Abstract
The polar bear has become the flagship species in the climate-change discussion. However, little is known about how past climate impacted its evolution and persistence, given an extremely poor fossil record. Although it is undisputed from analyses of mitochondrial (mt) DNA that polar bears constitute a lineage within the genetic diversity of brown bears, timing estimates of their divergence have differed considerably. Using next-generation sequencing technology, we have generated a complete, high-quality mt genome from a stratigraphically validated 130,000- to 110,000-year-old polar bear jawbone. In addition, six mt genomes were generated of extant polar bears from Alaska and brown bears from the Admiralty and Baranof islands of the Alexander Archipelago of southeastern Alaska and Kodiak Island. We show that the phylogenetic position of the ancient polar bear lies almost directly at the branching point between polar bears and brown bears, elucidating a unique morphologically and molecularly documented fossil link between living mammal species. Molecular dating and stable isotope analyses also show that by very early in their evolutionary history, polar bears were already inhabitants of the Artic sea ice and had adapted very rapidly to their current and unique ecology at the top of the Arctic marine food chain. As such, polar bears provide an excellent example of evolutionary opportunism within a widespread mammalian lineage.
Keywords: ancient DNA, Arctic, mammal evolution, next-generation sequencing, Svalbard
Rapid morphological evolution has long been recognized in mammals. For example, it is widely accepted that anatomically modern humans evolved in Africa around 200–160 thousand years ago (kya), expanded into most habitable parts of the Old World between 90 and 40 kya (1), and swiftly radiated in phenotypic traits readily recognized in modern populations. Ursine bears have themselves diverged recently into a number of different species (2, 3). The polar bear (Ursus maritimus Phipps 1774), which is the largest of the six extant ursine bear species, currently has a circumpolar distribution, their range determined by the extent of Arctic sea ice. The polar bear is closely related to the brown bear (Ursus arctos). Kurtén (4) suggested that polar bears branched off from brown bears that became isolated on Siberian coastal enclaves some time during the Mid to Late Pleistocene, perhaps as recently as 100–70 kya, with time becoming increasingly specialized carnivores that hunt solely on sea ice. Recent genetic studies have shown that polar bears evolved from within brown bears, and that a genetically unique clade of brown bear populations that live exclusively on the Admiralty, Baranof, and Chichagof (ABC) islands of southeastern Alaska’s Alexander Archipelago are more closely related to polar bears than to other brown bears (5, 6). Although fertile hybrids between the two species are well-known in captivity (7), wild hybrids are extremely rare and both species are well-adapted to their different, allopatric habitat requirements. With their distinctly different morphology, metabolism, and social and feeding behaviors, the polar and brown bears are classified as separate species (8).
Timing estimates for the divergence of polar bears from brown bears have differed considerably. In addition to the hypothesis of a very recent split, a divergence time of 250–200 kya has also been proposed (6), whereas based on comparisons of complete mitochondrial (mt) genomes the cladogenesis has been estimated to be as distant as 1,320 kya (9). Recently, using an extended dataset of 10 mt genomes, the split of brown and polar bears was estimated to be 1,170–660 kya (2). Sources of these inconsistent estimates may have included inappropriate, deep fossil calibrations as well as limited sampling, and in particular lack of data on brown bears from the ABC islands. Recognizing these shortcomings, internally calibrated substitution rates have been calculated from a dataset of short stretches of mt control-region sequences, including several radiocarbon-dated individuals, with the result that the divergence time between polar and ABC island bears was estimated to be as recent as 72–48 kya (10). However, the limited polymorphic characters within these very short stretches may contribute to erroneous dating in this case.
