Abstract
During the course of evolution, a massive reduction of the mitochondrial genome content occurred that was associated with transfer of a large number of genes to the nucleus. To further characterize factors that control the mitochondrial gene transfer/retention process, we have investigated the barriers to transfer of yeast COX2, a mitochondrial gene coding for a subunit of cytochrome c oxidase complex. Nuclear-recoded Saccharomyces cerevisiae COX2 fused at the amino terminus to various alternative mitochondrial targeting sequences (MTS) fails to complement the growth defect of a yeast strain with an inactivated mitochondrial COX2 gene, even though it is expressed in cells. Through random mutagenesis of one such hybrid MTS-COX2, we identified a single mutation in the first Cox2 transmembrane domain (W56 → R) that (i) results in the cellular expression of a Cox2 variant with a molecular mass indicative of MTS cleavage, which (ii) supports growth of a cox2 mutant on a nonfermentable carbon source, and that (iii) partially restores cytochrome c oxidase-specific respiration by the mutant mitochondria. COX2W56R can be allotopically expressed with an MTS derived from S. cerevisiae OXA1 or Neurospora crassa SU9, both coding for hydrophobic mitochondrial proteins, but not with an MTS derived from the hydrophilic protein Cox4. In contrast to some other previously transferred genes, allotopic COX2 expression is not enabled or enhanced by a 3′-UTR that localizes mRNA translation to the mitochondria, such as yeast ATP23′-UTR. Application of in vitro evolution strategies to other mitochondrial genes might ultimately lead to yeast entirely lacking the mitochondrial genome, but still possessing functional respiratory capacity.
Keywords: cytochrome coxidase, mitochondria, Saccharomyces cerevisiae
As the site of oxidative phosphorylation, mitochondria fulfill a key function in the energy metabolism of eukaryotic cells. Mitochondria also play an important role in programmed cell death, cell proliferation, and other critical cellular processes. This organelle is unusual in that it harbors its own genome, which, together with allotopically expressed nuclear genes, encodes those proteins required for mitochondrial function. To better understand the factors that control the nuclear or mitochondrial localization of genes that encode these proteins, we have begun to systemically analyze the roles of coding sequence, mitochondrial targeting sequences, and mRNA localization sequences on the allotopic expression of mitochondrial membrane proteins. Such efforts (1–3) should lead to an improved ability to control the expression of both endogenous and foreign genes in the mitochondria through established genetic methods. Currently mitochondrial genes are not amenable to standard reverse genetic techniques, the exception being the Saccharomyces cerevisiae mitochondrion that can be transformed by biolistic transformation. The ability to allotopically express mitochondrial genes involved in human disease might facilitate the use of gene therapy to treat both genetic diseases and age-related diseases associated with mitochondrial dysfunction. Ultimately it may even be possible to create synthetic organisms in which the mitochondrial genome is completely eliminated.
The biogenesis of mitochondria is a complex process that involves the concerted expression of both nuclear and mitochondrial genomes. It is well documented that during the course of evolution a majority of the genes that were originally encoded in the mitochondrial genome have been transferred to the nucleus, replaced by preexisting nuclear genes, or lost altogether (4). In the most extreme case of Apicomplexans, only three protein-coding genes have been retained in the mitochondrial genome that code for cytochrome c oxidase (COX) subunits I and III and apocytochrome b (5). The unusually high hydrophobicity of these three proteins may hinder their import into the mitochondria and may constitute a barrier that prevented relocation of their corresponding genes into the nucleus (6). In support of such hypothesis, these three genes are found in the mitochondrial genomes of almost all respiring eukaryotic cells but not in the nucleus (one exception being Chlamydomonad algae cox3) (7).
Together with COX1 and COX3, a large majority of respiring eukaryotes encode an additional COX subunit gene, COX2, in their mitochondrial genome. Cox2 is less hydrophobic than either Cox1 or Cox3 and consists of two transmembrane helices with a large hydrophilic domain at the C terminus (8). In addition to Apicomplexans, COX2 has been evolutionarily transferred to the nucleus in the green Chlamydomonad algae and legumes. In Apicomplexans and green algae, the original Cox2 coding sequence was split into two separate genes that can be harbored by either the mitochondrial or the nuclear genome. For example, in some green algae species (e.g., Scendesmus) the 3′ region of COX2 migrated to the nucleus, whereas the 5′ section remained in the mitochondrion (9). Alternatively, in other green algae genera (e.g., Chlamydomonas or Polytomella) both sections of the split COX2 were transferred to the nucleus (10). This type of transfer (in two fragments) but with the entire COX2 coding sequence being relocated occurred also in Apicomplexans (11). The evolutionary transfer of the COX2 gene in legumes, on the other hand, occurred without gene splitting and appears to be a relatively recent event as two full-length gene copies are present in some species, one in the nucleus and the other in the mitochondrion (12–14). Such cases may represent intermediate steps of gene transfer as some legume species transcribe both nuclear and mitochondrial COX2, whereas others transcribe only one or the other (15).
In the budding yeast S. cerevisiae, Cox2 is encoded in the mitochondrial genome and is cotranslationally inserted into the inner mitochondrial membrane through a complex maturation process, which includes the export of the large hydrophilic C-terminal domain into the mitochondrial intermembrane space (16–19). Because of the limited number of examples of successful COX2 transfer into the nucleus throughout evolution, it is not clear whether yeast Cox2 translated in the cytoplasm could be transported into the mitochondrial lumen and assembled into a functional COX complex. Indeed, we found that a recoded yeast COX2 transferred into the nucleus and fused to a mitochondrial targeting sequence (MTS) fails to produce a functional Cox2 when expressed in S. cerevisiae. However, we identified through random mutagenesis a single mutation in the first Cox2 transmembrane (TM) domain that enabled allotopic Cox2 expression and rescued the respiratory defect of the cox2 yeast mutant. The observed rescue depended on the MTS and resulted from the translocation of cytoplasmically expressed mutant Cox2 into the mitochondria and its assembly into a functional COX complex.
