
Let the probability that a cell exchanges its location with one of its two neighbors in the direction of the long side of the lattice be pl/2 (Fig. 1C). Then the probability of an exchange in the direction of the short side of the lattice is (1 - pl)/2. When cells are equally likely to move in the long-side and short-side directions of the lattice, pl = 0.5 and cell movement is isotropic (i.e., completely random). In anisotropic movement, pl > 0.5 represents long-side-biased movement and larger pl means a stronger long-side bias. In contrast, pl < 0.5 represents short-side-biased movement and smaller pl means a stronger short-side bias. pl - 0.5 indicates the magnitude of anisotropy in the direction of cell movement. In our simulations, we use pl = 0.5 unless otherwise indicated. At the lattice boundaries, cells exchange their locations with one of their two or three neighbors.
Fig. 1D shows the regulatory network of the her segmentation clock gene in zebrafish. Her protein suppresses the transcription of her and delta mRNA (6, 30). This negative feedback causes oscillatory expression of the her gene (6, 8, 30). Each cell interacts with neighboring cells through Delta–Notch signaling, which activates her gene transcription (6, 30). We modeled the dynamics of her mRNA, Her protein in cytoplasm, Her protein in nucleus, and Delta protein explicitly (Eq. 2 in the Appendix, and ref. 26). Note that the following results obtained by our model hold even if we adopt another model for her gene regulation proposed previously by Lewis (24) and adopt a more abstract phase dynamics model (34) (see SI Text for the models, and Figs. S1–S4).
We assumed that the reaction parameter values are the same in all cells in our model. In this article, we use a set of reaction parameter values with which the model (Eq. 2, Appendix) generates a stable limit cycle with cells perfectly synchronized, but we obtained similar results, as described below, with different sets of reaction parameter values (Fig. S5; see also Appendix and Table S1 for the procedure used to choose the parameter set). Below, we use “synchronized oscillation” to mean the limit cycle with all the cells perfectly synchronized. In general, diverse spatiotemporal patterns can appear in systems composed of coupled oscillators (35). In this study, however, we focused only on whether cells achieved global synchronization as is observed in vertebrate somitogenesis.
We assumed that when a cell arrives at a new location it immediately begins interacting with its new neighbors through Delta–Notch signaling and immediately stops interacting with its old neighbors. This is because at the present time, we have no information on how soon the effect of old neighbors on a focal cell disappears and when the interaction between the cell and its new neighbors begins. However, the results shown below still hold if it is assumed that it takes several minutes for cells to begin to interact with their new neighbors (see SI Text and Fig. S6 for details).
To ascertain the effect of random cell movement on the dynamics of the segmentation clock, we examined how global synchronization is recovered after an external perturbation. We introduced a perturbation as follows. First, we defined the phase (θ) on the limit cycle with cells perfectly synchronized whose value ranges from 0 to 2π and becomes 0 when the amount of her mRNA is maximal. Then, an initial phase randomly chosen from within the interval [-2π(α/2), 2π(α/2)] was allotted to each cell, where α is a parameter controlling the magnitude of the initial phase difference between cells. Next, we caused the intracellular variables in each cell to slightly deviate from the limit cycle (see Fig. 1E and Appendix). In the following analysis, for simplicity, we use α∈[0,1] as the magnitude of perturbation.
To quantify the degree of synchronization, we defined the following quantity, named the “synchronization index” [see Appendix, (36)]:
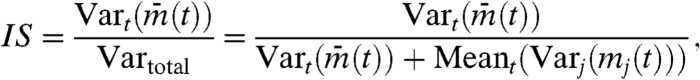 |
[1] |
where mj(t) is the time series of the her mRNA concentration in cell j, 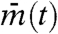 is the average of mj(t) over all cells, Vartotal is the total variance of mj(t) over time and all cells,
is the average of mj(t) over all cells, Vartotal is the total variance of mj(t) over time and all cells, 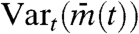 is the temporal variance of the time series
is the temporal variance of the time series 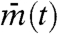 , Varj(mj(t)) is the between-cell variance of gene expression mj(t), and Meant(Varj(mj(t))) is the temporal average of Varj(mj(t)) (see Appendix). Note that IS is a function of time, and in the following analysis, we calculate the temporal average and variances during one period of oscillation. IS does not strongly depend on the length of the time interval used for calculating the temporal average and variances in Eq. 1.
, Varj(mj(t)) is the between-cell variance of gene expression mj(t), and Meant(Varj(mj(t))) is the temporal average of Varj(mj(t)) (see Appendix). Note that IS is a function of time, and in the following analysis, we calculate the temporal average and variances during one period of oscillation. IS does not strongly depend on the length of the time interval used for calculating the temporal average and variances in Eq. 1.
