Abstract
Modification of proteins by the addition of poly(ADP-ribose) is carried out by poly(ADP-ribose) polymerases (PARPs). PARPs have been implicated in a wide range of biological processes in eukaryotes, but no universal function has been established. A study of the Aspergillus nidulans PARP ortholog (PrpA) revealed that the protein is essential and involved in DNA repair, reminiscent of findings using mammalian systems. We found that a Neurospora PARP orthologue (NPO) is dispensable for cell survival, DNA repair and epigenetic silencing but that replicative aging of mycelia is accelerated in an npo mutant strain. We propose that PARPs may control aging as proposed for Sirtuins, which also consume NAD+ and function either as mono(ADP-ribose) transferases or protein deacetylases. PARPs may regulate aging by impacting NAD+/NAM availability, thereby influencing Sirtuin activity, or they may function in alternative NAD+-dependent or NAD+-independent aging pathways.
INTRODUCTION
Poly(ADP-ribose) polymerases (PARPs) are ADP-ribose transferases that catalyze the formation of both linear and branched polymers of ADP-ribose (PAR) on target proteins. PAR is covalently linked to the γ–carboxy group of glutamic acid residues at acceptor sites (BURZIO et al. 1979; RIQUELME et al. 1979). Poly(ADP-ribosylation) (PARylation) consumes nicotinamide adenine dinucleotide (NAD+) and generates nicotinamide (NAM). The addition of PAR to proteins is thought to have dramatic effects on their catalytic activities, as well as on potential protein-protein and protein-nucleic acid interactions (BURKLE 2000; D'AMOURS et al. 1999; KRAUS and LIS 2003). Recently a number of different proteins have been identified that bind to PAR both in vitro and in vivo, including proteins containing Macro domains and proteins containing novel poly(ADP-ribose)-binding zinc finger (PBZ) motifs (AHEL et al. 2008; KARRAS et al. 2005). In higher eukaryotes PARylation is reversible through the action of PAR glycohydrolases (PARG), which are active in a variety of subcellular compartments, and are thought to be important in regulation of cell death after DNA damage (AME et al. 2009a; AME et al. 2009b). Thus, the principle players in PARylation thus far identified are the PARPs, PARG and PAR binding proteins.
PARP homologs have been identified in plants, metazoans, protists and filamentous fungi, but not in the yeasts, while PARG homologs have been identified in all eukaryotes, excluding fungi. PARPs and PARylation impact a variety of biological processes including development, transcriptional regulation, chromatin structure, epigenetic phenomena, DNA repair, mitosis, genome stability, neuronal function, cell death and aging (BENEKE and BURKLE 2004; BENEKE and BURKLE 2007; BOUCHARD et al. 2003; BOULU et al. 2001; BURKLE 2000; BURKLE 2001a; BURKLE et al. 2005; CHIARUGI and MOSKOWITZ 2002; D'AMOURS et al. 1999; HERCEG and WANG 2001; HONG et al. 2004; JEGGO 1998; KIM et al. 2005; KRAUS and LIS 2003; PIEPER et al. 1999; SMULSON et al. 2000).
The canonical PARP enzyme from mammals, PARP-1, has been implicated in both double and single strand break repair (DSB and SSB), as well as base excision repair (BER) (BURKLE 2001b; DANTZER et al. 1999; MASUTANI et al. 2003). In human and mouse cells, the majority of PARylation involves auto-modification of PARP-1 in response to DNA damage and PARP-1 has been described as a DNA damage sensor (D'AMOURS et al. 1999; DE MURCIA et al. 1997; HULETSKY et al. 1989; OGATA et al. 1981). Residual PARylation is detectable in mouse embryonic fibroblast homozygous for PARP-1 null mutations (PARP-1−/−) (SHIEH et al. 1998) and this may reflect PARP-2, which has also been shown to PARylate in response to DNA damage (AME et al. 1999). Both PARP-1−/− and PARP-2−/− mice are viable, but are sensitive to DNA damaging agents, and PARP-1−/− mice have inherent genomic instability (DE MURCIA et al. 1997; MENISSIER DE MURCIA et al. 2003; TRUCCO et al. 1998; WANG et al. 1995; WANG et al. 1997). PARP-1−/−/PARP-2−/− mice die as embryos prior to E8.0, and PARP-1+/−/PARP-2−/− female mice exhibit X-chromosome instability, infertility, and higher levels of embryonic lethality (MENISSIER DE MURCIA et al. 2003). These results suggest that PARylation may be essential in higher eukaryotes.
A recent investigation using the filamentous fungus Aspergillus nidulans revealed the presence of a single PARP ortholog (PrpA) (SEMIGHINI et al. 2006). Disruption of the prpA gene was found to be lethal in haploid strains, and diploid strains carrying only a single copy of prpA had severe growth restrictions and were found to be sensitive to several mutagenic compounds (SEMIGHINI et al. 2006). These results suggest that the requirement of PARP for DNA repair and viability is conserved between animals and filamentous fungi.
In addition to evidence that PARPs and PARylation control diverse aspects of gene expression, DNA repair and genome stability, there are suggestions that PARP-1 is involved in controlling aging in metazoans. GRUBE and BURKLE (1992) found a strong positive correlation between lifespan and the degree of PARP activity in leukocytes of 13 mammalian species. Long-lived species had higher levels of PARylation, but similar levels of PARP protein, implying greater enzyme activity (GRUBE and BURKLE 1992). In addition, the WRN protein, which is defective in individuals with the premature aging disorder Werner’s syndrome, was found to physically and functionally interact with PARP-1 (LI et al. 2004; VON KOBBE et al. 2004).
Research using microorganisms as models for aging has been dominated by studies in Saccharomyces cerevisiae. Replicative lifespan in S. cerevisiae is measured by determining the number of daughter cells an individual mother cell can produce (MORTIMER and JOHNSTON 1959). Mutations in SIR (Silent Information Regulator) complex components were isolated in a genetic screen designed to identify genes that control this form of aging (KENNEDY et al. 1995). In particular, the NAD+-dependent histone deacetylase Sir2 was shown to be a key regulator, acting to suppress recombination between rDNA repeats, thereby blocking the formation of extrachromosomal rDNA circles (ERCs), which are antagonistic to long replicative lifespan in budding yeast (KAEBERLEIN et al. 1999; SINCLAIR and GUARENTE 1997).
Although Sir2-like proteins (Sirtuins) have been implicated in controlling lifespan in metazoans, regulation of ERC production is thought to be a yeast-specific aging mechanism (ROGINA and HELFAND 2004; TISSENBAUM and GUARENTE 2001). Like Sir2 itself, Sirtuins are NAD+-dependent enzymes. Some Sirtuins act as mono(ADP-ribose) transferases (ARTS), others function as protein deacetylases, and some have both activities (BELENKY et al. 2007). Genetic and biochemical investigations using S. cerevisiae have established that NAD+ and NAM levels impact replicative aging through regulation of Sir2 deacetylase activity (GALLO et al. 2004; SANDMEIER et al. 2002). Additional studies have shown that lifespan extension by calorie restriction (CR) in S. cerevisiae involves Sir2, as well as the NAD+-dependent deacetylase Hst2, and is thus regulated by NAD+ and NAM levels as well (ANDERSON et al. 2003; LAMMING et al. 2005; LIN et al. 2000; LIN et al. 2004). In addition, a yeast pathway for Sirtuin-independent lifespan extension by CR is also influenced by NAD+ and NAM availability (TSUCHIYA et al. 2006). The fact that CR extends lifespan in higher eukaryotes, and that Sirtuins have been implicated in controlling aging in flies and worms suggests that NAD+ and NAM metabolism may be of general importance in the regulation of lifespan. While Sirtuins are present in all eukaryotes including the yeasts, additional ARTS, along with PARPs and cADP-ribose synthases exist in metazoans and filamentous fungi (BELENKY et al. 2007). All of these enzymes are major consumers of NAD+, and might therefore be expected to impact aging. While aging studies in S. cerevisiae have provided many valuable insights, the involvement of certain key biological regulatory pathways that are common to many eukaryotic organisms, but absent from yeast, have not been adequately investigated. Research directed at understanding the roles of PARP and PARylation in aging of higher eukaryotes may be hindered by functional redundancy of multiple PARP enzymes and lethality of PARP mutants. Thus we chose to explore the function of PARP in the filamentous fungus N. crassa, which only has a single gene encoding this enzyme.
MATERIALS AND METHODS
Media and culturing conditions
N. crassa was cultured as described previously (DAVIS and DESERRES 1970). Strains were grown in liquid Vogel’s minimal media with 1.5% sucrose or 2% glucose and supplements were added where indicated. Solid media was the same but with 2% agar. Strains were grown on FGS (0.05% fructose, 0.05% glucose, 2% sorbose, 1X Vogel’s salts, 2% agar) to induce colonial growth. The concentrations of supplements were as follows: 1X alanine (1 mg/ml), 1X anthranillic acid (140 µg/ml ), 1X histidine (0.5 mg/ml), 1X lysine (0.6 mg/ml), 1X nicotinamide (10 µg/ml). Hygromycin was used in the range of 200 µg/ml-1.5 mg/ml. Crosses were carried out on synthetic crossing medium with glucose or sucrose concentrations at 0.5% or 2.0%.
Southern and northern blots
DNA was isolated from N. crassa and Southern blots performed as previously described (LUO et al. 1995; MIAO et al. 2000). RNA extraction and northern blots were performed as previously described (ROUNTREE and SELKER 1997).
Analysis of PARP, MacroD and zf-PARP sequences
Fungal PARP-like proteins, MacroD-like, zf-PARP-like and their ORFs were identified using the BLASTP and TBLASTN programs at NCBI (http://www.ncbi.nih.gov). The BLASTP program was also used at the Broad Neurospora Genome Project database (http://www.broad.mit.edu/annotation/genome/neurospora) to identify the NPO and MacroD proteins, and the NGP genome browser was used to identify their ORFs. Protein motifs and domains were verified in the SMART database (http://smart.embl-heidelberg.de/). All sequence alignments and analysis was performed with programs at the SDSC Biology Workbench. The BL2SEQ program was used to compare NPO with human PARP-1 and PARP-2. The npo gene and amino acid sequence shown in Figure 3 was generated with the Publish program using Genetics Computer Group (GCG) software.
FIGURE 3. Sequence of the npoRIP1 allele.
The NPO protein sequence is indicated beneath the upper case nucleotide coding sequence. Lower case nucleotides represent intron sequences. The boxed amino acids in the first exon indicate the BRCT domain and the boxed amino acids in the second exon indicate the WGR motif. Guanine residues mutated to adenines are highlighted, and the tryptophan codon that was mutated to a stop codon is boxed. The sequence of the npoRIP1 allele has been deposited in Genbank with the accession number EU869543.
Analysis of subcellular localization of GFP tagged proteins
PCR products of the hp1, rap1, mcd (MacroD) and npo genes amplified from wild type Neurospora were cloned into pMF272 (HONDA and SELKER 2009) to allow his-3 targeting of GFP tagged fusions expressed from the Neurospora ccg-1 promoter. These PCR products spanned the start and stop codons of these genes, excluding five prime and three prime UTR sequences. The details of all cloning steps are available upon request. The Neurospora yph1 and zfp (zf-PARP) genes were cloned, along with 2 kb of upstream sequences, as NotI-PacI PCR fragments into NotI-PacI digested pMF272. The NotI-PacI pMF272 restriction fragment lacks the ccg-1 promoter. All GFP fusion constructs were used to transform a his-3 targeting strain p49 (relevant genotype: his-3; inl; npo+) obtained by crossing the original npo KO strain (14-6-1-1A) with FGSC 7508. Conidia from transformants were imaged using a Zeiss LSM 510 confocal microscope at 630× magnification. Images were taken as Z-sections and analyzed using the program ImageJ. Individual slices were selected and saved as JPEGs.