Results
Design of Allotopic Expression Constructs for Yeast COX2.
The successful gene transfer of cox2 from the mitochondrion to the nucleus in legumes was accompanied by the acquisition of an unusually long N-terminal MTS (>120 amino acids) that could not be functionally replaced by another MTS in soybean (13). According to our current understanding, the MTS binds to receptors on the surface of the mitochondrial outer membrane, then facilitates protein entry into the translocation channel, and subsequently mediates interaction with mHsp 70 on the mitochondrial matrix side (20).
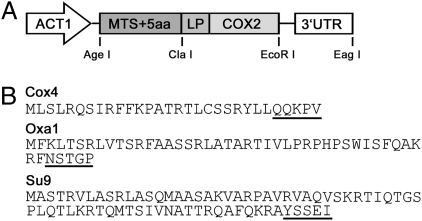
To examine if allotopic expression of S. cerevisiae COX2 could be enabled in a similar fashion by an MTS, we generated a set of expression constructs that code for yeast COX2 fused at its N terminus to various MTSs. The MTS was selected from each of three nucleus-encoded, inner mitochondrial membrane proteins: S. cerevisiae Cox4 and Oxa1 and Neurospora crassa Su9 (Fig. 1). The selected proteins differ in their overall length (Cox4, 155 aa; Oxa1, 402 aa; Su9, 147 aa), number of TM domains (Cox4, none; Oxa1, 5; Su9, 2), average total hydropathicity (Cox4, −0.492; Oxa1, −0.134; Su9, 1.004), and length of MTS (Cox4, 25 aa; Oxa1, 42 aa; Su9, 66 aa). In addition to these three MTS-COX2 fusions (COX4MTS-COX2, OXA1MTS-COX2, and SU9MTS-COX2), we also generated two COX2 constructs that contain duplicated OXA1 and SU9 MTSs (constructs labeled as 2×OXA1MTS-COX2 and 2×SU9MTS-COX2). In the case of the SU9 MTS such duplication was previously shown to improve allotopic expression of two nonrelated, inner mitochondrial membrane proteins (Atp8 and truncated Cob1) in S. cerevisiae (21, 22). Finally, we generated the control construct that lacked an MTS and coded for the COX2 gene only (labeled as nullMTS-COX2). These six COX2 variants allowed a broad examination of the role of the MTS in allotopic COX2 expression.
Fig. 1.
Construct design for allotopic COX2 expression. (A) The COX2 expression cassettes include the ACT1 yeast promoter, yeast ADH1 or ATP2 3′-UTR, and nuclear-recoded COX2 fused in frame to various MTSs. Three MTSs were derived from S. cerevisiae COX4 and OXA1 and N. crassa SU9; two additional MTSs were used that were generated by duplication of OXA1 and SU9 MTS. In the case of duplicated MTSs, two MTS repeats were fused through a 3-aa linker (ADK). In all five constructs, the fusion junction between MTS and COX2 included 5 aa that immediately follow the MTS processing site in the corresponding MTS source protein and a 15-aa-long COX2 N-terminal leader peptide (LP). One additional construct was created that lacked any MTS. Restriction sites used for the generation of expression constructs are also shown. (B) Amino acid sequences of Cox4, Oxa1, and Su9 MTSs. Five amino acid blocks that constitute the N termini of the corresponding mature proteins and that were fused to Cox2 LP are underlined.
Translation of a subset of the mitochondria-imported proteins is targeted to the organelle proximity by localization signals encoded in the 3′-UTR of the corresponding mRNAs (23). To examine if such mRNA targeting can enable or improve COX2 allotopic expression, all six generated MTS-COX2 fusions were appended with two alternative 3′-UTRs that were derived from the yeast ATP2 and ADH1 genes. The former sequence was previously shown to localize translation of the ATP2 mRNA to mitochondria-bound polysomes, whereas the ADH1 mRNA is translated in the cytoplasm. Finally, we placed all 12 (6 MTS-COX2-ADH13′-UTR and 6 MTS-COX2-ATP23′-UTR) constructs under control of the yeast ACT1 promoter (Fig. 1) and inserted the complete expression cassettes into the yeast multicopy YEp352 plasmid.
Cytoplasmic Expression of MTS-COX2 Does Not Rescue cox2 Phenotype.
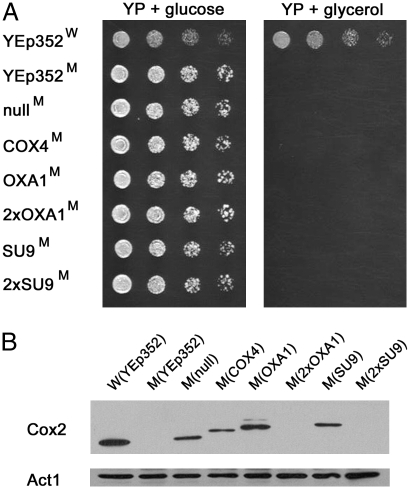
To quickly evaluate all 12 expression constructs for their ability to support allotopic COX2 expression, the constructs were introduced into the yeast cox2-60 mutant. This yeast strain contains a partial deletion in its mitochondrion-encoded COX2, and because it does not express Cox2, it does not assemble a functional COX complex (24). As a result of this defect, the mutant fails to grow on a nonfermentable carbon source (Fig. 2A), affording a simple evaluation of successful allotopic COX2 expression through the restoration of such growth.