Fossil remains of the polar bear are very rare (4, 11–13), because the animals mostly live and die over vast areas of sea ice, and when they die their remains are likely to be scavenged by other animals and disappear into the ocean. In light of this paucity of fossil finds, every new specimen is of interest. Recently, a lower jawbone (left mandible) was excavated in situ at Poolepynten on Prins Karls Forland, a narrow strip of land on the far western edge of the Svalbard Archipelago, Norway (11). Diagnostic polar bear traits and morphometric measurements of this well-preserved mandible, comparing it to brown bear and other available subfossil polar bear remains as well as a large collection of extant polar bears from Svalbard, proved that it falls within the range of modern polar bears and suggested that it belonged to an adult male (11). Accelerator mass spectrometry 14C age determination from a canine tooth attached to the jawbone dated it to older than 45 thousand years (ky) old (11). Based on long-term studies of the stratigraphy and depositional history of the Poolepynten area and infrared-stimulated luminescence of the sediments (14), the specimen was estimated to be 130–110 ky old, which is significantly older than any other known polar bear subfossils (i.e., partly fossilized specimens), none of which are older than possibly ∼70 ky (and most younger than 10 ky) (11). The discovery of this jawbone confirms that the polar bear was already a distinct species at least 110 kya, and as such any findings from genetic research based on this specimen could contribute to answering key questions on the evolutionary history of this species.
High-throughput sequencing-by-synthesis techniques (15) (a type of shotgun sequencing) have recently been applied to ancient DNA (16, 17). These methods have opened new possibilities with the vast number of short sequence reads that can be generated, not only from the mt genome but also the nuclear genome, using relatively small amounts of degraded DNA. This method, essentially metagenomic in that it samples DNA that is expected to contain elements from the environment in addition to the species at hand, circumvents amplification and cloning biases, because single DNA molecules are compartmentalized in a lipid vesicle before amplification. Furthermore, generation of high coverages of the mt genome makes it possible to efficiently deal with single-base errors caused by postmortem miscoding lesion damage. Using this “next-generation” sequencing technology, we generated a complete, multifold-coverage mt genome from the Poolepynten specimen, which is significantly older than other stratigraphically validated mammal subfossils from which mt genomes have been reported (18).
Results and Discussion
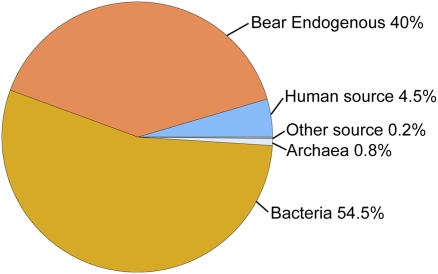
Initial diagnostic PCR amplifications of short stretches of the mt D loop and cytochrome b gene using DNA isolated from the attached canine tooth from the Poolepynten specimen confirmed the jaw’s polar bear identity. Although the molecular behavior of this subfossil specimen clearly matched what is expected from ancient DNA (19), it also exhibited the signature of well-preserved DNA. Consequently, using a Roche GS FLX sequencer (454 Life Sciences), 77 million base pairs (bp) were generated. A comparison of the 454 sequence reads against the dog genome suggested that ∼40% of the reads represent endogenous bear DNA (Fig. 1). Although 4.5% and the remaining ∼55% represent human and bacterial contamination, respectively, the read length and high percentage of endogenous, mostly nuclear sequence demonstrate remarkable DNA preservation. It is believed that due to their constant low temperatures, glacial ice and permafrost environments provide ideal conditions for long-term survival of DNA molecules (20), and indeed, some of the oldest authenticated ancient DNA results have been reported from ice and permafrost cores (20, 21). The quality of the DNA extracted from the Poolepynten specimen further suggests that environments like Svalbard may show great promise for the recovery of well-preserved ancient nucleic acids.
Fig. 1.
Metagenomic composition of the 454 sequence reads from the Poolepynten polar bear tooth. The pie chart shows the percentages of sequence reads assigned to the categories bacteria, bear endogenous, human, Archaea, and other sources. A comparison of the 454 sequence reads against the dog genome suggested that ∼40% of the reads represent endogenous bear DNA, whereas 4.5% and the remaining ∼55% represent human and bacterial contamination, respectively.