Knockout of npo by homologous replacement
The npo gene was amplified by PCR from wild type N. crassa using the following primers: 5’-CAAATGGACGAAAGAGGAGA-3’ and 5’-TGGTGAAAGGAAGGATGGAA-3’. The 6.5kb PCR product was digested with EcoRI and SacI, and cloned into pBluescript SK+. This construct was then digested with XhoI to remove the npo ORF and the hygromycin B-resistant gene (hph), derived from pCB1003 (CARROLL et al. 1994), was cloned in its place. The resulting plasmid (pP1) contains the hph gene flanked by 1.9 kb and 1.0 kb of npo upstream and downstream sequences, respectively. To knockout the npo gene, wild type N. crassa (74-OR31-14a) was transformed with an EcoRI –SacI fragment from pP1. Transformation was carried out by electroporation as described (NINOMIYA et al. 2004) and hygromycin resistant transformants were crossed to a wild type N. crassa strain of opposite mating type (74-OR31-16A) to render the integrations homozygous. Finally, npo knockout mutants were identified by PCR and Southern hybridization.
Cloning and mutation of npo by RIP
npo was cloned by PCR amplification from wild type N. crassa (N150, 74-OR23-IVA) using the following primers: (2653F) 5’-TCGAATTCATGCCGCCCAGACGAGCAAAG-3’; (2653R) 5’-CTGCGGCCGCTCATACGCAATGTACTCGTTG-3’. The PCR product was digested with EcoRI and NotI and cloned into pBM61 (MARGOLIN 1997) to generate pGK111. This construct was linearized with DraI, and targeted to the his-3 locus in strain N1674. Ten transformants were isolated, and correct integrations were confirmed by Southern hybridization. Four of the transformants (pGK111-T1, T2, T3, T4) were crossed with strain N1444 and DNAs from ten histidine prototrophic progeny from each of the four crosses were analyzed for evidence of mutation of npo. Probing of Southern blots of DpnII/Sau3A digested DNAs with npo sequences revealed RFLPs and heavy methylation in progeny 11 (P11), among others. P11 was obtained from a cross of strain pGK111-T2 with N1444. From here on this strain is referred to as N3180. The endogenous npo gene was cloned by PCR from strain N3180 using the following primers: (2653F2) 5’-CTTCACACACTTCACACCTTTGTTTC-3’; (2653R2) 5’-GCTATCTTGACACGGAAAAG-3’. Digestion of the PCR product with DpnII confirmed the presence of the RFLPs detected by Southern blot, and the PCR product was gel isolated and sent for sequencing using primer 2653F. The npo allele present in N3180 is designated npoRIP1.
Testing for genetic interactions between npoRIP1 and N. crassa Sirtuins (nsts)
For the purpose of isolating the npoRIP1 allele in a mat a background, and to look for possible genetic interaction between npo and nst-1, N3180 was crossed with N1983 (mat a; mtr col4; nst-1RIP1 trp-2). No obvious defects in growth or development were observed in double mutant progeny. Strain N3181 (mat a; npoRIP1; nst-1+) was obtained from this cross. To isolate npoRIP1 in a background with a TPE marker and both nst-1 and nst-3 mutations, N3181 was crossed with N2636 (mat A nst-3RIP1; mtr col4; telVR::hph::T; nst-1RIP1 trp-2). Numerous progeny were isolated from this cross and Southern blots were used to determine their genotypes. Among the progeny were P6 (nst-3+; npo+ telVR::hph::T; nst-1+), P80 (nst-3+; npoRIP1 telVR::hph::T; nst-1+) and P23 (nst-3RIP1; npo+ telVR::hph::T; nst-1+) which were tested for TPE (see Fig. 7) along with others. No obvious defects in growth or development were observed for triple mutant progeny.
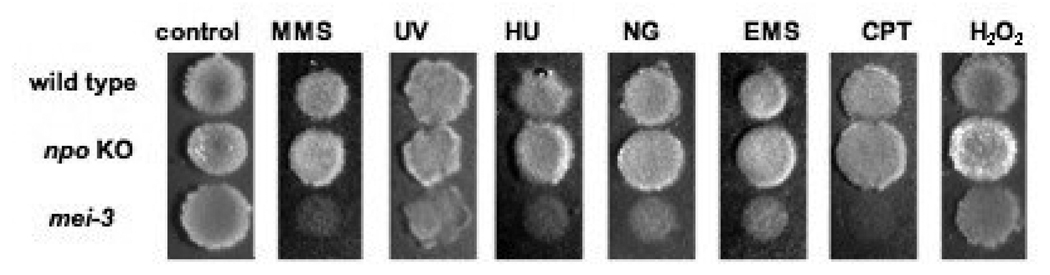
FIGURE 7. Mutagen sensitivity of the npo KO strain.
Spot-tests of conidia on FGS plates for wild type N. crassa (top), the npo (middle) and mel-3 strains (bottom) were done as described in Materials and Methods. The mel-3 strain was used as a positive control for mutagen sensitivity. Panels from left to right are as follows: no mutagen; 0.015% methyl methane sulfonate (MMS); conidia pretreated with 450 J/MF UV; 30 mM hydroxy urea (HU); 0.05µg/ml N-methyl-N-nitro-N-nitrosoguanidine (MNNG); 0.3% ethyl methane sulfonate (EMS); 0.3µg/ml camptothecin (CPT) and 0.0015% H2O2.
TPE assays
Progeny from the cross of N3181 with N2636 were spot-tested on hygromycin to assay the effects of mutation of npo, nst-1 and nst-3 on TPE in genetic backgrounds with telVR::hph::T (SMITH et al. 2008). All possible combinations of alleles were analyzed. Approximately 1000 conidia were spot-tested on FGS plates containing 600 µg/ml or 1.5 mg/ml hygromycin and supplemented with alanine, lysine, inositol and anthranillic acid. Spot-tests were also done on identical plates with no hygromycin as a control for growth.
Mutagen sensitivity assays
For mutagen sensitivity assays, progeny from the cross of N3181 with N2636 were spot-tested on the same media used in the TPE assays, but containing either MMS (0.03%), MNNG (0.5 µg/ml), EMS (0.3%) or CPT (0.3 µg/ml ). As with the TPE assay approximately 1000 conidia were spot-tested, and identical control plates with no mutagen were used as a control for growth. Mutagen sensitivity of the npo KO strain was tested as previously described (WATANABE et al. 1997).
PARylation assay
Crude N. crassa extracts were incubated in 50 mM Tri-HCl (pH8.0), 10 mM MgCl2, 1 mM dithiothreitol, 10 µM (74 KBq/nmol) 32P-NAD (Du Pont), 20 µg/ml activated DNA (Sigma) and 20 µg/ml calf thymus type II-A histones (Sigma H9250) at 25°C for 30 min. To stop the reaction, the PARP inhibitor 3-aminobenzamide was added to 5 mM and unincorporated NAD was removed using spin columns containing Sephadex G-50 resin (GE Healthcare). E. coli extracts expressing human recombinant PARP-1 (IKEJIMA et al. 1990) were used as positive controls. After centrifugation at 300×g for 4 min, the eluent containing 32P-PARylated proteins was treated with 0.1 M NaOH at 37°C for 30 min. to detach 32P-PAR, and the solution was neutralized by addition of Tri-HCl (pH7.5) to 50 mM and HCl to 0.1 N. After extraction with water-saturated phenol and chloroform-isoamyl alcohol (49:1 (v/v)), ammonium acetate was added to 2 M and 32P-PAR was ethanol-precipitated. After washing with 70% ethanol, the fraction was dried and dissolved in a loading dye containing urea (PANZETER and ALTHAUS 1990). The fraction was then analyzed by 20% polyacrylamide gel electrophoresis as described elsewhere (PANZETER and ALTHAUS 1990). The gel was exposed and analyzed with BAS2500 (Fuji Film). The radioactive area containing 32P-PAR was cut out and further analyzed. The gel fragments were rinsed with water and crushed. The radioactive material was eluted and digested by incubation overnight at 25°C in 100 µl of a PARG buffer containing 20 mM potassium phosphate (pH7.5), 10 mM β-mercaptoethanol, 0.05% triton X-100 (Sigma), 0.1% bovine serum albumin and rat PARG-conjugated with glutathione-S-transferase (GST-PARG) (SHIMOKAWA et al. 1999). Treatment with GST-PARG digested PAR to ADP-ribose, and the reaction mixture was treated with perchloric acid at 0.5 N on ice for 20 min and neutralized with 0.7 M glycyl-glycine-3 M potassium hydroxide and centrifuged at 15,000×g for 5 min at 4°C. The supernatant was subjected to high performance liquid chromatography (HPLC). HPLC was carried out using Develosil columns (C30-UG-5, Ø46×250 mm, Nomura Chemicals). UV absorbance was monitored at 254 nm (Toso, UV-8000). A linear gradient elution for 100 min. using buffer A (0.1 M ammonium acetate) and buffer B (50 mM ammonium acetate–50% acetonitrile) was performed, ranging from 2% to 100% buffer B at a flow rate of 0.5 ml/min. The retention time of ADP-ribose was 17–19 min. Each 0.5 ml fraction between 13–20 min. was concentrated and spotted on DE81 paper (Whatman) and analyzed by BAS2500.
Telomere erosion assay
Genomic DNAs from wild type and the npo strain were digested with ClaI and HindIII. Electrophoresis was carried out in a 2.5% agarose gel for 5 hrs at 50mV and Southern blots performed as previously described (LUO et al. 1995; MIAO et al. 2000). These blots were then probed with a non-isotopically labeled oligo composed of 7 direct tandem copies of the telomere repeat sequence [5’-CCCTAA-3’].