If cells are completely independent, different cells have different phases and averaging gene expression in many cells tends to cancel each other out; thus, their mean 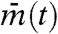 tends to stay constant. Hence, the variance over time of
tends to stay constant. Hence, the variance over time of 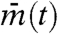 is very small. On the other hand, because cells have different values, between-cell variance Varj(mj(t)) is large. As a result, the synchronization index IS ≈ 0 when cells are independent. In contrast, suppose that cells are almost perfectly synchronized and keep oscillating. Then, the between-cell variance Varj(mj(t)) is close to zero, whereas
is very small. On the other hand, because cells have different values, between-cell variance Varj(mj(t)) is large. As a result, the synchronization index IS ≈ 0 when cells are independent. In contrast, suppose that cells are almost perfectly synchronized and keep oscillating. Then, the between-cell variance Varj(mj(t)) is close to zero, whereas 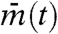 fluctuates. Hence, the synchronization index is close to unity (IS ≈ 1). In general cases, IS lies between 0 and 1 because the two terms in the denominator of the right-hand side of Eq. 1 are both positive.
fluctuates. Hence, the synchronization index is close to unity (IS ≈ 1). In general cases, IS lies between 0 and 1 because the two terms in the denominator of the right-hand side of Eq. 1 are both positive.
Results
Random Cell Movement Enhances the Restoration of Synchronization.
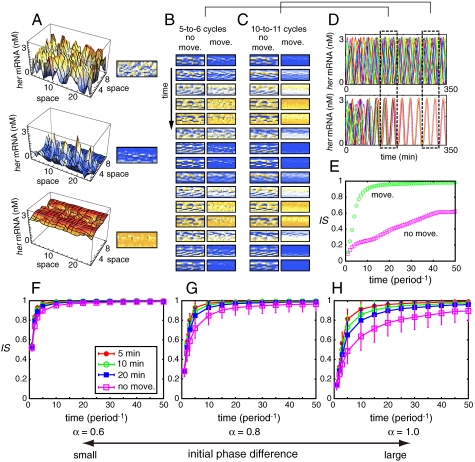
Fig. 2B and C (Left Columns) show the time courses of her mRNA level for N cells for the case in which all N cells were fixed in the lattice (see also Fig. 2A and D, Movie S1). At t = 0, we set the initial phase of each cell to a value randomly chosen from [-π,π] (i.e., α = 1). Even after 50 cycles, cells were unable to achieve globally synchronized oscillation of her gene expression; instead, spatially and temporally heterogeneous patterns of her gene expression appeared and persisted for a long time. Synchronization index IS increased with time, but its value was still small (≈0.6) after 50 cycles (Fig. 2E).
Fig. 2.
Random cell movement enhances the restoration of synchronization. (A) Correspondence between 3D plots of her mRNA concentration and density plots of that. In the density plots orange color indicates high concentration of her mRNA, and blue indicates low concentration. (B), (C) The time courses of her mRNA expression with cell movement (Right Columns; indicated by “move”) and without cell movement (Left Columns; indicated by “no move”) between (B) 124 min and 180 min, and (C) 268 min and 324 min. In right columns of (B) and (C), each cell experienced an exchange of its location with one of its neighbors within 10 min on average. (D) The time courses of her mRNA expression in 15 of N cells when all N cells are fixed in the lattice [Upper; corresponding to left columns in (B) and (C)] and when the cells moved randomly in the lattice [Lower; corresponding to right columns in (B) and (C)]. The 15 cells were chosen randomly among N cells. (E) The time courses of IS corresponding to (D). (F)–(H) The average time course of IS when the magnitude of the initial phase differences between cells was set to (F) α = 0.6, (G) α = 0.8, and (H) α = 1.0. Each cell experienced an exchange of its location with one of its neighbors within 5 min (Red Filled Circles), 10 min (Green Open Circles), or 20 min (Blue Filled Squares) on average. Pink open squares represent the case in which all N cells were fixed in the lattice. We used 20 different initial conditions. We ran 100 simulations for each initial condition in which we considered cell movement. We averaged all of the trials (2,000 runs for systems with cell movement and 20 runs for systems with cells fixed in the lattice). Error bars indicate standard deviations. The reaction parameter values used in Eq. 2 are listed in the Appendix as the standard parameter set.