Of the total of 482,364 sequence reads recovered from the Poolepynten specimen, 1290 mt-associated reads provided a 14× coverage of the mt genome (Fig. S1). The organization and length of the genome is comparable to that of extant bears, showing clear sequence similarity to both ABC bears and modern polar bears (Fig. 2). To investigate the precise relationship of the ancient polar bear specimen to modern bears, we similarly used shotgun sequencing to generate six high-quality mt genomes of extant bears: two polar bears from the Little Diomede and St. Lawrence islands, Alaska, three brown bears from the ABC islands, and a brown bear from Kodiak Island, Alaska (Table S1 and Fig. S2). These seven complete mt genomes from ancient and modern bears were aligned to previously published mt genomes, including two mt genomes each from polar bear and brown bear (3, 22, 23). The four modern polar bears included represent well the distinct genetic clusters of polar bear populations identified from microsatellite analyses (24).
Fig. 2.
Mitogenomic sequence variation and organization. Sequence differences found among 17 bear mitochondrial genomes with respect to a previously published polar bear genome (GenBank accession no. NC_003428). Each vertical bar depicts a nucleotide difference from this reference sequence (shown at the bottom illustrating the organization of the genome into the different regions). The ancient Poolepynten bear sequence is highlighted in red.
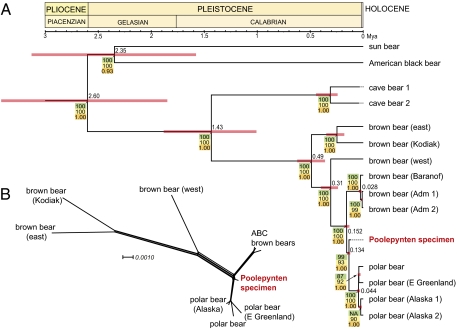
Comprehensive phylogenetic reconstruction analyses of this dataset resulted in trees of similar topology (Fig. 3A and Figs. S3 and S4) comparable to recently reported phylogenies based on complete mt genomes and nuclear sequences (2, 3, 25). The phylogenetic results clearly demonstrate, with high support, the close relationship of the subfossil specimen to modern polar bear (Fig. 3A). Intriguingly, however, this ancient polar bear, which exhibits a very short branch length, lies almost directly at the branching point between polar bear and the genetically unique clade of ABC brown bears (Fig. 3B). Thus, both cladistically and anagenetically, this ancient specimen existed very close to the most recent common ancestor of polar bears and brown bears.
Fig. 3.
Phylogenetic and chronographic reconstruction of polar bear evolution. (A) Maximum clade probability tree inferred from a BEAST analysis of complete mt genome sequences excluding the VNTR repeat in the D loop (Table S2). Numbers at selected nodes indicate mean ages in million years. The red bars at nodes illustrate the age width of the 95% highest posterior density interval. The posterior probability values of each clade are indicated in orange. An identical tree topology was obtained using maximum parsimony and maximum likelihood (bootstrap support values in green and yellow, respectively). For details on voucher information for the mtDNA genome sequences included in this study, see Table S1 and Figs. S3 and S4. The 2009 Geologic Time Scale with major relevant epochs is shown above the tree. (B) Phylogenetic network of complete mt genomes (excluding the VNTR repeat) of 11 polar and brown bears based on Neighbor-Net analysis with LogDet distances (see scale bar).
The robust phylogeny and the close position of the subfossil polar bear specimen to the polar/brown bear split offer an ideal opportunity to ultimately settle a time of origin for the polar bear. Our Bayesian analyses with different datasets returned a divergence date for the entire brown bear/polar bear lineage to a mean of less than 500 ky (Fig. 3A; see also Table S2), which is consistent with recent estimates using deeper fossil calibration in the Ursidae (3). Within this clade, we estimated the mean age of the split between the ABC bears and the polar bears to be 152 ky, and the mean age for all polar bears as 134 ky, near the beginning of the Eemian interglacial period and completely in line with the stratigraphically determined age of the Poolepynten subfossil (11). Analyses of an extended dataset of 39 mtDNA control-region sequence fragments from a number of carbon-dated brown bears (26) and modern polar bears, importantly including four from Svalbard, provided comparable, although slightly older (190 ky for the ABC/polar bear split), divergence time estimates (Fig. S5). Although mtDNA capture cannot be excluded to have happened between ABC bears and polar bears, these estimates nevertheless affirm with strong support a very recent divergence of polar bears from brown bears. Even more surprising, the age of the modern polar bear crown group (the clade containing the last common ancestor of all extant members) is estimated to be less than 45 ky, slightly older than the age of the ABC bears (Fig. 3A), a date that is also found with the expanded dataset of control-region sequence fragments (Fig. S5). These estimates suggest a very recent and rapid expansion of modern polar bear populations throughout the Arctic since the Late Pleistocene, perhaps following a climate-related population bottleneck, although data from more modern and Holocene polar bear specimens will be required to establish this.