RESULTS AND DISCUSSION
There are two classes of fungal PARP-like proteins
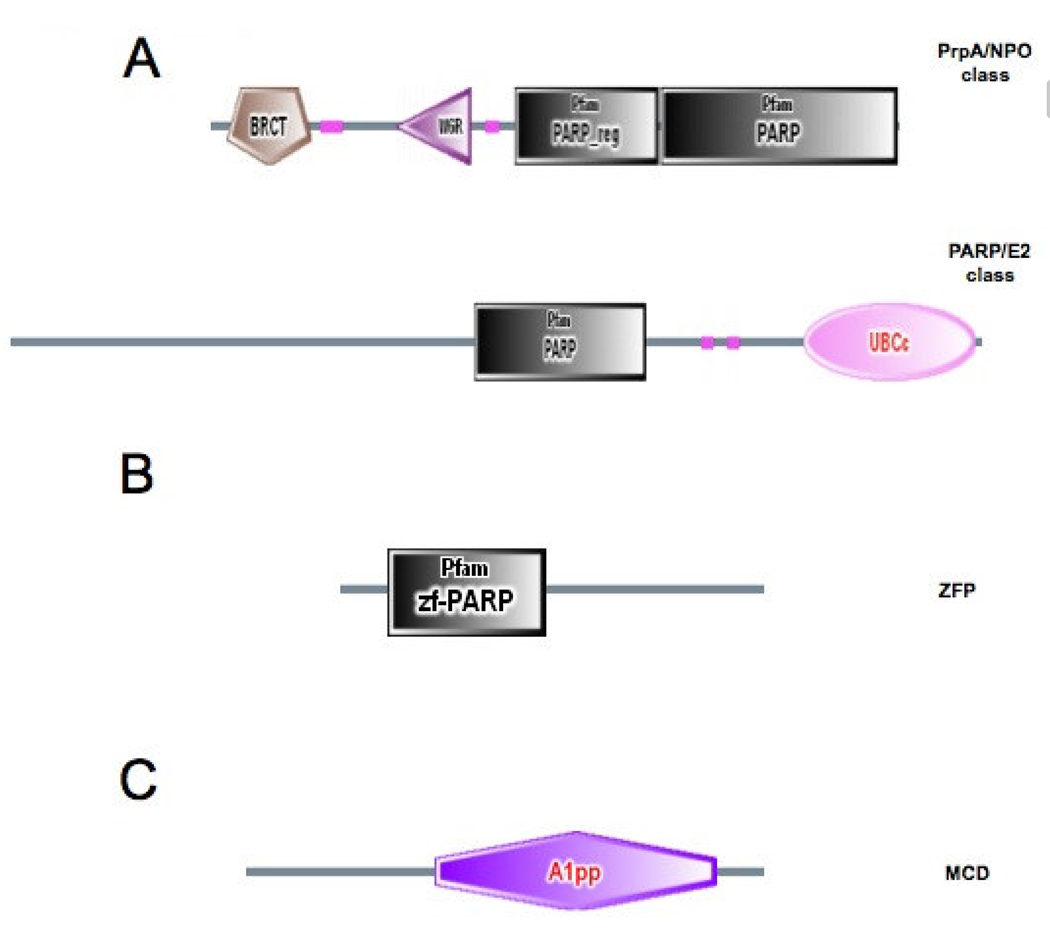
SEMIGHINI et al. (2006) observed that PARP homologs exist in fungi that have multicellular hyphae and sophisticated developmental structures, but lack a prominent yeast-like budding growth phase. The canonical PARP enzyme from mammals, PARP-1, contains an N-terminal zinc finger DNA binding domain (zf-PARP), a BRCT motif that is the major target for auto-modification, a WGR motif, and a core catalytic domain (AME et al. 2004; KIM et al. 2005). We performed TBLASTN searches through the NCBI (http://www.ncbi.nih.gov) fungal genome databases using the human PARP-1 catalytic domain as the query. We then analyzed the hits using the SMART database (http://smart.embl-heidelberg.de/) to confirm the presence of a PARP catalytic domain (pfam 00644). Our analysis revealed two classes of PARP-like proteins: 1. homologous to A. nidulans PrpA, containing BRCT (pfam 00533) and WGR motifs (pfam 05406), and 2. those with a catalytic domain most similar to mammalian PARP-6/PARP-8 family members and having a carboxyl terminal extension showing homology to the catalytic domain (SMART 00212) of ubiquitin conjugating enzyme E2 (Fig. 1A). This domain organization seems to be specific to filamentous fungi. We refer to this second class of fungal PARP-like proteins as PARP/E2. Like the PrpA class, the PARP/E2 proteins are broadly distributed in the euascomycetes. In fact, N. crassa is the only euascomycete represented in the NCBI fungal genome databases (25 species) that does not have a PARP/E2 homolog, raising the possibility that a N. crassa homolog is in a sequencing gap. Homologs in the PARP/E2 class were also found in the basidomycetes Coprinus cinereus (EAU83704.1) and Phanerochaete chrysosporium (unannotated protein, contig accession: AADS01000086, gi:46851846, approx. coordinates 71000–75000).
FIGURE 1. Domain organization of fungal PARP-like proteins and associated DNA and PAR binding proteins.
(A) Schematic representation of the domain organization of the two classes of fungal PARP proteins. The complete amino acid sequences of NPO. Neurospora crassa PARP ortholog [CAD21266] and Aspergillus nidulans AN0482.2 [xm_652994.1] were used as queries to search the SMART database (smart.embl-heidelberg.de). Searching with NPO identified BRCT [IPR001357], WGR [IPR008893]. PARP-regulatory [PF02877], and PARP-catalytic [PF00644] domains, defining the PrpA/NPO class. Searching with AN0482.2 identified PARP-catalytic [PF00644] and Ubiquitin-conjugating enzyme E2 catalytic domains [SM00212], defining the PARP/E2 class. (B) Domain organization of the Neurospora MacroD protein. The complete amino acid sequence encoded by Neurospora ORF NCU07925.3 was used to search the SMART database identifying the A1pp domain [SM00506]. (C) Domain organization of the Neurospora zf-PARP protein. The complete amino acid sequence of a Neurospora hypothetical protein [Broad coordinates LGl, contig 2:447973-449833+] was used to search the SMART database identifying a single zf-PARP Pam domain [PF00645].
N. crassa has a single PARP homolog of the PrpA class
Fungal PARP proteins of the PrpA class lack an amino terminal zinc finger DNA-binding domain (zf-PARP), but have both an N-terminal BRCT motif and a WGR motif (SEMIGHINI et al. 2006). A BLASTP search with PARP-1 sequences through the Broad Institute Neurospora genome database (http://www.broad.mit.edu/annotation/genome/neurospora) identified a single ORF encoding a predicted protein of 592 amino acids with a WGR motif, but lacking a BRCT motif (NCU08852.3, EAA31746, GI:157070000, accession AABX02000063.1). We feel that the most likely start codon for this ORF is 235 nucleotides upstream of that suggested by the Broad annotation, which would predict a protein of 670 amino acids with both WGR and BRCT motifs, as expected for a member of the PrpA class (Fig. 1A). We refer to this protein as NPO (Neurospora PARP Ortholog). The presence of BRCT motifs in the PrpA class of fungal PARPs, and their absence from PARP-2 homologs, suggests that proteins of the PrpA class are more closely related to PARP-1. However, comparison of NPO with human PARP-1 and PARP-2 using the BL2SEQ program suggests a closer relationship with PARP-2 [NPO:PARP-1, e =10−79, identities = 216/681 (31%), similarities = 335/681 (49%), gaps = 67/681 (9%); NPO:PARP-2, e = 3 × 10−82, identities = 187/464 (40%), similarities = 259/464 (55%), gaps = 39/464 (8%)]. In agreement with our analysis SEMIGHINI et al. (2006) observed that PrpA-like PARPs belong to a microbial clade more similar to PARP-2 than PARP-1.
Fungal zf-PARP proteins, Macro domain proteins, and nuclear localization of NPO
Fungal PARP-like proteins, including NPO, lack any obvious DNA binding domain, raising the question of whether these proteins are principally associated with chromatin, like their metazoan counterparts. NPO might contain a cryptic DNA-binding domain or could require a partner for DNA binding. Because PARP-1 contains a highly characteristic amino terminal zinc finger DNA-binding domain (zf-PARP), we sought to identify fungal proteins containing a similar motif. To this end we performed TBLASTN searches through the NCBI fungal genome database using the PARP-1 zinc finger as query. These searches identified a single Neurospora ORF encoding a protein of 404 amino acids, containing a single zinc finger of the zf-PARP class (Fig. 1B). We refer to this protein as ZFP. Although this ORF has not been annotated with an NCU number in the Broad database, we believe that it represents a functional gene, as a GFP tagged form, expressed via its own promoter, has a punctate nuclear staining pattern, similar to the heterochromatin associated protein, HP1 (Fig. 2) (FREITAG et al. 2004a). To determine the subcellular location of NPO we tagged the protein with GFP at its carboxyl terminus, and expressed the fusion protein in aerial hyphae and conidia using the developmentally regulated ccg-1 promoter (Fig. 2) (FREITAG et al. 2004b; LOROS et al. 1989; MCNALLY and FREE 1988). GFP tagging of ectopically expressed NPO verifies that this protein is also localized primarily to nuclei (Fig. 2), and thus has a functional nuclear localization signal. The subnuclear distribution of NPO seems to be essentially uniform, in comparison to proteins localized specifically to heterochromatin (HP1), telomeres (RAP1) and rDNA (YPH1) (Fig. 2).
FIGURE 2. Confocal Images of GFP-tagged Neurospora proteins.
Expression of heterochromatin protein 1 (HP1::GFP), a telomere repeat binding protein (RAP1::GFP), MacroD::GFP and NPO::GFP was driven by the ccg-1 promoter. An rDNA associated protein (YPH1::GFP) and zf-PARP::GFP were expressed via their endogenous promoters.
We have not been able to identify any PARG-like protein in any filamentous fungal database, and thus PAR may be a more stable posttranslational modification in fungi than in higher eukaryotes. Although we were unable to identify fungal proteins with the PAR-binding C2H2 zinc finger (PBZ) domain (AHEL et al. 2008), we did identified one ORF encoding a protein of 277 amino acids with a single Macrodomain (also designated as A1pp ) (Fig. 1C). Macrodomains have also been shown to bind PAR both in vivo and in vitro (KARRAS et al. 2005). The Neurospora Macrodomain protein is annotated in the Broad database as NCU07925.3, and we refer to it as MCD (Macrodomain). An over-expressed GFP tagged form of MCD has essentially uniform cytoplasmic and nuclear distributions, but is slightly more concentrated in nuclei than in cytoplasm (Fig. 2). Thus, while NPO has an autonomous nuclear localization signal, it may be brought to DNA via association with other proteins such as ZFP. Furthermore, while fungi are unlikely to remove PAR via a glycohydrolase activity, as mammals do, they are likely to recognize PAR via nuclear localized Macrodomain proteins such as MCD.
npo is a nonessential gene in N. crassa
After verifying the nuclear distribution of NPO we then isolated N. crassa stains with mutations in the npo gene using Repeat Induced Point mutation (RIP) (SELKER 1990) and made knockout strains by replacing the npo coding sequence with the bacterial hygromycin phosphotransferase gene (hph) (Fig. 3, Fig. 4A and B). Both homozygous and heterozygous crosses of strains carrying duplications of npo at the his-3 locus were fully fertile. These results suggest that npo is not required in the brief diploid phase for completion of meiosis, as heterozygous duplications would be expected to trigger meiotic silencing by unpaired DNA (MSUD) (ARAMAYO and METZENBERG 1996; SHIU et al. 2001). However, it is also possible that there is enough transcript or protein present in ascogenous hyphae to override the effect of MSUD during the diploid phase.