We examined the dependence of the time needed to recover synchronization from an initial heterogeneous state on the magnitude of the initial phase differences between cells (α) when cells do not move in the lattice (in Fig. 2F–H, Pink Squares). We calculated the synchronization index IS for 20 different initial conditions and averaged them. When the initial phase differences were not large (see Fig. 2F, where α = 0.6), the differences almost always disappeared and global synchronization was achieved with time (e.g., after 20 cycles). However, as the initial phase differences became large (e.g., Fig. 2G and H, where α = 0.8 and α = 1.0, respectively), global synchronization often failed to be achieved and spatially and temporally heterogeneous patterns of her gene expression were observed, as shown in Fig. 2B and C and Movie S1. These results are consistent with the observations of Tiedemann et al. (25).
Fig. 2B and C (Right Columns) show the time courses of her mRNA level for N cells for the case in which cells randomly moved in the lattice (see also Fig. 2D and Movie S1). Each cell experienced an exchange of its location with one of its neighboring cells within 10 min on average. pl = 0.5 was assumed. All reaction parameter values and initial values used in this case were the same as those used for the case of the cells being fixed in the lattice (Fig. 2B and C, Left Columns). Global synchronization was recovered much faster than when cells did not move in the lattice (Fig. 2E).
As shown in Fig. 2F–H, random cell movement shortened the time taken to recover global synchronization against initial phase differences between cells. When the differences between cells were not large (α = 0.6, Fig. 2F), the effect of random cell movement was relatively small; the synchronization index IS at each time point was almost the same between the cases when cells moved and when they remained fixed in the lattice. In contrast, cell movement clearly led to the rapid realization of synchronization when the initial phase differences were large (α = 0.8 ∼ 1.0, Fig. 2G and H). Figs. 2F–H also show that as cells moved more frequently in the lattice, the time taken to recover global synchronization became shorter.
The promotion of synchronization of the segmentation clock by random cell movement can be explained intuitively as follows. Each PSM cell interacts with its neighboring cells via Delta–Notch signaling. Because of this local coupling, cells can match oscillation phases only with their direct neighbors, but they cannot recognize the phases of cells located further away. As a consequence, cells tend to form local patches of synchronized cells, and between these patches the phase of oscillation may greatly differ. These patches can be robust and persist for a long time, retarding global synchronization. Random cell movement increases the chance that global synchronization will be achieved within a shorter time by breaking up this persistent heterogeneous spatial structure (Movie S1).
Random Cell Movement Expands the Parameter Range to Achieve Synchronization of the Segmentation Clock.
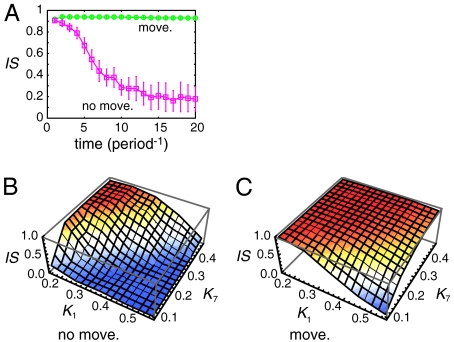
Even if the synchronized oscillation of the segmentation clock is stable, the basin of attraction may very narrow and convergence from a randomly chosen initial state may be extremely difficult (Fig. 3A). In Fig. 3A, we used a different value for the threshold constant for the suppression of her mRNA by Her protein (K1 = 0.6) from that used in Fig. 2 (K1 = 0.157) (see Appendix for details). The magnitude of the initial phase difference between cells was set to α = 0.2, which is much smaller than the values used above (cases shown in Fig. 2). When cells were fixed in the lattice (Fig. 3A, Pink Squares), from their initial synchronized state (IS ≈ 0.9) they became unsynchronized (IS ≈ 0.2) with time (see also Movie S2). This numerical observation indicates that, for the given parameter set, global synchronization is either unstable or it is locally stable with a very small basin of attraction; note that it is difficult to judge which is true because the number of dimensions of the system is very high. In contrast, when cells randomly moved in the lattice, global synchronization was stably maintained even after 20 cycles (Fig. 3A), clearly showing that cell movement can extend the range of parameter values that allow global synchronization to be stably maintained. This has an important biological meaning: Because chemical parameter values can be different between embryos and between cells, wider ranges of parameter values allowing achievement of global synchronization are desirable for robust development against such variability (37).
Fig. 3.