Stable isotope analyses of carbon and nitrogen have been used as a tool to evaluate trophic relationships, both in past and present environments (26, 27). Modern polar bears prey mainly on ringed seals and bearded seals (28), and stable isotopes have confirmed that they are marine predators that make little use of terrestrial food sources (29–31). To investigate the trophic relationships of the ancient Poolepynten specimen as compared with present-day polar bears from Svalbard, we analyzed the stable isotope content in the canine tooth. The stable isotope values for the ancient tooth (δ13C = −13.9‰ and δ15n = 19.4‰) were within the range found from extant polar bear teeth and other tissues and were reflective of marine feeding (26, 29–31). Importantly, these isotope values are distinct from those found in Late Pleistocene brown bears, including from the ABC brown bear lineage (26), as well as present-day coastal Alaskan brown bears (32). Thus, our results clearly demonstrate that the jaw is from an individual that had a feeding ecology similar to present-day polar bears, at the top of the Arctic marine food chain. Furthermore, analyses of the stratum containing the subfossil jawbone uncovered a bivalve and foraminifera fauna reflecting an arctic, open marine environment influenced by glacier input and advection of warm North Atlantic water as today (11, 14).
The stable isotope data, phylogenetic analysis, and the geological and molecular age estimates of the Poolepynten specimen indicate that ancient polar bears adapted extremely rapidly both morphologically and physiologically to their current and unique ecology within only 10–30 ky following their split from a brown bear precursor and, subsequently, within the course of ∼100 ky, spread to the full perimeter of the polar basin. As such, the polar bear is an excellent example of evolutionary opportunism within a widespread mammalian lineage (33). Moreover, the extreme proximity of the Poolepynten specimen to the polar bear ancestor provides a unique case of a morphologically and molecularly validated fossil link between living mammal species.
With recent announcements of plans for sequencing the genome from present-day polar bear (34) offering a necessary reference, future sequencing of all or a substantial fraction of the nuclear genome of this exceptionally well preserved Pleistocene polar bear specimen may be feasible. This genome would not only provide excellent markers to study extant polar bear populations but could also provide clues as to how polar bears rapidly evolved and subsequently survived through the last interglacial period. Such information is important for an assessment of how polar bears will be able to cope with the predicted changes of their main habitat: the Arctic sea ice.
Methods
Initial PCR and Cloning and Ancient DNA Validation.
We extracted and amplified DNA from the subfossil polar bear specimen at the Natural History Museum, University of Oslo, Norway, using standard ancient-DNA approaches and protocols (35); that is, all DNA extractions and setup of PCR reactions were performed in a building physically separated from molecular laboratory facilities where subsequent amplification, cloning, and sequencing of the products were performed. A Dremel tool was used at low speed to drill out powder of the dentine of the canine tooth attached to the jawbone after the tooth was first washed with 10% chlorine to remove any contamination at the surface. Approximately 0.1 g of bone powder was subsequently transferred to a 2.0-mL screw-capped centrifugation tube and DNA was extracted as previously published (35). Initial diagnostic genetic analyses targeted one short mtDNA segment in the cytochrome b gene and two overlapping segments in the D loop. Primer design, PCR amplifications, cloning, and sequencing were performed using standard procedures (35). Successful amplification and sequencing of these short stretches indicated with some certainty that the DNA fragments amplified came from the fossil polar bear remains, because characteristic C→T (deamination of cytosine) mutations were uncovered, and the positions of these mutations differed in separately bacterially cloned pieces of PCR-amplified DNA. Furthermore, experiments with different amplification conditions and fragment lengths strongly suggested a molecular behavior expected from ancient DNA (19), but also that the DNA was well-preserved. For example, amplification of products up to ∼300 bp was successful, whereas amplification over 600 bp yielded no products.