Fig. 2.
Cytoplasmic MTS-COX2 does not reverse the cox2-60 mutant growth defect on glycerol medium. (A) Wild-type (labeled with W) and cox2-60 (labeled with M) yeast strains harboring the indicated plasmids were serially diluted and stamped on solid media containing either glucose (YP + glucose) or glycerol (YP + glycerol) as the sole carbon source. None of six MTS-COX2-ADH13′-UTR expression plasmids (MTS constructs present in cells are indicated) were able to restore glycerol-dependent growth of the cox2-60 mutant. (B) Expression of Cox2 in yeast cells harboring MTS-COX2-ADH13′-UTR expression constructs. Amounts of yeast actin present in samples are also shown for comparison.
The 12 transformed cox2-60 strains were found to be phenotypically similar to the mutant transformed with the control YEp352 plasmid. They all grew on glucose-containing media, but did not generate colonies when replicated on media containing glycerol as the sole carbon source (Fig. 3). The absence of growth on glycerol media indicates that the COX2 expression constructs did not correct the mutant COX defect. To determine if these constructs result in Cox2 protein expression, the whole cell lysates from 12 transformants were evaluated for the presence of Cox2 by Western blotting. As expected, the cox2-60 parent strain does not contain detectable Cox2 (Fig. 2B) Transformants harboring nullMTS-, COX4MTS-, OXA1MTS-, and SU9MTS-COX2-ADH13′-UTR constructs expressed Cox2 at similar levels that were somewhat lower than that found in the wild-type yeast strain. With the exception of nullMTS-COX2-ADH13′-UTR, the other three constructs expressed a Cox2 that migrated as a higher molecular mass species, indicating that the MTS was not proteolytically removed. In the case of the 2×SU9MTS-COX2-ADH13′-UTR and 2×OXA1MTS-COX2-ADH13′-UTR constructs, the Cox2 expression levels were very low and difficult to detect (Fig. 2B). Identical results were also obtained with the cox2-60 transformants harboring the equivalent MTS-COX2-ATP23′-UTR constructs.
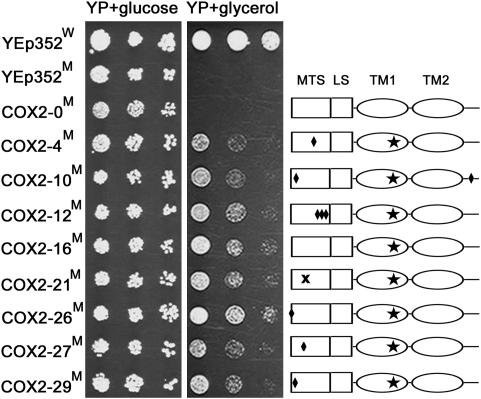
Fig. 3.
Growth on glycerol of a cox2-60 yeast mutant harboring cytoplasmically expressed 2×SU9MTS-COX2W56R–ADH13′-UTR alleles. Wild-type (indicated with W) and cox2-60 (indicated with M) yeast strains harboring the indicated plasmids were serially diluted and stamped on solid media containing either glucose (YP + glucose) or glycerol (YP + glycerol) as the sole carbon source. COX2-0 corresponds to the YEp352-based expression vector coding for 2×SU9MTS-COX2-ADH13′-UTR. COX2-X (X = 4, 10, etc.) refer to the mutant 2×SU9MTS-COX2W56R-ADH13′-UTR expression plasmids described in detail in Table S1. The right side shows the positions of mutations within the complementing 2×SU9MTS-COX2W56R alleles. A solid star inside TM1 indicates W56R replacement and solid diamonds depict the other missense mutations present in the individual 2×SU9MTS-COX2W56R alleles. The stop codon is shown as “x”. Identical results were also obtained with the cox2-60 transformants harboring the equivalent MTS-COX2-ATP23′-UTR constructs.
A Single Mutation in Cox2 TM1 Enables Growth of the cox2 Mutant on a Nonfermentable Carbon Source.
To determine whether amino acid mutations could be identified that would enable allotopic COX2 expression, we performed random mutagenesis of the 2×SU9MTS-COX2 ORF by mutagenic PCR. A library of ∼20,000 mutant 2×SU9MTS-COX2 alleles in a YEp352-based expression vector was generated and then transformed into the cox2-60 host. Approximately 120,000 transformants were replicated on YPG plates to select for cells with restored COX function. In total, 60 YPG-growing transformants were identified and then subsequently used for plasmid DNA extraction. The recovered plasmid DNAs were retransformed into the cox2-60 mutant, and the resulting transformants were again tested for the ability to grow on YPG plates. Plasmids from 12 of the original 60 YPG-growing transformants were found to rescue the mutant phenotype again, thus establishing a linkage between these 12 plasmids and correction of the cox2-60 respiratory defect. Sequencing of the recovered 12 2×SU9MTS-COX2 alleles revealed that they all contained a common mutation W198R (W56R when numbering from the Cox2 start codon), which maps to Cox2 TM1 (amino acids 30–62, the TM1 assignment based on homology with bovine Cox2) (8). Five of the 12 recovered alleles harbored only this single amino acid replacement, whereas the other 7 alleles contained additional mutations. All but one of these secondary mutations, A324V (A182V when counting from the Cox2 start codon), mapped to the 2×SU9 MTS (Fig. 3 and Table S1).