Cell movement extends the parameter range that allows cells to maintain synchronization. (A) The time courses of the average IS when the threshold constant for the suppression of her mRNA by Her protein K1 = 0.6. Other parameters were set to the standard values listed in Appendix. Each cell experienced an exchange of its location with one of its neighbors within 10 min on average (Green Open Circles). Pink open squares represent the case in which all N cells were fixed in the lattice. The magnitude of the initial phase differences between cells was set to α = 0.2. Error bars indicate standard deviations. (B), (C) The average IS after 10 cycles for different values of the threshold constant for the suppression of her mRNA by Her protein (K1) and that for the suppression of Delta protein by Her protein (K7). Other parameters were set to the standard values listed in Appendix. (B) Cells were fixed in the lattice, or (C) each cell experienced an exchange of its location with one of its neighbors within 10 min on average. The magnitude of the initial phase differences between cells was set to α = 0.6. In (A–C) we used 10 different initial conditions for each parameter set. We ran 10 simulations for each initial condition when we considered cell movement. Then we averaged all trials (100 runs for systems with cell movement and 10 runs for systems with cells fixed in the lattice).
Fig. 3B and C show the ranges of the threshold constant for the suppression of her mRNA by Her protein (K1) and of that for the suppression of Delta protein by Her protein (K7) that allow global synchronization to be maintained after 10 cycles. Cells maintained a synchronized state when K1 was small and K7 was large regardless of their mobility. However, as K1 increased or K7 decreased, the maintenance of synchronization was impossible when cells were fixed in the lattice (Fig. 3B). In contrast, when cells moved randomly, the system maintained global synchronization with high probability even when K1 was large and K7 was small (Fig. 3C). We obtained similar results even when we changed the values of other parameters from the standard values given in the Appendix (Fig. S7).
To confirm that cell movement generally promotes the achievement of global synchronization, we analyzed systems using 50 different parameter sets randomly generated from plausible parameter ranges (Fig. S5; see Appendix for the procedure used to choose reaction parameters). We found one parameter set with which random cell movement made the degree of synchronization worse (Fig. S5, Panel 39). However, with almost all of the parameter sets, cell movement facilitated the synchronization of the segmentation clock. Moreover, with several parameter sets, global synchronization was lost with time (IS decreased) without cell movement but it was maintained with cell movement (for example Fig. S5, Panels 1, 8, 17, and 21). Thus, we conclude that random cell movement extends the range of parameter values that allow global synchronization to be stably maintained.
Optimal Magnitude of Anisotropy in Cell Movement Depends on the Aspect Ratio of the Lattice.
Although cells often change the direction of their movement over time, a previous experimental study observed a medial-to-lateral flow of cells in the tailbud region of the zebrafish PSM (12). Anisotropy in the direction of movement might contribute to the synchronization of the segmentation clock. Here we study the effect of anisotropic cell movement by changing the parameter pl, larger values of which mean that cells are more likely to move in the long-side direction of the lattice (see Model and Fig. 1C).
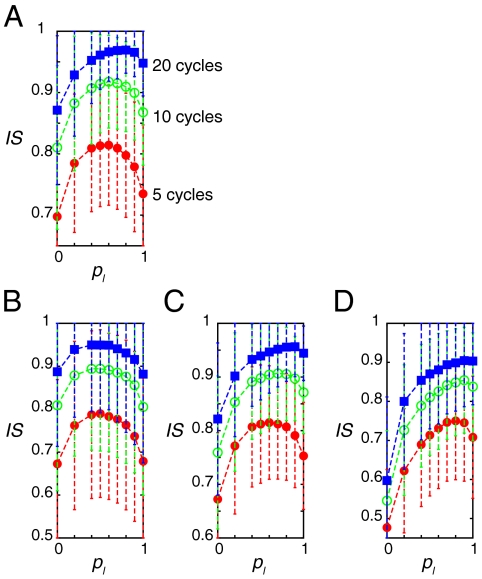
In the 25 × 10 lattice, cells realized synchronized oscillation more quickly when they engaged in long-side-biased movement than when they engaged in short-side-biased movement (Fig. 4A). However, to realize global synchronization the most quickly, cells should not move only in the long-side direction. For example, after each cycle of oscillations, IS was larger when pl was approximately 0.7 than when pl = 1 (Fig. 4A). This indicates that there is an optimal magnitude of anisotropy concerning random cell movement, say between pl = 0.6 and pl = 0.9 in this case, that allows cells to achieve synchronization the most quickly.
Fig. 4.