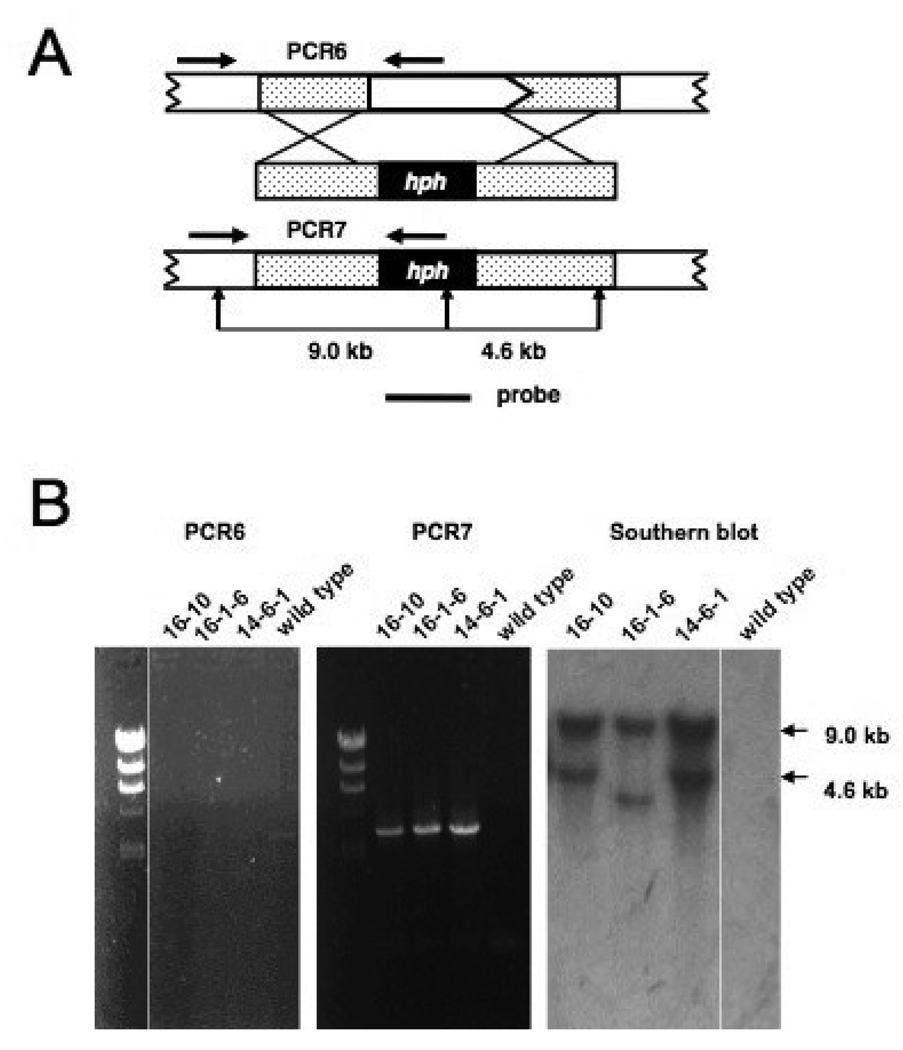
FIGURE 4. Disruption of the N. crassa npo gene by homologous recombination.
(A) Schematic illustration of knockout strategy for the npo gene. A white arrow box represents the npo gene and shows the direction of transcription. Stippled boxes indicate immediate flanking sequences. The knockout construct is shown below the genomic sequence with the E. coli hph gene represented by a black box. The genomic sequence with the E. coli hph gene represented by a black box. The genomic sequence resulting from correct replacement is shown beneath the knockout construct. Horizontal black arrows indicate the positions of PCR primers used to analyze the transformants. Vertical black arrows indicate restriction sites used to characterize the transformants by Southern hybridization A horizontal black line represents the probe used in the Southern blot.(B) The images labeled PCR6 and PCR7 are ethidium bromide-stained agarose gels with size markers run in the left-most lanes. The next three lanes contained PCR products that had been amplified from template DNAs isolated from the indicated transformants. The right-most lanes show PCR products amplified from wild type N.crasse DNA, as controls. The position of primers for the PCR6 and PCR7 reactions are shown in panel A. The right-most image shows an autoradiograph of a Southern blot probed with hph sequences. DNAs from the indicated transformations were digested with Ncol. DNA from wild type N.crassa was run in the right-most lane as a control.
We confirmed the presence of mutations by RIP in progeny from these crosses by Southern hybridization and DNA sequencing. Clear evidence of RIP was detected by Southern hybridization in six out of forty progeny. None of the six progeny exhibited any gross morphological or developmental phenotypes. Sequencing of a PCR product amplified from progeny number 11 (P11) identified 21 C:G to T:A transition mutations in a 591 base pair segment of the endogenous npo gene (Fig. 2). All G to A mutations were found on the coding strand, spanning the first and second exons. The 21 mutations affected 15 codons, with five mutations occurring in the intron. Of the 16 mutation occurring in codons, four were in 3rd position and silent (V53, L106, L203and Q205), seven were in 1st position, resulting in conservative substitutions (V59→I, E101→K, D121→N, D152→N, V181→M, V197→I and D200→N), two were in 1st position resulting in nonconservative substitutions (G78→R and A105→T) and one was in second position producing a stop codon (W209→stop). The conservative substitution at position 185 (V185→I) resulted from G to A transitions in both the 1st and 3rd positions. The introduction of a stop codon at W209 is very likely to eliminate NPO function, as it occurs in the amino terminal region of the WGR motif, upstream of the PARP catalytic domain (Fig. 3). We refer to this allele as npoRIP1 and the original progeny harboring the allele (P11) as N3180.
All tested strains carrying npoRIP1 were fully fertile as males or females, and homozygous crosses appeared normal as well. Knockouts of npo were made by homologous replacement of the npo coding region with hph in a wild type background. Proper replacements were confirmed by PCR analysis and Southern blots (Fig. 4A and B). Strains with the npo KO, like strains with the npoRIP1 allele, did not exhibit gross morphological or developmental phenotypes, and were fully fertile in heterozygous and homozygous crosses. We conclude that npo is a nonessential gene in N. crassa and is not required for normal growth or development. These results stand in contrast to what was reported for a prpA knockout in Aspergillus, which was lethal in haploid strains, and produced severe growth restrictions and developmental phenotypes in heterozygous diploid strains (ΔprpA/+), described as haplo-insufficient (SEMIGHINI et al. 2006).
NPO is a PAR-polymerase
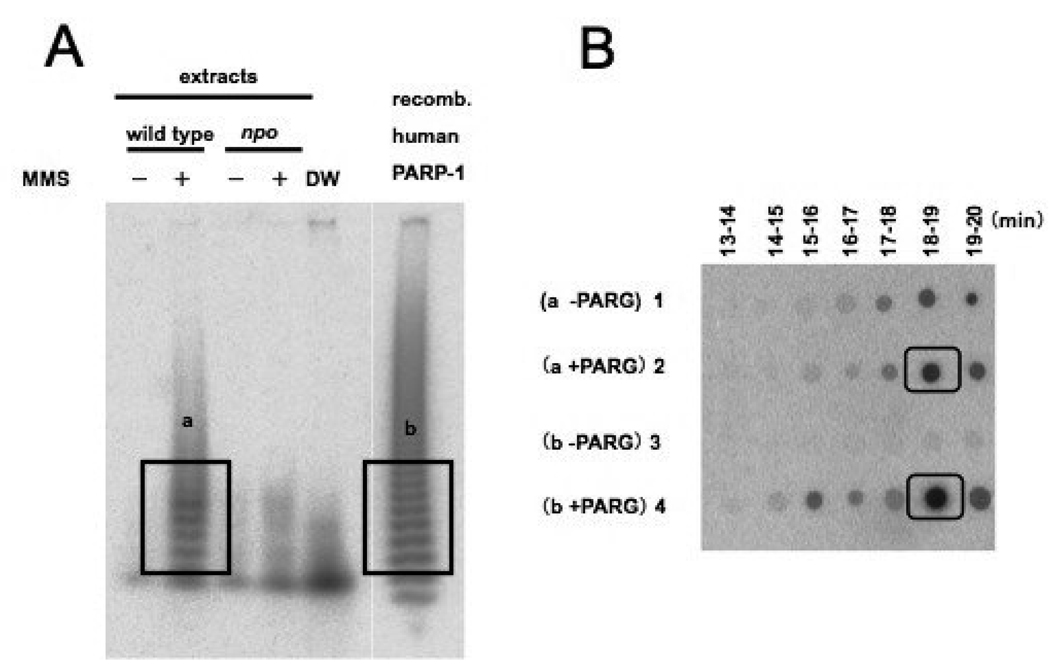
We assayed PARylation activity in extracts from both wild type and npo KO strains to determine if NPO functions as a protein PAR-polymerase. To our knowledge, results from PARylation assays have only been reported for mammalian systems. To assay PARylation, crude extracts from wild type and the npo KO strain were prepared from conidia that had either been treated or not treated with MMS for 60 min. The crude extracts were incubated with 32P-NAD, sheared DNA and histones. As a positive control, an assay was also performed on extracts from E. coli cells expressing recombinant human PARP-1. PAR was detached from proteins by alkaline treatment and analyzed on 20% PAGE. As shown in Fig. 5A, the MMS-treated wild type strain produced a PAR-ladder like the human PARP-1 control (right-most lane), but the npo KO strain did not. To confirm that the ladder observed with the MMS-treated wild type strain reflected PAR, the radioactive material was eluted from the gel, digested with PARG (PAR-glycohydrolase), which specifically cleaves PAR into ADP-ribose, and analyzed by HPLC. As shown in Fig. 5B, the radioactivity that eluted at the retention time of ADP-ribose, namely at 18–19 min., is higher in the PARG-treated sample than in the untreated control. It is possible that the radioactivity detected at 18–19 min in the PARG untreated control is due to degradation of PAR to ADP-ribose during PARG-treatment or due to unrelated products generated during the 32P-NAD incorporation reaction. The control extract containing human PARP-1 also showed high intensity spots at 18–19 min., corresponding to ADP-ribose. We conclude that N. crassa PARylation increases in response to MMS treatment, and that this activity depends on NPO, which is likely responsible for most or all PARylation in N. crassa.
FIGURE 5. Verification of NPO PARylation activity.
(A) Autoradiogram of a 20% polyacrylamide gel showing 32P-PAR ladder. PARylation reactions with extracts from wild type N.crassa cells treated (+) or not treated (−) with MMS were run alongside reactions with extracts from npo KO cells treated (+) or not treated (−) with MMS. The lane labeled DW is a negative control reaction using distilled water in place of extract. The right most lane contains a positive control reaction with recombinant human PARP-1 expressed in E. coli. The boxed regions labeled a and b were excised and the radioactivity was eluted for analysis by HPLC.(B)An autoradiogram (BAS2500) of fractions from HPLC blotted onto DE81 paper (Whatman) with retention times indicated above and sample designations on the left. Eluents of 32P-PAR from these gel slices were either treated with recombinant PARG, or not, and fractionated by HPLC as described in Materials and Methods. The boxed regions show the peak signals eludated at the retention time for ADP-ribose.
npo transcription is induced by MMS treatment
It is well established that auto-modification of mammalian PARP-1 increases dramatically with the binding of the protein to double and single-strand DNA breaks. Although the transcriptional response of the mammalian PARP-1 gene to DNA damaging agents has not been reported, plant PARP-1 and PARP-2 gene transcription is highly induced by DNA damage (DOUCET-CHABEAUD et al. 2001). In addition, SEMIGHINI et al. (2006) found prpA steady-state transcript levels increased in response to MMS, BLM and 4-NQO treatments. Our results from PARylation assays demonstrated a dramatic increase in NPO activity in response to MMS treatment. To determine if this reflected a change in npo transcript levels or enzyme activity, we performed northern blots of RNAs isolated from wild type and npo KO strains that had either been treated or not treated with MMS. The blots were probed with npo sequences, and cox-5 sequences as a control for the loading. In untreated wild type cells, npo transcripts were undetectable by northern blot, but a large accumulation of npo transcript was detected 30 min. after treatment of wild type cells with MMS, and high levels of transcript were still detectable 120 min. after treatment (Fig. 6). As expected, no npo transcripts were detectable in the npo KO strain (Fig. 6). Thus npo transcription is likely to be regulated in response to DNA damage, like the A. nidulans prpA gene.