Even though the cox2-60 transformants harboring any of eight unique complementing alleles had a corrected cox2-60 phenotype, they all produced smaller colonies on YPG plates than the isogenic wild-type strain (Fig. 3), which indicates a slower growth of the complemented mutants on glycerol-containing medium. Additionally, only one allele from among those with secondary mutations, M1V, W198R (W56R), supported an improved growth on YPG medium as compared to the W198R (W56) allele (Fig. 3).
The W56R Mutation Results in Allotopic Cox2 Expression and Partially Restores Cellular Respiration of the cox2-60 Mutant.
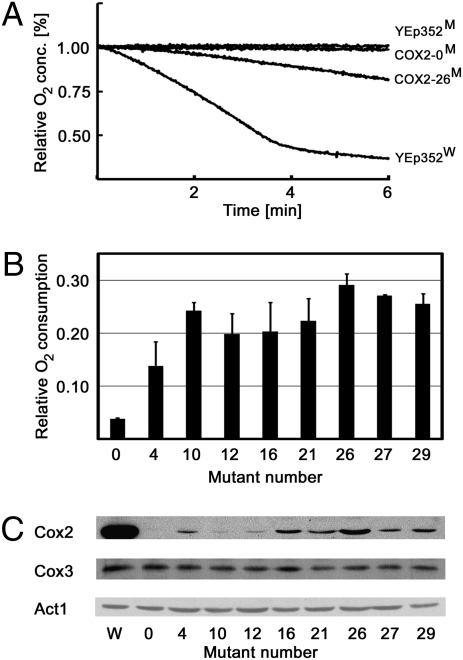
To test whether the correction of cox2-60 growth phenotype by the eight complementing 2×SU9MTS-COX2 alleles is the consequence of restored cellular respiration, we measured oxygen consumption capacity of the corresponding cox2-60 transformants and relevant control strains. As expected, the wild-type yeast culture exhibited robust respiration with rapid depletion of oxygen from the assay chamber. In comparison, cox2-60 transformants harboring YEp352 alone or 2×SU9MTS-COX2-ADH13′-UTR did not show any detectable respiration. In agreement with the observed growth on YPG medium, the cox2-60 strains transformed with any of eight complementing 2×SU9MTS-COX2 alleles exhibited plasmid-dependent oxygen consumption (Fig. 4A). Using the oxygen consumption rate normalized to that of the wild-type strain, the respiration of complemented strains varied from ∼15% to ∼30%, depending on the allele present in cells (Fig. 4B). In general, the observed variations in mutant respiration rates showed a good correlation with the mutant growth rates on the YPG plates, with 2×SU9M1V,MTS-COX2W56R supporting both the most vigorous growth and the highest respiratory rate from among all complementing alleles. We further confirmed by Western blotting that the observed cellular respiration was associated with the expression of a Cox2 variant that had a similar molecular mass to that of the mitochondrially expressed protein, which was distinctly smaller than the Cox2 species produced by the original, nonmutated 2×SU9MTS-COX2 gene. In agreement with both the glycerol growth rates and cellular respiratory capacity, all transformants harboring 2×SU9MTS-COX2W56R alleles also expressed detectable, although low levels of the smaller Cox2 species (Fig. 4C).
Fig. 4.
2×SU9MTS-COX2W56R alleles support yeast cellular respiration. (A) Cellular respiration by wild-type (W) and cox2-60 (M) yeast strains harboring the indicated plasmids (plasmid labeling code described in Fig. 3 legend). Stationary yeast cultures grown in SC −Ura medium containing raffinose were placed into an oxygen sensor chamber and the time course of oxygen depletion was monitored. (B) Oxygen consumption rates of the cox2-60 strains harboring the indicated 2×SU9MTS-COX2W56R-ADH13′-UTR expression plasmids (see Table S1 for details). Oxygen consumption rates are expressed relative to those of the wild-type strain. Averages of three independent measurements (n = 3) and standard deviations are shown. (C) Cellular Cox2 expression levels supported by the individual 2×SU9MTS-COX2W56R alleles. For comparison, amounts of Cox3 and actin present in samples are also shown.
Allotopic COX2 Expression Restores the Enzymatic Activity of COX.
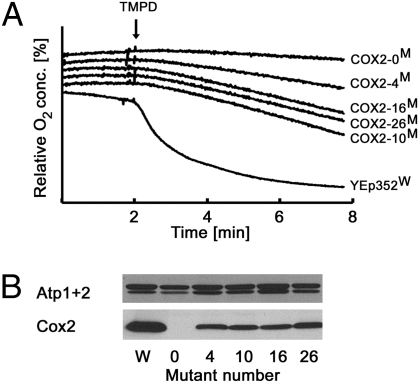
To directly assess if COX activity was restored in cox2-60 cells allotopically expressing COX2, we purified mitochondria from the wild-type strain and cox2-60 transformants harboring different complementing 2×SU9MTS-COX2 alleles. COX activity in the mitochondrial preparations was determined using TMPD, an artificial electron donor that can serve as a substrate for COX, but not the other S. cerevisiae respiratory complexes (type II NADH dehydrogenase, complex II, and complex III). As expected, the addition of TMPD to wild-type mitochondria resulted in a rapid, cyanide-sensitive depletion of oxygen from the reaction mixture (Fig. 5A). On the other hand, mitochondria isolated from the cox2-60 strain harboring nonmutated 2×SU9MTS-COX2-ADH13′-UTR were not able to oxidize TMPD, indicating an absence of functional COX. In agreement with the previous experiments, which strongly suggest the assembly of enzymatically active COX in the mitochondria upon expression of 2×Su9MTS-Cox2W56R mutant proteins, cox2-60 transformants harboring any of the selected complementing alleles displayed partially restored TMPD oxidation (Fig. 5A). With the exception of the 2×SU9T107A,MTS-COX2W56R transformant, which also possessed the lowest cellular respiratory capacity, mitochondrial preparations from cells expressing other Cox2W56R variants displayed a similar extent of the TMPD-dependent oxygen consumption. To normalize respiratory capacity to the amounts of mitochondrial proteins in the samples, we probed by Western blot the mitochondrial preparations for Atp1, Atp2 (both subunits of mitochondrial F1F0-ATP synthase), and Cox2. All isolated mitochondria with the exception of the preparation from the nonmutated 2×SU9MTS-COX2 transformant contained approximately equal amounts of Atp1 and Atp2. The mitochondrial membranes prepared from cox2-60 cells harboring individual 2×SU9MTS-COX2 complementing alleles also displayed similar levels of Cox2 protein. In summary, the above experiments clearly indicate that cytoplasmic 2×SU9MTS-COX2W56R expression leads to the assembly of a functional COX complex in the mutant mitochondria.