The aspect ratio of a lattice determines the optimal magnitude of anisotropy in the direction of cell movement for quick recovery of synchronization. (A)–(D) Average IS over 2,000 simulations of 5, 10, and 20 cycles when each cell exchanged its location with one of its neighbors with each pl in a (A) 25 × 10 lattice, (B) a 16 × 16 lattice, (C) a 32 × 8 lattice, and (D) a 64 × 4 lattice. In (A)–(D), each cell experienced an exchange of its location with one of its neighbors within 5 min on average. The magnitude of the initial phase differences between cells was set to α = 1.0. We used 20 different initial conditions and ran 100 simulations for each initial condition. Error bars indicate standard deviations. Reaction parameter values in Eq. 2 were the standard parameter set listed in Appendix.
We studied the dependence of the optimal magnitude of anisotropy in the direction of cell movement on the aspect ratio of the lattice (i.e., tissue geometry) by adopting 16 × 16, 32 × 8, 64 × 4, and 128 × 2 lattices. Note that the total cell number is equal among these lattices (N = 256).
As the aspect ratio of lattices increased, the optimal magnitude of anisotropy also increased (Fig. 4B–D). Cells in the 16 × 16 lattice achieved synchronization the most quickly when cell movement was completely random (i.e., cells achieved synchronization the fastest when pl was between 0.4 and 0.6, Fig. 4B). In the 32 × 8 lattice, the optimal magnitude of anisotropy in the direction of cell movement was between pl = 0.6 and pl = 0.8 (Fig. 4C). In the 64 × 4 and 128 × 2 lattices, global synchronization was achieved the most quickly when the cells moved only in the long-side direction of the lattice (Fig. 4D and Fig. S8D).
We also confirmed that the optimal magnitude of anisotropy in cell movement does not depend on the total cell number N or on whether the aspect ratio of the lattice is an integer (Fig. S8). We conclude that the aspect ratio of the lattice and the magnitude of anisotropy in the direction of cell movement influence the synchronization of the segmentation clock.
Random Cell Movement in Other Segmentation Clock Models.
We confirmed that the results obtained by our model as described above held even if we adopted the model for her gene regulation proposed by Lewis (24) (SI Text and Figs. S1–S3) or the more abstract phase dynamics model proposed by Sakaguchi et al. (34). In the case of the latter model, we also confirmed that the synchronized oscillation was enhanced by random cell movement in the presence of external noise other than an initial phase perturbation, and in the presence of cell-to-cell variability of parameter values (see SI Text and Fig. S4 for details).
Discussion
In this study, we discovered that dynamic cell movement, or rearrangement of the relative positions of cells, enhances the synchronization of the segmentation clock in vertebrate somitogenesis: Synchronization is recovered faster after external perturbation and is achieved with a wider range of parameter values than in the case without cell movement. This enhancement occurs because random cell movement increases the chance that global synchronization will be achieved within a short time by breaking up spatially heterogeneous oscillatory patterns. In living organisms, cells constantly proliferate during somitogenesis, which can be a major source of noise (30). Our results clearly show that dynamic cellular movement has an important biological function in achieving robust development against such noise.
Similar results obtained with different models (Eqs. S1 and S2) suggest that the effect of random cell movement does not depend on the concrete molecular mechanisms causing the oscillation of gene expression or on those causing cell–cell interactions. This suggests that the enhancement of synchronization by cell movement is not restricted to the synchronization of the segmentation clock in vertebrate somitogenesis but may be adopted in broader situations in which cells must synchronize their oscillation of gene expressions by coupling between nearest neighbors. For example, in chick limb development, synchronized oscillation of hairy2 is observed in some regions although its role remains unknown at the present time (38).
In this paper, we modeled cell movement as cells exchanging positions in the lattice with their neighbors at random times, because at the present time, little information on the movement of cells in the PSM is available. Quantitative measurement of cell movement, as well as that of the phase distributions of gene expression (12), will enable a more realistic analysis of the effect of cell movement on the dynamics of segmentation clock genes.
We showed that there is an optimal magnitude of anisotropy in the direction of cell movement that depends on the aspect ratio of the lattice for synchronization of the segmentation clock to be realized the most quickly. This result suggests that both the shape of the PSM and the anisotropy in the direction of cell movement influence the synchronization of the segmentation clock. Whether cells actually move in the PSM in a way that enables them to promote synchronization of the segmentation clock is an interesting question. Close monitoring of cell movement and precise determination of the shape of the PSM will provide an answer to this question.
Appendix
Equations for the Regulation of her Gene Expression.