DNA Library Construction and 454 Sequencing.
High-throughput sequencing-by-synthesis techniques (15) have recently been applied to ancient DNA (16, 17, 36). After confirming that DNA was well-preserved in the canine tooth attached to the jawbone, library construction and shotgun sequencing were performed using the 454 Life Sciences standard GS FLX protocol and a Roche GS FLX sequencer (454 Life Sciences) at the Center for Comparative Genomics and Bioinformatics, Pennsylvania State University. Two 454 GS FLX fragment runs were performed for the ancient polar bear sample, altogether producing 77 million bp of DNA sequence data. One was a full-panel 454 GS FLX fragment run, which produced 347,660 reads with an average length of 156 bp. The other was a quarter-panel 454 GS FLX fragment run, which produced 134,704 reads with an average length of 168 bp.
To exclude the possibility of contamination in the analysis of the ancient sample, DNA library constructions of modern samples were performed after all analyses of the ancient sample. DNA extractions of the modern polar and brown bear tissue samples were done at the University at Buffalo, using a DNeasy tissue kit (Qiagen). The modern polar bear samples were obtained from the U.S. Fish and Wildlife Marine Region 7 Mammal Management Office and harvested in 1998, 2001, and 2002 (Table S1), that is, before listing of the species in the Endangered Species Act. The modern bear samples were sequenced in one 454 GS FLX fragment run with multiplex identifiers (MIDs) according to the 454 Life Sciences MID GS FLX protocol. For each sample, a unique 10-bp nucleotide identifier was attached to each DNA fragment to distinguish them in the same fragment run. A total of 10 modern bear samples were sequenced, labeled from MID1 to MID10, 6 of which generated sufficient mt reads for assembly of mtDNA genomes. A total of 624,921 reads, with a mean length of 186 bp, were produced in this MID fragment run. The total 116 million bp of sequence data are unevenly distributed among the 10 samples (Table S1).
Composition Analysis of Sequenced Reads.
The reads produced for the ancient polar bear sample were analyzed to determine their composition. Only reads longer than 100 bp were considered, to minimize erroneous hits commonly seen when using short reads as queries. These reads were first searched against the National Center for Biotechnology Information (NCBI) nonredundant peptide sequence (NR) database (April 22, 2009 version) using BLASTx (37), with the threshold E value = e−4. The same set of reads was then searched against the NCBI nucleotide sequence (NT) database (April 22, 2009 version) using BLASTn (37), with the threshold E value = e−10. For each query, the top 10 best hits were recoded. Both results were analyzed and compared in MEGAN v. 3.3.5 (38).
Fraction of the Reads That Are from the Bear.
We first determined which reads represent human contamination, by aligning reads to the human genome sequence, scoring matches 1 and mismatches –3. This step estimated that about 4.5% of the sequences are human. To estimate the fraction of sequence data that comes from the polar bear genome, we aligned the remaining reads with the dog genome, scoring matches 1 and mismatches –2, noting that about 15% of the reads aligned. This underestimates the fraction that are from the bear genome because it misses sequences in the following categories: (i) inserted in the bear genome since the bear-dog split (e.g., recent families of interspersed repeats), (ii) deleted in the dog lineage since the bear-dog split, (iii) missing from the current dog genome assembly, or (iv) not alignable to dog because ancient DNA is broken into small pieces. We estimated the combined effect of these factors by taking reads from a carefully prepared modern bear sample, which we assume consists entirely of bear DNA, artificially shortening those reads to make the length distribution match that of the ancient polar bear sample, and aligning to the dog genome sequence using the same thresholds. This led to the estimation that the true amount of bear nuclear DNA is over 2.5 times as much as what aligns to dog under these conditions, which in turn led to the estimation that around 40% of the sequences in our sample are from the polar bear genome.
Mitochondrial Genome Assembly.
The short reads produced by 454 sequencing runs were first filtered by size at a threshold of 100 bp to ensure the specificity of assembled reads. The filtered reads were then screened for those associated with bear mtDNA using BLASTn (37) against the polar bear mt reference genome from NCBI (accession no. NC003428), with the threshold E value = e−20. The mt genome assembly was done using SeqMan 8.0 (DNASTAR), allowing minimal 80% identity. The assembly was then manually inspected to ensure the correctness of the assembly, which eliminated all ambiguities in the consensus sequence.