Fig. 5.
Allotopic expression 2×SU9MTS-COX2W56R restores COX activity in mitochondrial membranes. (A) Mitochondrial membranes isolated from the wild-type (W) and cox2-60 (M) yeast strains harboring the indicated plasmids (same as Fig. 4A) were assayed for COX activity by measurement of oxygen consumption. Mitochondrial membranes were resuspended in a buffer containing ascorbic acid and placed into the respiration chamber. After 2 min of equilibration, respiration was initiated by addition of TMPD, a COX-specific substrate. The time course of oxygen consumption is shown. (B) Mitochondrial protein content of membranes is shown for Cox2, Atp1, and Atp2.
Allotopic COX2W56R Expression Is Dependent on the MTS but Not on the Mitochondria-Targeting 3′-UTR.
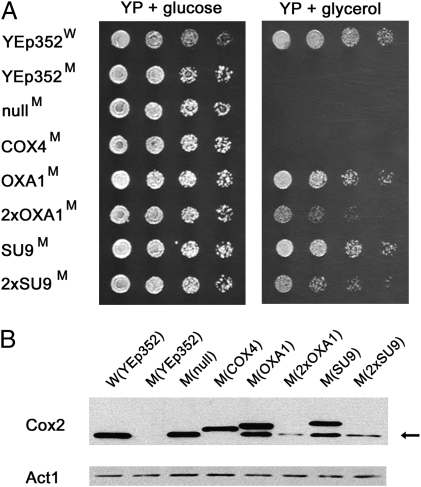
During gene transfer from the mitochondria to the nucleus, soybean Cox2 acquired a 124-aa-long MTS that is critical for expression of functional protein and that could not be successfully replaced with a shorter, 41-aa-long MTS derived from the alternative oxidase protein Aox1 (25). To better understand the MTS requirements associated with allotopic expression of S. cerevisiae COX2W56R, we replaced the wild-type COX2 allele in the six MTS-COX2-ADH13′-UTR expression constructs with COX2W56R. Newly generated expression plasmids were introduced into the cox2-60 mutant, and the resulting transformants were tested for their ability to grow on YPG medium. Whereas cells harboring COX2W56R constructs without a MTS or with the COX4 MTS were not able to use glycerol for growth, the 2×OXA1MTS-COX2W56R fusion supported glycerol-dependent growth to the same extent as the original 2×SU9MTS-COX2W56R construct. Interestingly, constructs coding for the shorter SU9MTS-COX2W56R and OXA1MTS-COX2W56R fusions supported a more vigorous glycerol-dependent growth than the constructs with duplicated MTSs and also formed colonies on YPG plates that were comparable in size to those of the wild-type strain (Fig. 6A). To determine whether the YPG growth phenotype supported by these three additional MTS-COX2W56R constructs was also associated with the appearance of the lower molecular mass Cox2 species, whole cell lysates from all six MTS-COX2W56R-ADH13′-UTR transformants were analyzed by Western blot. In good agreement with the YPG growth experiment, cells harboring the SU9MTS-COX2W56R and OXA1MTS-COX2W56R constructs expressed an increased amount of the faster migrating Cox2 species than that found in the 2×SU9 MTS-COX2W56R or 2×OXA1MTS-COX2W56R cells. As expected, cells expressing COX4MTS-COX2W56R did not contain a detectable quantity of the lower molecular mass Cox2 species (Fig. 6B).
Fig. 6.
Multiple MTSs support allotopic expression of COX2W56R. (A) Wild-type (W) and cox2-60 (M) yeast strains harboring the indicated plasmids were serially diluted and stamped on solid media containing either glucose (YP + glucose) or glycerol (YP + glycerol). Four MTS-COX2W56R-ADH13′-UTR expression plasmids (MTSs OXA1, 2×OXA1, SU9, and 2×SU9) were able to restore glycerol-dependent growth of the cox2-60 mutant. (B) Expression of Cox2W56R in yeast cells harboring MTS-COX2W56R-ADH13′-UTR expression constructs. Amounts of yeast actin present in samples are shown for comparison. Cox2 species migrating at the position of the mature, mitochondria-coded Cox2 are indicated with an arrow.
To assess a possible role for mRNA localization at the mitochondria on allotopic expression of COX2W56R, we further generated six MTS-COX2W56R-ATP23′-UTR expression constructs by replacing ADH13′-UTR in the above six plasmids. cox2-60 transformants containing these plasmids were characterized by their ability to grow on YPG plates as described above. It was found that the growth of individual transformed strains was the same as the growth observed for the equivalent MTS-COX2W56R-ADH13′-UTR constructs. Similarly, Cox2W56R expression driven by the constructs with the ATP2 3′-UTR was the same as that observed for cells harboring equivalent ADH13′-UTR constructs.