Let mj, yj, zj, and wj be the concentrations of her mRNA, Her protein in cytoplasm, Her protein in nucleus, and Delta protein in cell j (j = 1,2,…,N), respectively (Fig. 1D). We consider the following model (26):
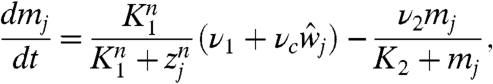 |
[2a] |
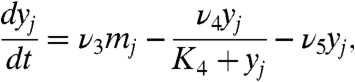 |
[2b] |
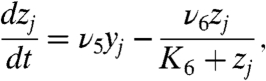 |
[2c] |
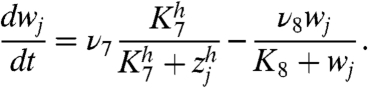 |
[2d] |
Eq 2a describes the time evolution of her mRNA. The first term represents the transcription of her mRNA. ν1 is the basal transcription rate, and νc is the activation rate by Delta–Notch signaling. We assume that 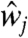 in Eq. 2a is the average of the Delta protein concentrations in the four neighboring cells. For cells on the boundaries,
in Eq. 2a is the average of the Delta protein concentrations in the four neighboring cells. For cells on the boundaries, 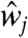 is the average of the concentrations in the two or three neighbors. Her protein in nucleus suppresses the transcription of its gene, as shown in the denominator of the first term on the right-hand side. The second term represents the degradation of her mRNA.
is the average of the concentrations in the two or three neighbors. Her protein in nucleus suppresses the transcription of its gene, as shown in the denominator of the first term on the right-hand side. The second term represents the degradation of her mRNA.
Eq. 2b describes the time evolution of Her protein in cytoplasm. The first term represents the synthesis of Her protein by translation. The second term represents the degradation of Her protein in cytoplasm. The third term represents the decrease in Her protein in cytoplasm caused by its transport from cytoplasm to nucleus. Eq. 2c describes the time evolution of Her protein in nucleus. The first term represents the increase in Her protein in nucleus caused by transport from the cytoplasm. The second term represents the degradation of Her protein in nucleus.
Eq. 2d describes the time evolution of Delta proteins that are expressed on the cell membrane. The first term represents the synthesis of Delta protein. Her protein suppresses Delta protein synthesis by inhibiting delta mRNA transcription (30). The second term represents the degradation of Delta protein.
We use the following reaction parameter values as the standard parameter values throughout this paper: ν1 = 0.0135 nM min-1, ν2 = 0.6 nM min-1, ν3 = 0.575 min-1, ν4 = 0.851 nM min-1, ν5 = 0.021 min-1, ν6 = 0.162 nM min-1, ν7 = 2.465 nM min-1, ν8 = 9.583 nM min-1, νc = 0.263 min-1, K1 = 0.157 nM, K2 = 0.104 nM, K4 = 0.142 nM, K6 = 0.13 nM, K7 = 0.49 nM, K8 = 9.72 nM, and n = h = 2. We chose this parameter set as follows: We set m1 = m2 = ·· = mN = m, y1 = y2 = ·· = yN = y, z1 = z2 = ·· = zN = z, and w1 = w2 = ·· = wN = w in Eq. 2 and obtained 4-variable differential equations. We analyzed the linear stability of the equilibrium of the 4-variable model and found a limit cycle with parameter values randomly chosen from the ranges listed in Table S1 (see (26) for how we determined the range of each parameter). Then we checked the local stability of the limit cycle in Eq. 2. To this end, we introduced small initial phase differences between cells, as described in the main text, at t = 0. If the phase differences disappeared and the system converged to the limit cycle after a sufficiently long time, we judged that the limit cycle was locally stable. The above parameter set satisfied these conditions (26). Following the above procedure, we generated 50 parameter sets with which Eq. 2 produced locally stable synchronized oscillation (Fig. S5).
Initial Conditions.
After we allotted an initial phase to each cell as described in the main text, we deviated the values of intracellular variables in each cell from the limit cycle with cells perfectly synchronized by adding a vector perpendicular to the tangent vector of the limit cycle at each point. The norm of the vector was set to 10% of the distance between the centroid of the limit cycle and the point on the limit cycle closest to the centroid.
Spatiotemporal Pattern Statistics.
To quantify the degree of global synchrony, we defined IS by Eq. 1, in which
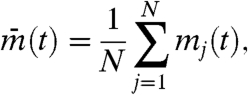 |
[3a] |
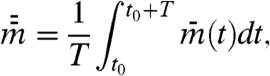 |
[3b] |
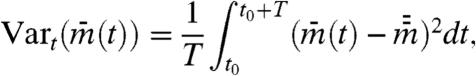 |
[3c] |
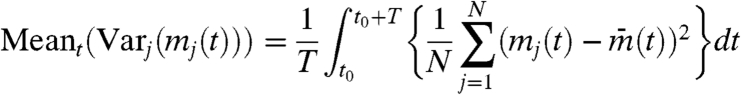 |
[3d] |
where t0 = 0,T,2T,3T,…. In the calculation of IS, we set T = 28.73 min, the synchronized oscillation period produced with the standard parameter set given above.