The ursine mt genomes contain variable-number tandem repeats (VNTR) in the control region. The average VNTR length is ∼500 bp, which exceeds the maximum length of 454 fragment run reads. To resolve the correct number of repeats for each assembled mt genome, PCR amplification of the regions containing VNTR were done for each successfully assembled sample (primers: 5′-CGCCACTAAATCGAACGAAC-3′, 5′-GGGGGTTTGATTAAGCTAAGTT-3′). PCR products were purified using a QIAquick PCR purification kit (Qiagen) and analyzed on an Agilent 2100 bioanalyzer with the DNA 7500 kit (Agilent Technologies). The final VNTR repeat number of the mt genomes was corrected based on the bioanalyzer data (Fig. S6).
Phylogenetic Reconstruction.
Phylogenetic analyses were performed using a comprehensive maximum parsimony (MP) approach with a variety of tree space exploration techniques and bootstrap resampling as implemented in TNT (39) and maximum likelihood (ML) with a rapid bootstrap algorithm as implemented in RAxML (40), and network construction was analyzed using the Neighbor-Net algorithm (41) with LogDet distances (42) in SplitsTree v. 4.10 (43).
Estimations of Divergence Times.
Dating of divergence events within the brown bear/polar bear lineage were estimated using a Bayesian “relaxed molecular clock” approach (44) in the software BEAST (45) v. 1.4.8 using variable parameter sets and datasets (Table S2). Following suggestions to use calibration points as close as possible to dates being estimated when timing recent divergences (46), a mean age of 120 ky for the Poolepynten subfossil polar bear was used in addition to data from carbon-dated specimens (2, 3, 26). Results were assessed with Tracer (47) v. 1.4.1, and maximum clade credibility trees were produced with TreeAnnotator v. 1.4.8.
Isotope Analyses.
Stable isotope analyses of carbon and nitrogen have been used as a tool for evaluating trophic relationships, both in past and present environments (26, 27). Stable isotope analyses of the canine dentine material were performed at the Institute for Energy Technology (IFE), Kjeller, Norway. From 3.1 to 3.6 mg were weighed into tin capsules. The samples were combusted in the presence of O2 and Cr2O3 at 1,700 °C in a Eurovector element analyzer. Reduction of NOx to N2 was done in a Cu oven at 650 °C. H2O was removed in a chemical trap of KMnO4 before separation of N2 and CO2 on a packed 2-m PoraPLOT Q GC column (Varian). N2 and CO2 were directly injected on-line to a Nu Horizon Stable Isotope Ratio Mass Spectrometer (Nu Instruments) for determination of δ13C and δ15N. The C and N content were quantified on the basis of the mass 44 and mass 28 peak areas, respectively, and accuracy and precision of the analyses were measured by replicate analysis using an internal standard (IFE trout) and international standards (IAEA-N-1 and IAEA-N-2 calibrated against atmospheric N2 for 15N, and USGS-24 calibrated against PeeDee Belemnite, Vienna, for 13C) (48). The stable isotope compositions are reported using the standard δ notation.
Supplementary Material
Acknowledgments
We thank Victor A. Albert for critical reading and encouragement throughout this project, Joseph Cook for accessioning tissues of extant bears, the US Fish and Wildlife Service Region 7 Marine Mammals Management for obtaining the modern polar bear samples, and Yeting Zhang for producing the PCR and bioanalyzer data. Natalie Dawson, Kevin Sage, and Yixing Shi assisted with sample preparation. This study was supported by the College of Arts and Sciences, University at Buffalo, the Natural History Museum of the University of Oslo, and the US Geological Survey Alaska Science Center. S.C.S. is supported by the Gordon and Betty Moore Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. GU573485‒GU573491).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914266107/DCSupplemental.