Discussion
According to the hydrophobicity hypothesis, mitochondrial gene transfer to the nucleus during evolution was not completed due to a subset of the mitochondrion-encoded proteins whose highly hydrophobic nature interfered with their transport from the cytoplasm into the mitochondria (22, 26, 27). An examination of the requirements for allotopic expression of Cox2 in legumes, some species of which harbor both the nuclear and mitochondrial Cox2 genes, revealed that modification of a local protein property (a decrease in the hydrophobicity of TM1) can overcome this transport barrier. Interestingly, on the basis of this metric, S. cerevisiae COX2 should also be compatible with allotopic expression (13).
In this study, we show that despite the decreased hydrophobicity of the S. cerevisiae Cox2 TM1, the expression in the S. cerevisiae cytoplasm of the wild-type Cox2 protein does not result in complementation of a mitochochondrial cox2 yeast mutant. This failure is not the consequence of using an MTS incompatible with intramitochondrial delivery of a highly hydrophobic protein such as Cox2. N. crassa SU9 and 2×SU9 MTSs were previously shown to support allotopic expression in S. cerevisiae of both ATP8 (21) and various truncated forms of COB1 (22), which comprised up to four TM domains of this highly hydrophobic protein. Additionally, extensive mutagenesis of the 2×SU9MTS-COX2 ORF failed to yield 2×SU9MTS mutations allo-topically active with wild-type Cox2. The failure of Cox2 allotopic expression is also not the consequence of low cellular levels of cytoplasmically expressed Cox2. Two of the MTS-COX2 expression constructs generated in this report (OXA1MTS-COX2 and SU9MTS-COX2) accumulate Cox2 at a level similar to that of wild-type, intramitochondrially translated Cox2. Additionally, two other constructs (2×OXA1MTS-COX2 and 2×SU9MTS-COX2), even though they express Cox2 at a very low level, are able to support allotopic protein expression when present in the context of the COX2W56R allele. And finally, the inclusion into the MTS-COX2-3′-UTR constructs of S. cerevisiae ATP23′-UTR, which were shown to localize the ATP2 transcript to the mitochondria and to enhance the mitochondrial localization of some other nuclear-encoded mitochondrial proteins (23), did not result in wild-type Cox2 allotopic expression. In summary, the presented data strongly suggest that the expression of wild-type S. cerevisiae Cox2 in the cytoplasm is incompatible with its assembly into functional COX complex in the inner mitochondrial membrane. Consistent with this conclusion, the Cox2 proteins accumulated in cells display molecular masses equivalent to those calculated for the fusion Cox2 species not processed by the mitochondrial signal peptidase.
Through random mutagenesis of 2×SU9MTS-COX2 we identified a single mutation, W56R in the Cox2 TM1 that results in the functional expression of cytoplasmically translated Cox2 fusion protein. We showed that such expression allows for the rescue of cox2-60 mutant growth on a nonfermentable carbon source (glycerol) and leads to an accumulation of small amounts of cellular Cox2 with a molecular mass similar to that of the mitochondrion-encoded Cox2. Additionally, the cytoplasmic expression of 2×Su9-Cox2W56R results in a partial restoration of cellular respiration and partial recovery of COX enzymatic activity in the isolated mutant mitochondria. Together, these observations clearly indicate that 2×Su9-Cox2W56R is transported into mitochondria, its MTS is cleaved, and the protein is assembled into an enzymatically active COX complex. It is not clear from our study if additional alleles might exist besides W56R that would allow the allotopic expression of Cox2. In total, we recovered 12 independent complementing plasmids (assessment of independence based on the presence of nonidentical silent mutations in the 2×SU9MTS-COX2W56R ORF), but the allotopic activity of all of them was mediated by the same, W56R mutation. Considering that a strong mutational pressure was applied (two nucleotide changes per 1 kb as confirmed through the number of silent mutations identified in the recovered plasmids) and that a large number of mutants were screened, the number of single amino acid substitutions supporting allotopic Cox2 expression might be very limited.