Supplementary Material
Acknowledgments.
We thank T. Hirashima, N. Honda, and A. Mochizuki for their very helpful comments. This work was supported by a Grant-in-Aid from the Japan Society for the Promotion of Science (Y.I.) and a PRESTO project of the Japan Science and Technology Agency (Y.M.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907122107/DCSupplemental.
References
- 1.Bessho Y, Miyoshi G, Sakata R, Kageyama R. Hes7: A bHLH-type repressor gene regulated by Notch and expressed in the presomitic mesoderm. Genes Cells. 2001;6:175–185. doi: 10.1046/j.1365-2443.2001.00409.x. [DOI] [PubMed] [Google Scholar]
- 2.Holley SA, Geisler R, Nusslein-Volhard C. Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wave-front activity. Genes Dev. 2000;14:1678–1690. [PMC free article] [PubMed] [Google Scholar]
- 3.Jouve C, et al. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development. 2000;127:1421–1429. doi: 10.1242/dev.127.7.1421. [DOI] [PubMed] [Google Scholar]
- 4.Oates AC, Ho RK. Hairy/E(spl)-related (Her) genes are central components of the segmentation oscillator and display redundancy with the Delta/Notch signaling pathway in the formation of anterior segmental boundaries in the zebrafish. Development. 2002;129:2929–2946. doi: 10.1242/dev.129.12.2929. [DOI] [PubMed] [Google Scholar]
- 5.Palmeirim I, Henrique D, Ish-Horowicz D, Pourquie O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- 6.Giudicelli F, Ozbudak EM, Wright GJ, Lewis J. Setting the tempo in development: An investigation of the zebrafish somite clock mechanism. PLoS Biol. 2007;5:e150. doi: 10.1371/journal.pbio.0050150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirata H, et al. Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science. 2002;298:840–843. doi: 10.1126/science.1074560. [DOI] [PubMed] [Google Scholar]
- 8.Holley SA, Julich D, Rauch GJ, Geisler R, Nusslein-Volhard C. her1 and the notch pathway function within the oscillator mechanism that regulates zebrafish somitogenesis. Development. 2002;129:1175–1183. doi: 10.1242/dev.129.5.1175. [DOI] [PubMed] [Google Scholar]
- 9.Maroto M, Dale JK, Dequeant ML, Petit AC, Pourquie O. Synchronised cycling gene oscillations in presomitic mesoderm cells require cell–cell contact. Int J Dev Biol. 2005;49:309–315. doi: 10.1387/ijdb.041958mm. [DOI] [PubMed] [Google Scholar]
- 10.Masamizu Y, et al. Real-time imaging of the somite segmentation clock: Revelation of unstable oscillators in the individual presomitic mesoderm cells. Proc Natl Acad Sci USA. 2006;103:1313–1318. doi: 10.1073/pnas.0508658103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozbudak EM, Lewis J. Notch signalling synchronizes the zebrafish segmentation clock but is not needed to create somite boundaries. PLoS Genet. 2008;4:e15. doi: 10.1371/journal.pgen.0040015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mara A, Schroeder J, Chalouni C, Holley SA. Priming, initiation and synchronization of the segmentation clock by deltaD and deltaC. Nat Cell Biol. 2007;9:523–530. doi: 10.1038/ncb1578. [DOI] [PubMed] [Google Scholar]
- 13.Riedel-Kruse IH, Muller C, Oates AC. Synchrony dynamics during initiation, failure, and rescue of the segmentation clock. Science. 2007;317:1911–1915. doi: 10.1126/science.1142538. [DOI] [PubMed] [Google Scholar]
- 14.Kerszberg M, Wolpert L. A clock and trail model for somite formation, specialization and polarization. J Theor Biol. 2000;205:505–510. doi: 10.1006/jtbi.2000.2085. [DOI] [PubMed] [Google Scholar]
- 15.Mazzitello KI, Arizmendi CM, Hentschel HG. Converting genetic network oscillations into somite spatial patterns. Phys Rev E: Stat, Nonlinear, Soft Matter Phys. 2008;78:021906. doi: 10.1103/PhysRevE.78.021906. [DOI] [PubMed] [Google Scholar]
- 16.Baker RE, Schnell S, Maini PK. A mathematical investigation of a Clock and Wavefront model for somitogenesis. J Math Biol. 2006;52:458–482. doi: 10.1007/s00285-005-0362-2. [DOI] [PubMed] [Google Scholar]