FIGURE 6. Analysis of npo transcription by northern blot.
The left panel shows an autoradiogram of a northern blot of RNAs extracted from wild type N. crassa after the indicated duration of MMS treatment. The upper panel shows results of probing the blot with npo sequence and in the lower panel shows results of probing with cox-5 sequences as a control for loading. The right panel shows the same for the npo strain.
npo mutant strains are not sensitive to DNA damaging agents
Genetic and biochemical studies of mammals established roles for PARP-1 in DNA repair and genome stability (MASUTANI et al. 2003; WATANABE et al. 2004). The fact that the steady-state transcript levels of npo were regulated by exposure to MMS suggested that NPO may play a role in a DNA damage response. We tested the effects of a number of DNA damaging agents on N. crassa strains carrying either the npoRIP1 allele or the npo KO. We tested CPT, EMS, H2O2, HU, MMS, MNNG and UV (Fig. 7), as well as BLM (data not shown). Neither mutant showed sensitivity to any of these compounds. SEMIGHINI et al. (2006) found the haplo-insufficient ΔprpA/+ mutant to be extremely sensitive to both phleomycin (PLM), which induces double-strand breaks, and the UV-mimetic agent 4-NQO. While we did not test PLM, the npo mutants were not sensitive to BLM, which also induces DNA double-strand breaks (POVIRK et al. 1977). Because npo transcript levels increase in response to MMS, it is likely that NPO function is connected with a DNA damage response. The fact that npo mutants are not sensitive to DNA damaging agents suggests the function may be redundant, or it may impact a nonessential aspect of repair. Alternatively, NPO may function in related processes such as regulating expression of genes controlled by DNA damage. The fact that A. nidulans PrpA is necessary for normal repair reveals divergence in DNA damage response pathways between Neurospora and Aspergillus.
NPO is not a global regulator of TPE
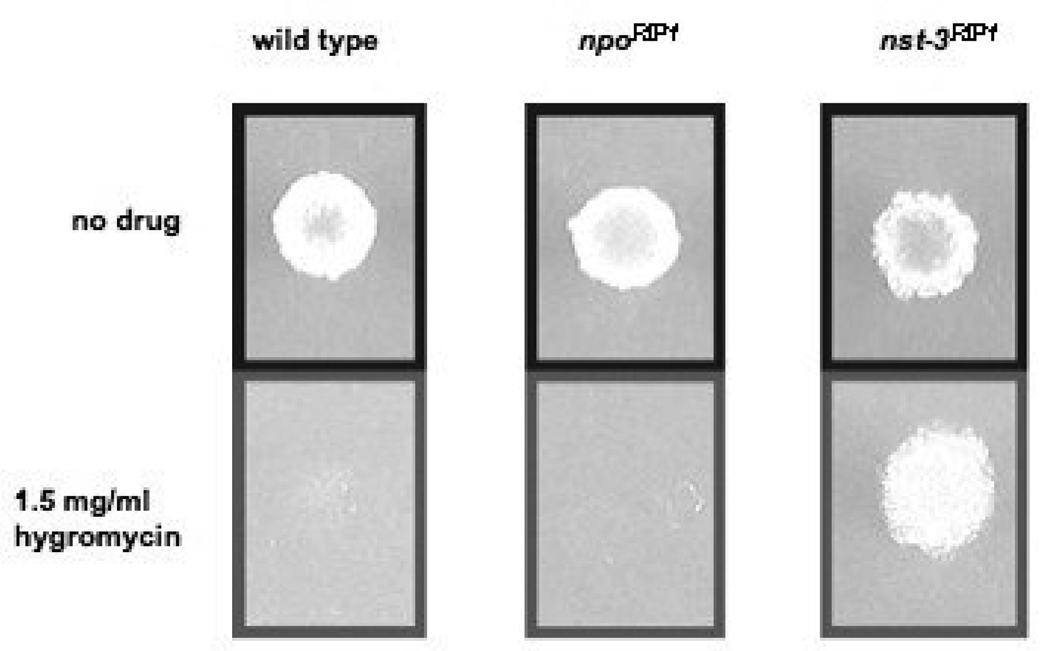
In metazoans, PARP enzymes are involved in chromatin-mediated regulation of transcription (KRISHNAKUMAR et al. 2008). Although considerable progress has been made in understanding the role of PARPs in regulating chromatin structure, simple genetic studies to test their possible involvement in epigenetic position effects, such as Telomere Position Effect (TPE), are lacking. We recently developed N. crassa strains with markers at subtelomeric positions to examine TPE (SMITH et al. 2008). This system allowed us to identify factors that control TPE, including several Sirtuins, termed NSTs (Neurospora Sirtuins). To analyze the effect of mutations in npo on TPE, we crossed the npoRIP11 allele into a background with the E. coli hph gene targeted to telomere VR (telVR::hph::T). We found significant derepression of hph at telomere VR in a strain with mutations in the N. crassa Sirtuin gene nst-3 (nst-3RIP1), but not in a strain with npoRIP1 (Fig. 8).
FIGURE 8. Telomere position effect assay.
Spot-tests of conidia on FGS plates for wild type N. crassa, npoRIP1, and nst-3RIP1 strains on media with 1.5 mg/ml hygromycin or no hygromycin, as described in Materials and Methods.
NPO is not involved in DNA methylation or DNA methylation-dependent silencing
In mammals it has been reported that PARP-1 is antagonistic to DNA methylation. Treatment of mouse fibroblasts with the competitive PARP inhibitor 3-aminobenzamide (3-AB) resulted in DNA hypermethylation and PAR has been shown to inhibit the activity of the maintenance DNA methylase, DNMT1 (REALE et al. 2005). N. crassa is the simplest genetically tractable system used to study DNA methylation. In N. crassa virtually all DNA methylation occurs in transposons that have been mutated by RIP (SELKER et al. 2003) and this methylation is not confined to symmetrical positions (SELKER et al. 1993). Numerous viable N. crassa mutants with reduced methylation have been described, including dim-2, dim-5, and hpo. Mutation in any of these genes completely abolishes all detectable DNA methylation (FREITAG et al. 2004a; KOUZMINOVA and SELKER 2001; TAMARU and SELKER 2001). Although no mutants with hypermethylation have been described in N. crassa thus far, strains of Ascobolus immersus carrying silenced copies of the histone H1 gene (hH1) were shown to have elevated levels of DNA methylation (BARRA et al. 2000). This hypermethylated DNA could be detected globally on ethidium bromide stained agarose gels, as higher molecular weight fragments after digestion with methylation sensitive restriction enzymes.
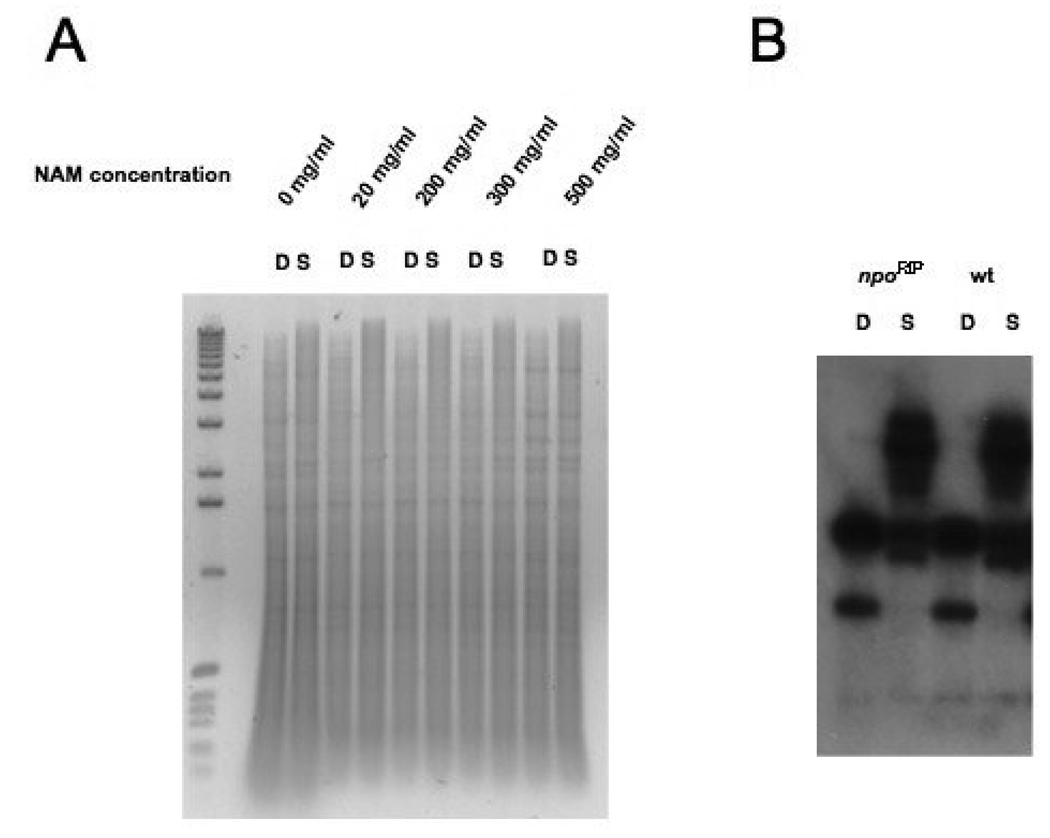
As a first test of whether inhibition of NPO effects DNA methylation we treated wild type N. crassa cells with high concentrations of nicotinamide (NAM) and looked at global DNA methylation by analyzing Sau3A- and DpnII-digested DNAs on agarose gels (Fig. 9A). NAM acts as a strong noncompetitive inhibitor of both Sirtuins and PARPs, and we had previously shown that treatment of N. crassa with NAM dramatically reduces silencing of telVR::hph, but has no effect on silencing of the methylated transgene amRIP::hph::amRIP (SMITH et al. 2008). No effect on global DNA methylation was observed after NAM treatment (Fig. 9A). The fact that NAM treatment did not relieve silencing of amRIP::hph::amRIP, suggests that neither NPO nor NSTs are involved in methylation-dependent silencing at this locus. Because it was conceivable that NPO is resistant to NAM, we also tested if mutation of npo would affect DNA methylation. Southern blots of Sau3A- and DpnII-digested DNAs from progeny with mutations by RIP in the npo gene, including strain P11, which is likely to be a null mutant, revealed heavy DNA methylation when probed with npo sequences (data not shown). We probed the same Southern blots with Ψ63 sequences, which are normally methylated (MARGOLIN et al. 1998), and did not see any change in DNA methylation at this locus (Fig. 9B). We also looked at global DNA methylation levels by ethidium bromide staining in N. crassa strains with the npoRIP1 allele and saw no effect (data not shown). In addition, presence of the npoRIP1 allele, or quelling experiments with npo sequences, had no effect on silencing of amRIP::hph::amRIP (data not shown), indicating that NPO is not involved in methylation-dependent silencing. We conclude that npo is not involved in DNA methylation.
FIGURE 9. Absence of effects of NAM treatment and npo mutation on DNA methylation.