The allotopic expression of COX2W56R is absolutely dependent on the presence of the MTS. The nullMTS-COX2W56R construct, which does not code for an MTS, is not able to restore respiratory growth of the cox2-60 mutant, even though it promotes accumulation of significant amounts of cellular Cox2. At the same time, there are less stringent MTS requirements associated with allotopic expression of yeast COX2W56R than was reported for the soybean Cox2 gene. This conclusion is based on our observation that MTSs derived from two nonrelated yeast genes (OXA1 and SU9) as well as their duplicated forms appear to be capable of delivering Cox2W56R into mitochondria with subsequent apparent assembly into a functional COX complex. In the case of soybean Cox2, which is translated in the cytoplasm as a fusion protein containing a 124-aa MTS at its N terminus, the MTS could not be replaced by a 41-aa MTS derived from soybean mitochondrial alternative oxidase (Aox1). Additionally, a chimeric MTS consisting of a 41-aa Aox1 MTS fused to Δ1-72 soybean Cox2 MTS was similarly incapable of supporting soybean Cox2 allotopic expression (25). Nevertheless, the fact that a shorter, 25-aa-long MTS derived from hydrophilic Cox4 does not support allotopic COX2W56R expression indicates that the MTS in the context of the mutant Cox2W56R may fulfill other functions in addition to targeting the protein to the mitochondrial translocation pore. For example, the initial transport stage of the mitochondrial protein import is driven by the inner membrane proton-motive force and is enhanced by an increase in the MTS net positive charge (28). As the MTS of a protein in transit is captured on the matrix side by mHsp70, the chaperone generates ATP-dependent locomotive force and facilitates unfolding of the cytoplasm-facing C-terminal domains of the imported protein (29). Both of these processes occur more efficiently with longer MTSs (assuming constant positive charge density of MTS during the first transport stage) and could account for a failure of a short, Cox4 MTS to deliver Cox2W56R into the mitochondrial matrix. The discovery that replacement of the hydrophobic tryptophan in TM1 by a positively charged arginine residue is necessary for yeast Cox2 allotopic expression correlates well with the observation that a decrease in soybean Cox2 TM1 hydrophobicity is essential for its gene transfer to the nucleus. However, it is also possible that the W56R mutation mediates allotopic Cox2 expression through another, more specific mechanism than hydrophobicity decrease. One such possibility could be a reduction in stability of local Cox2 structure such that it can be unfolded during transport into the mitochondria. Such a mechanism, for example, was reported to enable mitochondrial import of barnase mutants (30). It may also be possible that Arg56 is critical for a proper insertion of cytoplasmically translated Cox2 into the inner mitochondrial membrane. Whereas the experiments presented here do not favor any of the above or other possible mechanisms, these hypotheses can now be directly tested by using in vitro translated MTS-Cox2W56R proteins in transport experiments with purified mitochondria. The fact that a single mutation in Cox2 allows its allotopic expression suggests that nuclear expression of other essential mitochondrial genes may be facilitated by applying approaches similar to that described here.
In summary, the reported experiments indicate that the yeast COX2 gene was probably not prevented from relocation to the nucleus during evolution by its hydrophobicity or other amino acid sequence features incompatible with allotopic expression. In comparison with soybean nuclear Cox2, the yeast gene could be readily evolved into a form compatible with its allotopic expression. Only one mutation is needed in the yeast gene versus two in soybean Cox2. In addition, the MTS has less stringent requirements in the case of yeast protein. Further characterization of Cox2W56R through subcellular localization studies, additional mutagenesis, and in vitro mitochondrial import will allow development of a more detailed mechanistic understanding of requirements associated with allotopic expression of mitochondrial genes. Ultimately we hope to apply the insights gained from these experiments to other mitochondria-encoded genes to determine if respiring yeast can be evolved that completely lack the mitochondrial genome, which no doubt represents a significant challenge for the genes encoding the two most hydrophobic mito-chondrial proteins, Cox1 and apocytochrome b.
Materials and Methods
S. cerevisiae strains used in this study were NB80 (MATa lys2 leu2-3,112 ura3-52 his3ΔHinDIII arg8::hisG [rho+]) and its isonuclear derivative NB97 (MATa lys2 leu2-3,112 ura3-52 his3ΔHinDIII arg8::hisG [cox2-60 mit−]). Yeast cultivation, transformation, and mitochondria isolation were performed according to the published procedures (31, 32). Oxygen consumption experiments were conducted with a single-channel fiber-optic oxygen monitor (Instech Laboratories) model FO/110, using either stationary yeast cultures or isolated mitochondria. Standard techniques were used for PCR and DNA cloning, transformation, and plasmid DNA purification from Escherichia coli. Random mutagenesis of the 2×SU9-COX2 gene was performed by mutagenic PCR. Briefly, two 50-μL PCR reactions, which contained (in addition to other PCR standard components) three dNTPs at 200 μM concentration each (dATP + dGTP + TTP and dATP + dCTP + dGTP) and the corresponding fourth dNTP (dCTP or TTP) at 2 μM concentration, were amplified using 30 PCR cycles. The resulting PCR products were cloned into the pCRII-TOPO vector (Invitrogen). Details are described in SI Text.
Supplementary Material
Acknowledgments
The yeast strains were kindly provided by Dr. Nathalie Bonnefoy (Centre de Génétique Moléculaire, Paris). Dr. David Mueller (Rosalind Franklin University of Medicine and Science, North Chicago, IL) kindly provided the antibody against yeast ATP synthase. We thank Dr. Mary Sever (The Scripps Research Institute, La Jolla, CA) for critical reading of the manuscript. This work was supported by National Institutes of Health Grant GM62159.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000735107/DCSupplemental.