References
- 1.Powell A, Shennan S, Thomas MG. Late Pleistocene demography and the appearance of modern human behavior. Science. 2009;324:1298–1301. doi: 10.1126/science.1170165. [DOI] [PubMed] [Google Scholar]
- 2.Krause J, et al. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol Biol. 2008;8:220. doi: 10.1186/1471-2148-8-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bon C, et al. Deciphering the complete mitochondrial genome and phylogeny of the extinct cave bear in the Paleolithic painted cave of Chauvet. Proc Natl Acad Sci USA. 2008;105:17447–17452. doi: 10.1073/pnas.0806143105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurtén B. The evolution of the polar bear, Ursus maritimus Phipps. Acta Zool Fenn. 1964;108:1–30. [Google Scholar]
- 5.Shields GF, et al. Phylogeography of mitochondrial DNA variation in brown bears and polar bears. Mol Phylogenet Evol. 2000;15:319–326. doi: 10.1006/mpev.1999.0730. [DOI] [PubMed] [Google Scholar]
- 6.Talbot SL, Shields GF. Phylogeography of brown bears (Ursus arctos) of Alaska and paraphyly within the Ursidae. Mol Phylogenet Evol. 1996;5:477–494. doi: 10.1006/mpev.1996.0044. [DOI] [PubMed] [Google Scholar]
- 7.Gray AP. Mammalian Hybrids. A Check-List with Bibliography. Slough, UK: Commonwealth Agricultural Bureaux; 1972. [Google Scholar]
- 8.Stirling I, Guravich D. Polar Bears. Ann Arbor: University of Michigan Press; 1998. [Google Scholar]
- 9.Yu L, Li Y-W, Ryder OA, Zhang Y. Analysis of complete mitochondrial genome sequences increases phylogenetic resolution of bears (Ursidae), a mammalian family that experienced rapid speciation. BMC Evol Biol. 2007;7:198–209. doi: 10.1186/1471-2148-7-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho SYW, Saarma U, Barnett R, Haile J, Shapiro B. The effect of inappropriate calibration: Three case studies in molecular ecology. PLoS One. 2008;3:e1615. doi: 10.1371/journal.pone.0001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ingólfsson Ó, Wiig Ø. Late Pleistocene fossil find in Svalbard: The oldest remains of a polar bear (Ursus maritimus Phipps, 1744) ever discovered. Polar Res. 2008;28:455–462. [Google Scholar]
- 12.Laidre KL, et al. Quantifying the sensitivity of Arctic marine mammals to climate-induced habitat change. Ecol Appl. 2008;18:S97–S125. doi: 10.1890/06-0546.1. [DOI] [PubMed] [Google Scholar]
- 13.Harington CR. The evolution of arctic marine mammals. Ecol Appl. 2008;18:S23–S40. doi: 10.1890/06-0624.1. [DOI] [PubMed] [Google Scholar]
- 14.Andersson T, Forman SL, Ingólfsson Ó, Manley WF. Late Quaternary environmental history of central Prins Karls Forland, western Svalbard. Boreas. 1999;28:292–307. [Google Scholar]
- 15.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poinar HN, et al. Metagenomics to paleogenomics: Large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 17.Miller W, et al. The mitochondrial genome sequence of the Tasmanian tiger (Thylacinus cynocephalus) Genome Res. 2009;19:213–220. doi: 10.1101/gr.082628.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho SYW, Gilbert MTP. Ancient mitogenomics. Mitochondrion. 2010;10:1–11. doi: 10.1016/j.mito.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 19.Cooper A, Poinar HN. Ancient DNA: Do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 20.Willerslev E, Hansen AJ, Poinar HN. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol Evol. 2004;19:141–147. doi: 10.1016/j.tree.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Willerslev E, et al. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science. 2003;300:791–795. doi: 10.1126/science.1084114. [DOI] [PubMed] [Google Scholar]
- 22.Delisle I, Strobeck C. Conserved primers for rapid sequencing of the complete mitochondrial genome from carnivores, applied to three species of bears. Mol Biol Evol. 2002;19:357–361. doi: 10.1093/oxfordjournals.molbev.a004090. [DOI] [PubMed] [Google Scholar]