(A) Approximately 1 µg samples of chromosomal DNA, isolated from wild type N. crassa (N 150) grown for 3 days in Vogels minimal media with the indicated concentrations of NAM, were digested either with DpnII (D) or Sau3A (s) and fractionated on a 1× TAE/0.8% agarose gel containing 1 µg/ml ethidium bromide. The left-most lane contains 0.5 µg of 1 kilobase DNA ladder (Invitrogen). (B) A Southern blot of chromosomal DNAs from wild type and npoRP mutant strains digested with DpnII (D) or Sau3A (S), as described for panel A and in Materials and Methods. The Southern blot was probed with ψ63 sequences.
The npo knockout causes acceleration of replicative aging
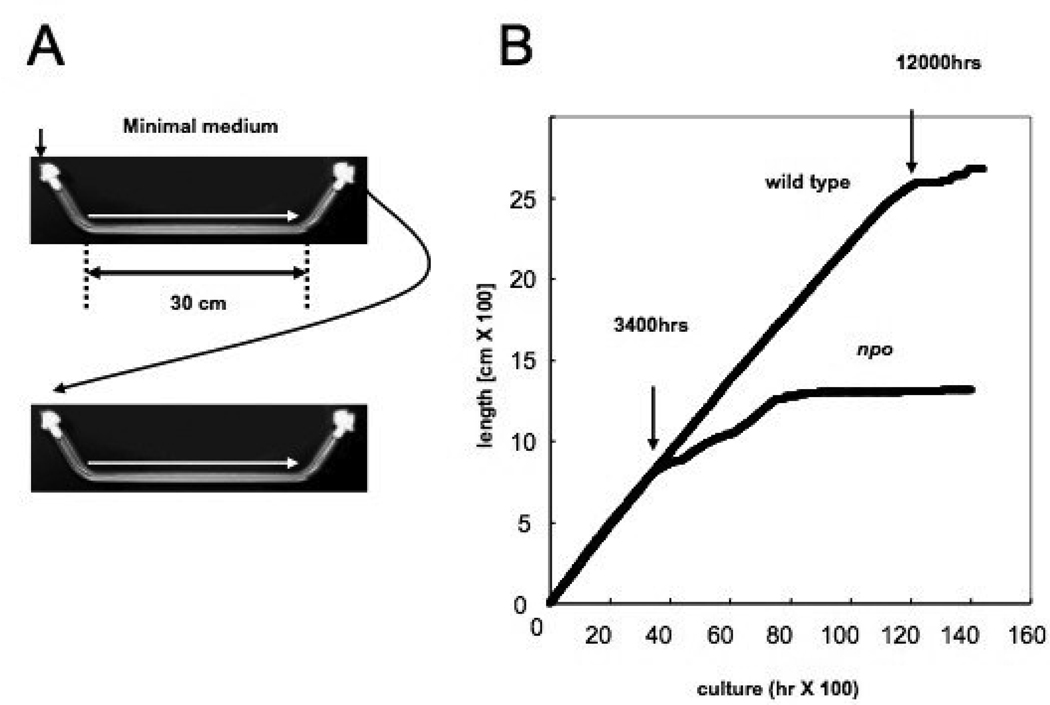
Studies of aging in filamentous fungi have focused largely on replicative aging associated with mitochondrial DNA (mtDNA) rearrangements triggered by mitochondrial plasmid/intron mobilizations (OSIEWACZ 2002). Replicative lifespan is a measure of the number of mitotic divisions a cell undergoes before senescence. Analogous to the ERC situation in yeast, these mechanisms seem to be specific to filamentous fungi. BARRA et al. (2000) reported that strains of Ascobolus immersus with silenced copies of the hH1 gene exhibited a decreased replicative lifespan, along with DNA hypermethylation. Such strains were found to initiate growth normally, but to senesce between 6 and 13 days after germination, whereas strains with unsilenced hH1 continued with a linear rate of growth for up to 40 days. We observed a similar phenotype for our npo KO strain, although the replicative lifespan of N. crassa mycelia is considerably longer than that of A. immersus (500 days versus 35–40 days, respectively). We grew both wild type and npo KO strains on minimal medium in 30 cm race tubes at 34°C with 12 hr. dark/light cycles, and were careful to transfer only mycelial fragments upon inoculation (Fig. 10A). The npo KO strain had a linear growth rate indistinguishable from wild type for the first 140 days of growth (6 cm/day), at which point the growth rate started to decrease gradually, culminating in senescence at around 300 days (Fig. 10B).
FIGURE 10. Method and results of senescence assay.
(A) Schematic of race tube strategy for measuring long-term linear extension rate. (B) Plot of growth (cm/hr) for wild type N. crassa and npo strain. Arrows at 3,400 hrs and 12,000 hrs indicate entry into senescence for npo and wild type N. crassa strains, respectively.
Telomere erosion does not occur in the npo knockout strain
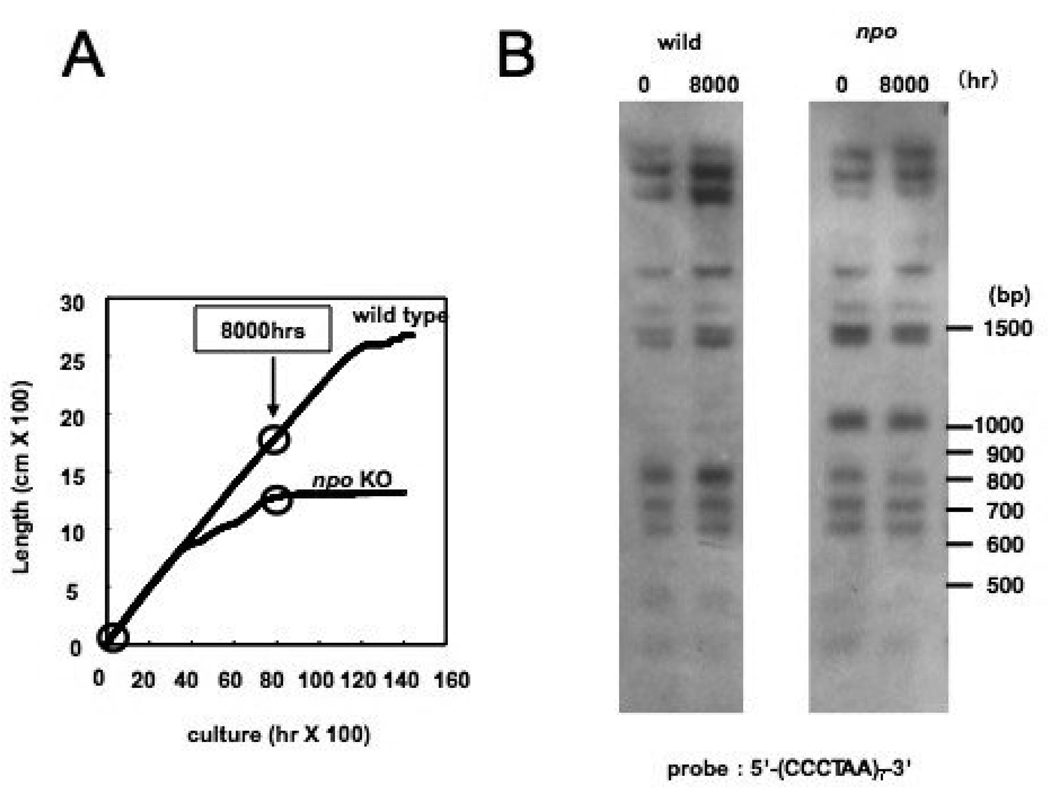
Eukaryotic microorganisms must maintain telomere length in every proliferating cell type, either by telomerase activity or by recombination. We were therefore interested to test if the increased replicative aging observed in the npo strain reflected defective maintenance of telomeres. To determine if mutation of npo affects telomere length in N. crassa, DNA was isolated from young cultures (~80 hours) and old cultures (~8000 hours) of both wild type and npo KO strains. The DNAs were digested with ClaI and HindIII and Southern blots were probed with telomere repeat sequences. The 8000 hour time point was chosen because this is when the npo KO strain begins to senesce (Fig. 11A). The Southern blots did not reveal any obvious change in the length of the npo KO telomeres, even after 8000 hours of culture time (Fig. 11B). Therefore, regulation of telomere length does not appear to be a factor in lifespan reduction for the npo KO strain.
FIGURE 11. Telomere stability in wild type and npo mutant strains.
(A) Arrows on plot shows time points used in telomere erosion assay. (B) Chromosomal DNAs were isolated from wild type and the npo mutant at the time points indicated in panel A. The DNAs were digested with HaeIII, blotted as described in Materials and probed with telomere repeat sequences.
PARylation is not universally required for viability or DNA repair
PARP orthologs have been identified in all eukaryotes, excluding yeast. Both plants and animals typically have multiple PARP orthologs, making genetic characterization difficult. Lethality of PARP-1−/−/PARP-2−/− mice and evidence linking PARylation with DNA repair and genomic stability support a view that PARylation impacts nuclear functions essential for higher eukaryotic development or survival. Unsuccessful attempts to generate PARP-1/PARP-2 double knockouts in mouse embryonic fibroblasts (MEDER et al. 2005) suggest that these functions may be critical for cellular survival. Mutation of dPARP in Drosophila results in larval lethality at the second instar stage, with disruption of heterochromatin organization and elimination of nucleoli (TULIN et al. 2002), again supporting the hypothesis that PARPs provide nuclear functions essential to the cell. Recent work on PARP in the filamentous fungus A. nidulans extends this view to PARylating lower eukaryotes (SEMIGHINI et al. 2006). Our work in N. crassa stands in contrast to what has been found for mammals and A. nidulans, as N.crassa npo mutants are viable and do not show sensitivity to mutagens, establishing that PARylation is dispensable for both viability and DNA-repair in certain eukaryotes with PARP orthologs. The fact that transcription of PARP genes is induced by DNA damage in both plants (DOUCET-CHABEAUD et al. 2001) and filamentous fungi (SEMIGHINI et al. 2006) does support the idea that there is a universal function for PARylation in DNA repair, but this function may be redundant in N. crassa, but not A. nidulans.
PARylation is not required for heterochromatin formation in N. crassa
Two major heterochromatin silencing pathways described in N. crassa are TPE (SMITH et al. 2008) and cytosine methylation (SELKER 2004). Our analysis indicates that neither pathway is significantly affected by mutation of npo. The histone H3 K9 methylase, DIM-5 (TAMARU and SELKER 2001), and the HP1 ortholog, HPO (FREITAG et al. 2004a), are necessary for silencing at all tested N. crassa telomeres (SMITH et al. 2008), as well as for all detectable DNA methylation. It is formally possible, however, that NPO might regulate TPE at telomeres other than VR or DNA methylation at a subset of unanalyzed genomic loci, although we have no reason to expect this to be so. We have shown that treatment of N. crassa with NAM dramatically reduces silencing of telVR::hph, but has no effect on silencing of the methylated transgene amRIP::hph::amRIP. Before our analysis of the effect of the npoRIP1 allele on silencing of telVR::hph, we could not fully interpret these data. Our genetic studies now suggest that the mechanism of action of NAM on TPE involves inhibition of NSTs, but not NPO. The fact that NAM treatment did not relieve silencing of amRIP::hph::amRIP, strongly suggests that neither NPO nor NSTs are involved in methylation or methylation-dependent silencing at this locus. The observation that PARP-1 activity impacts DNA methylation in mammals implies divergence in pathways that regulate methylation between mammals and filamentous fungi. This is not surprising considering that the activity of DNMT1, which is the primary maintenance methylase in mammals, is inhibited by PAR (REALE et al. 2005). N. crassa lacks this form of maintenance methylation, which acts specifically on hemimethylated CpG dinucleotides in conjunction with DNA replication. In N. crassa, both maintenance and de novo methylation are carried out by a single methyltransferase, DIM-2 (KOUZMINOVA and SELKER 2001), which does not require a symmetrical sequence (SELKER et al. 1993). It would be interesting to know whether PARP inhibitors or silencing of a PARP ortholog affect DNA methylation in A immersus, as this species may have a maintenance methylation system that is more similar to that in mammals.