- 17.Armstrong NJ, Painter KJ, Sherratt JA. Adding adhesion to a chemical signaling model for somite formation. B Math Biol. 2009;71:1–24. doi: 10.1007/s11538-008-9350-1. [DOI] [PubMed] [Google Scholar]
- 18.Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphogenesis. J Theor Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- 19.Zeiser S, Muller J, Liebscher V. Modeling the Hes1 oscillator. J Comput Biol. 2007;14:984–1000. doi: 10.1089/cmb.2007.0029. [DOI] [PubMed] [Google Scholar]
- 20.Santillan M, Mackey MC. A proposed mechanism for the interaction of the segmentation clock and the determination front in somitogenesis. PLoS ONE. 2008;3:e1561. doi: 10.1371/journal.pone.0001561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Gonzalez JG, Santillan M, Fowler AC, Mackey MC. The segmentation clock in mice: Interaction between the Wnt and Notch signalling pathways. J Theor Biol. 2007;248:37–47. doi: 10.1016/j.jtbi.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Goldbeter A, Pourquie O. Modeling the segmentation clock as a network of coupled oscillations in the Notch, Wnt and FGF signaling pathways. J Theor Biol. 2008;252:574–585. doi: 10.1016/j.jtbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 23.Cinquin O. Repressor dimerization in the zebrafish somitogenesis clock. PLoS Comput Biol. 2007;3:e32. doi: 10.1371/journal.pcbi.0030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis J. Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Curr Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- 25.Tiedemann HB, et al. Cell-based simulation of dynamic expression patterns in the presomitic mesoderm. J Theor Biol. 2007;248:120–129. doi: 10.1016/j.jtbi.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 26.Uriu K, Morishita Y, Iwasa Y. Traveling wave formation in vertebrate segmentation. J Theor Biol. 2009;257:385–396. doi: 10.1016/j.jtbi.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Jaeger J, Goodwin BC. Cellular oscillators in animal segmentation. In Silico Biol. 2002;2:111–123. [PubMed] [Google Scholar]
- 28.Kaern M, Menzinger M, Hunding A. Segmentation and somitogenesis derived from phase dynamics in growing oscillatory media. J Theor Biol. 2000;207:473–493. doi: 10.1006/jtbi.2000.2183. [DOI] [PubMed] [Google Scholar]
- 29.Morelli LG, et al. Delayed coupling theory of vertebrate segmentation. HFSP J. 2009;3:55–66. doi: 10.2976/1.3027088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horikawa K, Ishimatsu K, Yoshimoto E, Kondo S, Takeda H. Noise-resistant and synchronized oscillation of the segmentation clock. Nature. 2006;441:719–723. doi: 10.1038/nature04861. [DOI] [PubMed] [Google Scholar]
- 31.Jiang YJ, et al. Notch signalling and the synchronization of the somite segmentation clock. Nature. 2000;408:475–479. doi: 10.1038/35044091. [DOI] [PubMed] [Google Scholar]
- 32.Delfini MC, Dubrulle J, Malapert P, Chal J, Pourquie O. Control of the segmentation process by graded MAPK/ERK activation in the chick embryo. Proc Natl Acad Sci USA. 2005;102:11343–11348. doi: 10.1073/pnas.0502933102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulesa PM, Fraser SE. Cell dynamics during somite boundary formation revealed by time-lapse analysis. Science. 2002;298(5595):991–995. doi: 10.1126/science.1075544. [DOI] [PubMed] [Google Scholar]
- 34.Sakaguchi H, Shinomoto S, Kuramoto Y. Local and global self-entrainments in oscillator lattices. Prog Theor Phys. 1987;77:1005–1010. [Google Scholar]
- 35.Kuramoto Y. Chemical oscillations, waves, and turbulence. Berlin: Springer–Verlag; 1984. [Google Scholar]
- 36.Satake A, Iwasa Y. Spatially limited pollen exchange and a long-range synchronization of trees. Ecology. 2002;83:993–1005. [Google Scholar]
- 37.Hirata H, et al. Instability of Hes7 protein is crucial for the somite segmentation clock. Nat Genet. 2004;36:750–754. doi: 10.1038/ng1372. [DOI] [PubMed] [Google Scholar]
- 38.Pascoal S, et al. A molecular clock operates during chick autopod proximal-distal outgrowth. J Mol Biol. 2007;368:303–309. doi: 10.1016/j.jmb.2007.01.089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.