- 23.Arnason U, et al. Mammalian mitogenomic relationships and the root of the eutherian tree. Proc Natl Acad Sci USA. 2002;99:8151–8156. doi: 10.1073/pnas.102164299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paetkau D, et al. Genetic structure of the world’s polar bear populations. Mol Ecol. 1999;8:1571–1584. doi: 10.1046/j.1365-294x.1999.00733.x. [DOI] [PubMed] [Google Scholar]
- 25.Pagès M, et al. Combined analysis of fourteen nuclear genes refines the Ursidae phylogeny. Mol Phylogenet Evol. 2008;47:73–83. doi: 10.1016/j.ympev.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Barnes I, Matheus P, Shapiro B, Jensen D, Cooper A. Dynamics of Pleistocene population extinction in Beringian brown bears. Science. 2002;295:2267–2270. doi: 10.1126/science.1067814. [DOI] [PubMed] [Google Scholar]
- 27.Kelly JF. Stable isotopes of carbon and nitrogen in the study of avian and mammalian trophic ecology. Can J Zool. 1999;78:1–27. [Google Scholar]
- 28.Derocher AE, Wiig Ø, Andersen M. Diet composition of polar bears in Svalbard and the western Barents Sea. Polar Biol. 2002;25:448–452. [Google Scholar]
- 29.Bentzen TW, et al. Variation in winter diet of southern Beaufort Sea polar bears inferred from stable isotope analysis. Can J Zool. 2007;87:596–608. [Google Scholar]
- 30.Hobson KA, Stirling I, Andriashek DS. Isotopic homogeneity of breath CO2 from fasting and berry-eating polar bears: Implications for tracing reliance on terrestrial foods in a changing Arctic. Can J Zool. 2009;87:50–55. [Google Scholar]
- 31.Ramsay MA, Hobson KA. Polar bears make little use of terrestrial food webs: Evidence from stable-carbon isotope analysis. Oecologia. 1991;86:598–600. doi: 10.1007/BF00318328. [DOI] [PubMed] [Google Scholar]
- 32.Ben-David M, Titus K, Beier LR. Consumption of salmon by Alaskan brown bears: A trade-off between nutritional requirements and the risk of infanticide? Oecologia. 2004;138:465–474. doi: 10.1007/s00442-003-1442-x. [DOI] [PubMed] [Google Scholar]
- 33.Stanley SM. Macroevolution, Pattern and Processes. San Francisco: W.H. Freeman; 1979. [Google Scholar]
- 34.GenomeWeb News. BGI plans to sequence polar bears, penguins, Tibetan antelope. GenomeWeb Daily News. April 27, 2009 Available at http://www.genomeweb.com/sequencing/bgi-plans-sequence-polar-bears-penguins-tibetan-antelope. [Google Scholar]
- 35.Lindqvist C, et al. The Laptev Sea walrus Odobenus rosmarus laptevi: An enigma revisited. Zool Scr. 2008;38:113–127. [Google Scholar]
- 36.Green RE, et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell. 2008;134:416–426. doi: 10.1016/j.cell.2008.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huson DH, Auch A, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goloboff PA, Farris JS, Nixon KC. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- 40.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 41.Bryant D, Moulton V, Spillner A. Consistency of the Neighbor-Net algorithm. Algorithms Mol Biol. 2007;2:8. doi: 10.1186/1748-7188-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lockhart PJ, Steel MA, Hendy MD, Penny D. Recovering evolutionary trees under a more realistic model of sequence evolution. Mol Biol Evol. 1994;11:605–612. doi: 10.1093/oxfordjournals.molbev.a040136. [DOI] [PubMed] [Google Scholar]
- 43.Huson D, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 44.Drummond A, Ho S, Phillips M, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond A, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho SYW, Larson G. Molecular clocks: When times are a-changin’. Trends Genet. 2006;22:79–83. doi: 10.1016/j.tig.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 47.Rambaut A, Drummond A. 2007 Tracer v. 1.4. Available at http://beastbioedacuk/Tracer. [Google Scholar]
- 48.Søreide JE, Tamelander T, Hop H, Hobson KA, Johansen I. Sample preparation effects on stable C and N isotope values: A comparison of methods in Arctic marine food web studies. Mar Ecol Prog Ser. 2006;328:17–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.