The NPO aging pathway does not involve telomere length maintenance
Some current models for regulation of aging in humans consider telomere maintenance potentially important, as somatic human cells lack telomerase activity, and thus have a finite replicative lifespan (CAMPISI 2005; VERDUN and KARLSEDER 2007). Recently, SIRT6 has been shown to function as a telomere-specific histone H3 K9 deacetylase, which is necessary for normal telomere maintenance and for prevention of premature cellular senescence in human fibroblasts (MICHISHITA et al. 2008). In addition to playing a role in replicative cellular aging, SIRT6 has also been shown to impact chronological aging in mice, as SIRT6−/− animals exhibit phenotypes characteristic of progeroid disorders (MOSTOSLAVSKY et al. 2006). We did not observe any effect on telomere length in an npo mutant strain. These results do not rule out the possibility, however, that mutation of npo might affect other aspects of telomere maintenance or stability. In fact, the aberrations seen at telomeres in SIRT6 knockdown fibroblasts are similar to those seen in Werner syndrome cells, such as telomere deletions, duplications and fusions, with no obvious effect on the length of intact telomeres (MICHISHITA et al. 2008). Importantly, N. crassa has a homolog of SIRT6, termed NST-7 (Neurospora Sirtuin 7), not found in either S. cerevisiae or S. pombe (SMITH et al. 2008). It would be interesting to know whether NST-7 functions in the same aging pathway as NPO, and whether maintenance of telomere integrity is involved.
The NPO aging pathway and histone H1
The replicative aging phenotype that we observed in the npo mutant is novel for N. crassa but similar to that reported for a strain of the filamentous fungus Ascobolus immerses carrying a silenced epi-allele of the histone H1 (hH1) gene, that confers a DNA hypermethylation phenotype (BARRA et al. 2000). Although N. crassa hH1 mutants do not display hypermethylation (FOLCO et al. 2003), it would be interesting to know whether Neurospora hH1 mutants show a decreased replicative lifespan, and if so, whether this involves NPO. Conversely, one could ask whether PARP inhibitors or mutation/silencing of a PARP ortholog would affect replicative aging in A. immersus, and if so, whether the pathway is independent of the established hH1 pathway and/or DNA methylation. KIM et al. (2004) showed that PARP-1 associates with chromatin in a manner very similar to hH1: PARP-1 increases the nucleosome repeat length and competes with hH1 in nucleosome assembly reactions. Like hH1, binding of PARP-1 to chromatin in vitro triggers condensation and transcriptional repression. Unlike hH1, however, PARP-1 dissociates from chromatin in the presence of NAD+, and it has been suggested that localized NAD+ levels in nuclei might control chromatin structure and transcription (KIM et al. 2004). Results of ChIP-chip experiments have shown that actively transcribed promoters have high levels of PARP-1 and low levels of hH1, and that hH1 occupancy is excluded by PARP-1 binding (KRISHNAKUMAR et al. 2008). An attractive hypothesis is that PARPs and hH1 provide related functions associated with nuclear NAD+ levels, genome stability and aging. Consistent with this possibility, dramatic loss of hH1 accompanies cellular senescence of human fibroblasts (FUNAYAMA et al. 2006). While it is intriguing that both PARP and hH1 orthologs have been implicated in replicative aging in filamentous fungi, there is currently no evidence that fungal PARPs of the PrpA class have the linker histone-like properties of PARP-1. Furthermore, they lack an amino terminal DNA binding domain, which is required for PARP-1 chromatin association. It remains possible, however, that fungal PARPs interact with DNA binding proteins that target them to chromatin.
NPO might regulate aging in a pathway with Sirtuins
The possible function of NSTs in regulation of lifespan in N. crassa has not been investigated. If PARPs impact aging exclusively through indirect effects on the activity of Sirtuins, then our observation that NPO is necessary for normal replicative lifespan in N. crassa is difficult to reconcile with current models on how Sirtuins regulate aging in yeast and higher organisms. Current models from yeast that assume Sirtuins function exclusively to promote longevity would predict that when NAD+ is limiting, PARylation would inhibit long lifespan, because NAD+-dependent deacetylation and PARylation both consume NAD+ and produce NAM. Thus, an important question is whether localized NAD+ levels in nuclei are in fact limiting. If they are not, then NSTs and NPO could presumably act in the same or parallel pathways, with both functioning to promote longevity. ANDERSON et al. (2002) found that increasing the levels of NAD+ salvage pathway proteins in S. cerevisiae increased telomere and rDNA silencing in a Sir2-dependent manner. Although sir2 deletion mutants were not found to have elevated levels of total cellular NAD+, the authors argue that most or all of NAD+ salvage in S. cerevisiae occurs in nuclei, and that nuclear NAD+ salvage pathway flux is important in regulation of Sir2 deacetylase activity (ANDERSON et al. 2002).
Unlike Sir2, PARP-1 can dramatically reduce total cellular NAD+ levels in response to DNA damage (ZONG et al. 2004). If NPO is as robust as PARP-1, and if NAD+ availability within nuclei is limiting in N. crassa, then NSTs and NPO may compete for NAD+, and thus function antagonistically in the same aging pathway. However, if it is also assumed that Sirtuins act exclusively to promote longevity, as some models suggest, then PARylation should have a negative affect on lifespan, and mutation of npo should increase longevity, which is contrary to our observations.
Recently FABRIZIO et al. (2005) have shown that while Sir2 has a positive impact on replicative lifespan in S. cerevisiae, it actually has a negative impact on chronological lifespan, which is a measure of how long a non-dividing cell or organism survives. In addition, while it is generally accepted that Sirtuins positively regulate longevity in metazoans, SIRT1 may actually function in a pro-aging pathway (FABRIZIO et al. 2005), as sirt1−/− mice manifest many phenotypes of long-lived IGF-I-deficient dwarf mice (MCBURNEY et al. 2003). Furthermore, SIRT1 represses the DAF-16 homolog FOXO3 (MOTTA et al. 2004), and this is presumably antagonistic to longevity (LIN et al. 1997). If the activities of NSTs negatively regulate replicative lifespan in N. crassa, then competition between NSTs and NPO for NAD+ could occur, with NPO acting to promote longevity through inhibition of NSTs.
Regardless of whether Sirtuins promote or inhibit longevity, the general observation that NAD+-dependent deacetylases impact aging in both yeast and metazoans suggests conservation of this role during evolution. It is therefore reasonable to expect that NSTs may play a role in N. crassa as well. Until such a role has been definitively established, however, it is not possible to draw conclusions about the involvement of NSTs in the NPO pathway. Analysis of the aging phenotypes of nst mutants, individually and in combination with each other and the npo mutant, would provide an answer to these mechanistic questions.
TABLE 1.
NEUROSPORA CRASSA STRAINS USED IN THIS STUDY
| Strain number | Genotype | Source |
|---|---|---|
| N150 | mat A | FGSC 2489 |
| N1444 | mat a his-3; am132 | this study |
| N1674 | mat A his-3; lys-1 am132 inl; amRIP::hph::amRIP | (HAYS et al. 2002) |
| N1983 | mat a; mtr col4; nst-1RIP1 trp-2 | this study |
| N2636 | mat A nst-3RIP1; mtr col-4; telVR::hph::T; nst-1 RIP1 trp-2 | (SMITH et al. 2008) |
| N3180 | mat A his-3::npoRIP0; am132 npoRIP1 | this study |
| N3181 | mat a; npoRIP1 | this study |
| 74-OR31-16A | mat A al-2; pan-2;cot-1 | (DE SERRES 1980) |
| 74-OR31-14a | mat a al-2; pan-2; cot-1 | (DE SERRES 1980) |
| MKI-1411A | mat A al-2; pan-2; cot-1; npoKO | this study |
| MKI-1414a | mat a al-2; pan-2; cot-1; npoKO | this study |
| 14-6-1-1A | mat A al-2; pan-2; cot-1; npoKO | this study |
| G1 | mat A his-3 cyh-1 al-1; mtr; inl | FGSC 7508 |
| P49 | mat A his-3 cyh-1 al-1; inl | this study |
ACKNOWLEDGEMENTS
We would like to thank Melissa Hemphill for helping to analyze npo/nst genetic interaction and Wendy Hanna-Rose for comments on the manuscript. Thanks to Melissa Rolls for expert advice in confocal imaging. This work was supported by grant GM025690 from the National Institutes of Health to EUS and by Rational Evolutionary Design of Advanced Biomolecules, Saitama Prefecture Collaboration of Regional Entities for the Advancement of Technological Excellence, Japan Science and Technology Agency to HI.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, et al. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Ame JC, Fouquerel E, Gauthier LR, Biard D, Boussin FD, et al. Radiation-induced mitotic catastrophe in PARG-deficient cells. Cell Sci. 2009a;122:1990–2002. doi: 10.1242/jcs.039115. [DOI] [PubMed] [Google Scholar]
- Ame JC, Hakme A, Quenet D, Fouquerel E, Dantzer F, et al. Detection of the Nuclear Poly(ADP-ribose)-Metabolizing Enzymes and Activities in Response to DNA Damage. Methods Mol Biol. 2009b;464:267–283. doi: 10.1007/978-1-60327-461-6_15. [DOI] [PubMed] [Google Scholar]
- Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274:17860–17868. doi: 10.1074/jbc.274.25.17860. [DOI] [PubMed] [Google Scholar]
- Ame JC, Spenlehauer C, de Murcia G. The PARP superfamily. Bioessays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature. 2003;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo R, Metzenberg RL. Meiotic transvection in fungi. Cell. 1996;86:103–113. doi: 10.1016/s0092-8674(00)80081-1. [DOI] [PubMed] [Google Scholar]
- Barra JL, Rhounim L, Rossignol JL, Faugeron G. Histone H1 is dispensable for methylation-associated gene silencing in Ascobolus immersus and essential for long life span. Mol Cell Biol. 2000;20:61–69. doi: 10.1128/mcb.20.1.61-69.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belenky P, Bogan KL, Brenner C. NAD+ metabolism in health and disease. Trends Biochem Sci. 2007;32:12–19. doi: 10.1016/j.tibs.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Beneke S, Burkle A. Poly(ADP-ribosyl)ation, PARP, and aging. Sci Aging Knowledge Environ. 2004;2004:re9. doi: 10.1126/sageke.2004.49.re9. [DOI] [PubMed] [Google Scholar]
- Beneke S, Burkle A. Poly(ADP-ribosyl)ation in mammalian ageing. Nucleic Acids Res. 2007;35:7456–7465. doi: 10.1093/nar/gkm735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard VJ, Rouleau M, Poirier GG. PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol. 2003;31:446–454. doi: 10.1016/s0301-472x(03)00083-3. [DOI] [PubMed] [Google Scholar]
- Boulu RG, Mesenge C, Charriaut-Marlangue C, Verrecchia C, Plotkine M. [Neuronal death: potential role of the nuclear enzyme, poly (ADP-ribose) polymerase] Bull Acad Natl Med. 2001;185:555–563. discussion 564-555. [PubMed] [Google Scholar]