References
- 1.Nagley P, et al. Assembly of functional proton-translocating ATPase complex in yeast mitochondria with cytoplasmically synthesized subunit 8, a polypeptide normally encoded within the organelle. Proc Natl Acad Sci USA. 1988;85:2091–2095. doi: 10.1073/pnas.85.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manfredi G, et al. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat Genet. 2002;30:394–399. doi: 10.1038/ng851. [DOI] [PubMed] [Google Scholar]
- 3.Bonnet C, et al. Allotopic mRNA localization to the mitochondrial surface rescues respiratory chain defects in fibroblasts harboring mitochondrial DNA mutations affecting complex I or v subunits. Rejuvenation Res. 2007;10:127–144. doi: 10.1089/rej.2006.0526. [DOI] [PubMed] [Google Scholar]
- 4.Adams KL, Palmer JD. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol Phylogenet Evol. 2003;29:380–395. doi: 10.1016/s1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 5.Feagin JE. The extrachromosomal DNAs of apicomplexan parasites. Annu Rev Microbiol. 1994;48:81–104. doi: 10.1146/annurev.mi.48.100194.000501. [DOI] [PubMed] [Google Scholar]
- 6.Seeber F, Limenitakis J, Soldati-Favre D. Apicomplexan mitochondrial metabolism: A story of gains, losses and retentions. Trends Parasitol. 2008;24:468–478. doi: 10.1016/j.pt.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 7.Pérez-Martínez X, et al. Unusual location of a mitochondrial gene. Subunit III of cytochrome C oxidase is encoded in the nucleus of Chlamydomonad algae. J Biol Chem. 2000;275:30144–30152. doi: 10.1074/jbc.M003940200. [DOI] [PubMed] [Google Scholar]
- 8.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 9.Nedelcu AM, Lee RW, Lemieux C, Gray MW, Burger G. The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome. Genome Res. 2000;10:819–831. doi: 10.1101/gr.10.6.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Martínez X, et al. Subunit II of cytochrome c oxidase in Chlamydomonad algae is a heterodimer encoded by two independent nuclear genes. J Biol Chem. 2001;276:11302–11309. doi: 10.1074/jbc.M010244200. [DOI] [PubMed] [Google Scholar]
- 11.Funes S, et al. A green algal apicoplast ancestor. Science. 2002;298:2155. doi: 10.1126/science.1076003. [DOI] [PubMed] [Google Scholar]
- 12.Nugent JM, Palmer JD. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell. 1991;66:473–481. doi: 10.1016/0092-8674(81)90011-8. [DOI] [PubMed] [Google Scholar]
- 13.Daley DO, Clifton R, Whelan J. Intracellular gene transfer: Reduced hydrophobicity facilitates gene transfer for subunit 2 of cytochrome c oxidase. Proc Natl Acad Sci USA. 2002;99:10510–10515. doi: 10.1073/pnas.122354399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covello PS, Gray MW. Silent mitochondrial and active nuclear genes for subunit 2 of cytochrome c oxidase (cox2) in soybean: Evidence for RNA-mediated gene transfer. EMBO J. 1992;11:3815–3820. doi: 10.1002/j.1460-2075.1992.tb05473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adams KL, et al. Intracellular gene transfer in action: Dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc Natl Acad Sci USA. 1999;96:13863–13868. doi: 10.1073/pnas.96.24.13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiumera HL, Broadley SA, Fox TD. Translocation of mitochondrially synthesized Cox2 domains from the matrix to the intermembrane space. Mol Cell Biol. 2007;27:4664–4673. doi: 10.1128/MCB.01955-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rojo EE, Guiard B, Neupert W, Stuart RA. N-terminal tail export from the mitochondrial matrix. Adherence to the prokaryotic “positive-inside” rule of membrane protein topology. J Biol Chem. 1999;274:19617–19622. doi: 10.1074/jbc.274.28.19617. [DOI] [PubMed] [Google Scholar]
- 18.Herrmann JM, Koll H, Cook RA, Neupert W, Stuart RA. Topogenesis of cytochrome oxidase subunit II. Mechanisms of protein export from the mitochondrial matrix. J Biol Chem. 1995;270:27079–27086. doi: 10.1074/jbc.270.45.27079. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann JM, Bonnefoy N. Protein export across the inner membrane of mitochondria: The nature of translocated domains determines the dependence on the Oxa1 translocase. J Biol Chem. 2004;279:2507–2512. doi: 10.1074/jbc.M310468200. [DOI] [PubMed] [Google Scholar]
- 20.Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 21.Galanis M, Devenish RJ, Nagley P. Duplication of leader sequence for protein targeting to mitochondria leads to increased import efficiency. FEBS Lett. 1991;282:425–430. doi: 10.1016/0014-5793(91)80529-c. [DOI] [PubMed] [Google Scholar]
- 22.Claros MG, et al. Limitations to in vivo import of hydrophobic proteins into yeast mitochondria. The case of a cytoplasmically synthesized apocytochrome b. Eur J Biochem. 1995;228:762–771. [PubMed] [Google Scholar]
- 23.Margeot A, et al. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002;21:6893–6904. doi: 10.1093/emboj/cdf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonnefoy N, Fox TD. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol Gen Genet. 2000;262:1036–1046. doi: 10.1007/pl00008646. [DOI] [PubMed] [Google Scholar]
- 25.Daley DO, et al. Gene transfer from mitochondrion to nucleus: Novel mechanisms for gene activation from Cox2. Plant J. 2002;30:11–21. doi: 10.1046/j.1365-313x.2002.01263.x. [DOI] [PubMed] [Google Scholar]
- 26.Popot JL, de Vitry C. On the microassembly of integral membrane proteins. Annu Rev Biophys Biophys Chem. 1990;19:369–403. doi: 10.1146/annurev.bb.19.060190.002101. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne G. Why mitochondria need a genome. FEBS Lett. 1986;198:1–4. doi: 10.1016/0014-5793(86)81172-3. [DOI] [PubMed] [Google Scholar]
- 28.Huang S, Ratliff KS, Matouschek A. Protein unfolding by the mitochondrial membrane potential. Nat Struct Biol. 2002;9:301–307. doi: 10.1038/nsb772. [DOI] [PubMed] [Google Scholar]
- 29.Matouschek A, et al. Active unfolding of precursor proteins during mitochondrial protein import. EMBO J. 1997;16:6727–6736. doi: 10.1093/emboj/16.22.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilcox AJ, Choy J, Bustamante C, Matouschek A. Effect of protein structure on mitochondrial import. Proc Natl Acad Sci USA. 2005;102:15435–15440. doi: 10.1073/pnas.0507324102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schiestl RH, Gietz RD. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 32.Yaffe MP. In: Guide to Yeast Genetics and Molecular Biology. Guthrie C, Fink GR, editors. San Diego: Academic Press; 1991. pp. 627–643. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.