- Burkle A. Poly(ADP-ribosyl)ation: a posttranslational protein modification linked with genome protection and mammalian longevity. Biogerontology. 2000;1:41–46. doi: 10.1023/a:1010089924898. [DOI] [PubMed] [Google Scholar]
- Burkle A. PARP-1: a regulator of genomic stability linked with mammalian longevity. Chembiochem. 2001a;2:725–728. doi: 10.1002/1439-7633(20011001)2:10<725::AID-CBIC725>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Burkle A. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays. 2001b;23:795–806. doi: 10.1002/bies.1115. [DOI] [PubMed] [Google Scholar]
- Burkle A, Diefenbach J, Brabeck C, Beneke S. Ageing and PARP. Pharmacol Res. 2005;52:93–99. doi: 10.1016/j.phrs.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Burzio LO, Riquelme PT, Koide SS. ADP ribosylation of rat liver nucleosomal core histones. J Biol Chem. 1979;254:3029–3037. [PubMed] [Google Scholar]
- Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- Carroll AM, Sweigard JA, Valent B. Improved vectors for selecting resistance to hygromycin. Fungal Genetics Newsl. 1994;41:22. [Google Scholar]
- Chiarugi A, Moskowitz MA. Cell biology. PARP-1--a perpetrator of apoptotic cell death? Science. 2002;297:200–201. doi: 10.1126/science.1074592. [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- Dantzer F, Schreiber V, Niedergang C, Trucco C, Flatter E, et al. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69–75. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- Davis RD, DeSerres FJ. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 1970;17:79–143. [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol Genet Genomics. 2001;265:954–963. doi: 10.1007/s004380100506. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, et al. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Folco HD, Freitag M, Ramon A, Temporini ED, Alvarez ME, et al. Histone H1 Is required for proper regulation of pyruvate decarboxylase gene expression in Neurospora crassa. Eukaryot Cell. 2003;2:341–350. doi: 10.1128/EC.2.2.341-350.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU. HP1 is essential for DNA methylation in neurospora. Mol Cell. 2004a;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 2004b;41:897–910. doi: 10.1016/j.fgb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Funayama R, Saito M, Tanobe H, Ishikawa F. Loss of linker histone H1 in cellular senescence. J Cell Biol. 2006;175:869–880. doi: 10.1083/jcb.200604005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo CM, Smith DL, Jr, Smith JS. Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol. 2004;24:1301–1312. doi: 10.1128/MCB.24.3.1301-1312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grube K, Burkle A. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc Natl Acad Sci U S A. 1992;89:11759–11763. doi: 10.1073/pnas.89.24.11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herceg Z, Wang ZQ. Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat Res. 2001;477:97–110. doi: 10.1016/s0027-5107(01)00111-7. [DOI] [PubMed] [Google Scholar]
- Honda S, Selker E. Tools for Fungal Proteomics: Multifunctional Neurospora Vectors for Gene Replacement, Protein Expression and Protein Purification. Genetics. 2009 doi: 10.1534/genetics.108.098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci. 2004;25:259–264. doi: 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Huletsky A, de Murcia G, Muller S, Hengartner M, Menard L, et al. The effect of poly(ADP-ribosyl)ation on native and H1-depleted chromatin. A role of poly(ADP-ribosyl)ation on core nucleosome structure. J Biol Chem. 1989;264:8878–8886. [PubMed] [Google Scholar]
- Ikejima M, Noguchi S, Yamashita R, Ogura T, Sugimura T, et al. The zinc fingers of human poly(ADP-ribose) polymerase are differentially required for the recognition of DNA breaks and nicks and the consequent enzyme activation. Other structures recognize intact DNA. J Biol Chem. 1990;265:21907–21913. [PubMed] [Google Scholar]
- Jeggo PA. DNA repair: PARP - another guardian angel? Curr Biol. 1998;8:R49–R51. doi: 10.1016/s0960-9822(98)70032-6. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, et al. The macro domain is an ADP-ribose binding module. Embo J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Jr, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae. Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Kouzminova E, Selker EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. Embo J. 2001;20:4309–4323. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, et al. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Latorre-Esteves M, Medvedik O, Wong SN, Tsang FA, et al. HST2 mediates SIR2-independent life-span extension by calorie restriction. Science. 2005;309:1861–1864. doi: 10.1126/science.1113611. [DOI] [PubMed] [Google Scholar]
- Li B, Navarro S, Kasahara N, Comai L. Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J Biol Chem. 2004;279:13659–13667. doi: 10.1074/jbc.M311606200. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros JJ, Denome SA, Dunlap JC. Molecular cloning of genes under control of the circadian clock in Neurospora. Science. 1989;243:385–388. doi: 10.1126/science.2563175. [DOI] [PubMed] [Google Scholar]
- Luo Z, Freitag M, Sachs MS. Translational regulation in response to changes in amino acid availability in Neurospora crassa. Mol Cell Biol. 1995;15:5235–5245. doi: 10.1128/mcb.15.10.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin BS, Freitag M, Selker EU. Improved plasmids for targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genetics Newsletter. 1997;47:112. [Google Scholar]
- Margolin BS, Garrett-Engele PW, Stevens JN, Fritz DY, Garrett-Engele C, et al. A methylated Neurospora 5S rRNA pseudogene contains a transposable element inactivated by repeat-induced point mutation. Genetics. 1998;149:1787–1797. doi: 10.1093/genetics/149.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani M, Nakagama H, Sugimura T. Poly(ADP-ribose) and carcinogenesis. Genes Chromosomes Cancer. 2003;38:339–348. doi: 10.1002/gcc.10250. [DOI] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally MT, Free SJ. Isolation and characterization of a Neurospora glucose-repressible gene. Curr Genet. 1988;14:545–551. doi: 10.1007/BF00434079. [DOI] [PubMed] [Google Scholar]
- Meder VS, Boeglin M, de Murcia G, Schreiber V. PARP-1 and PARP-2 interact with nucleophosmin/B23 and accumulate in transcriptionally active nucleoli. J Cell Sci. 2005;118:211–222. doi: 10.1242/jcs.01606. [DOI] [PubMed] [Google Scholar]
- Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. Embo J. 2003;22:2255–2263. doi: 10.1093/emboj/cdg206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao VP, Freitag M, Selker EU. Short TpA-rich segments of the zeta-eta region induce DNA methylation in Neurospora crassa. J Mol Biol. 2000;300:249–273. doi: 10.1006/jmbi.2000.3864. [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci U S A. 2004;101:12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata N, Ueda K, Kawaichi M, Hayaishi O. Poly(ADP-ribose) synthetase, a main acceptor of poly(ADP-ribose) in isolated nuclei. J Biol Chem. 1981:4135–4137. [PubMed] [Google Scholar]
- Osiewacz HD. Aging in fungi: role of mitochondria in Podospora anserina. Mech Ageing Dev. 2002;123:755–764. doi: 10.1016/s0047-6374(01)00421-3. [DOI] [PubMed] [Google Scholar]
- Panzeter PL, Althaus FR. High resolution size analysis of ADP-ribose polymers using modified DNA sequencing gels. Nucleic Acids Res. 1990;18:2194. doi: 10.1093/nar/18.8.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper AA, Verma A, Zhang J, Snyder SH. Poly (ADP-ribose) polymerase, nitric oxide and cell death. Trends Pharmacol Sci. 1999;20:171–181. doi: 10.1016/s0165-6147(99)01292-4. [DOI] [PubMed] [Google Scholar]
- Povirk LF, Wubter W, Kohnlein W, Hutchinson F. DNA double-strand breaks and alkali-labile bonds produced by bleomycin. Nucleic Acids Res. 1977;4:3573–3580. doi: 10.1093/nar/4.10.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reale A, Matteis GD, Galleazzi G, Zampieri M, Caiafa P. Modulation of DNMT1 activity by ADP-ribose polymers. Oncogene. 2005;24:13–19. doi: 10.1038/sj.onc.1208005. [DOI] [PubMed] [Google Scholar]
- Riquelme PT, Burzio LO, Koide SS. ADP ribosylation of rat liver lysine-rich histone in vitro. J Biol Chem. 1979;254:3018–3028. [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rountree MR, Selker EU. DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes Dev. 1997;11:2383–2395. doi: 10.1101/gad.11.18.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier JJ, Celic I, Boeke JD, Smith JS. Telomeric and rDNA silencing in Saccharomyces cerevisiae are dependent on a nuclear NAD(+) salvage pathway. Genetics. 2002;160:877–889. doi: 10.1093/genetics/160.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EU. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Selker EU. Genome defense and DNA methylation in Neurospora. Cold Spring Harb Symp Quant Biol. 2004;69:119–124. doi: 10.1101/sqb.2004.69.119. [DOI] [PubMed] [Google Scholar]
- Selker EU, Fritz DY, Singer MJ. Dense nonsymmetrical DNA methylation resulting from repeat-induced point mutation in Neurospora. Science. 1993;262:1724–1728. doi: 10.1126/science.8259516. [DOI] [PubMed] [Google Scholar]
- Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, et al. The methylated component of the Neurospora crassa genome. Nature. 2003;422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
- Semighini CP, Savoldi M, Goldman GH, Harris SD. Functional characterization of the putative Aspergillus nidulans poly(ADP-ribose) polymerase homolog PrpA. Genetics. 2006;173:87–98. doi: 10.1534/genetics.105.053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, et al. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273:30069–30072. doi: 10.1074/jbc.273.46.30069. [DOI] [PubMed] [Google Scholar]
- Shimokawa T, Masutani M, Nagasawa S, Nozaki T, Ikota N, et al. Isolation and cloning of rat poly(ADP-ribose) glycohydrolase: presence of a potential nuclear export signal conserved in mammalian orthologs. J Biochem. 1999;126:748–755. doi: 10.1093/oxfordjournals.jbchem.a022512. [DOI] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith KM, Kothe GO, Matsen CB, Khlafallah TK, Adhvaryu KK, et al. The fungus Neurospora crassa displays telomeric silencing mediated by multiple sirtuins and by methylation of histone H3 lysine 9. Epigenetics and Chromatin. 2008;1 doi: 10.1186/1756-8935-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulson ME, Simbulan-Rosenthal CM, Boulares AH, Yakovlev A, Stoica B, et al. Roles of poly(ADP-ribosyl)ation and PARP in apoptosis, DNA repair, genomic stability and functions of p53 and E2F-1. Adv Enzyme Regul. 2000;40:183–215. doi: 10.1016/s0065-2571(99)00024-2. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya M, Dang N, Kerr EO, Hu D, Steffen KK, et al. Sirtuin-independent effects of nicotinamide on lifespan extension from calorie restriction in yeast. Aging Cell. 2006;5:505–514. doi: 10.1111/j.1474-9726.2006.00240.x. [DOI] [PubMed] [Google Scholar]
- Tulin A, Stewart D, Spradling AC. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Harrigan JA, Schreiber V, Stiegler P, Piotrowski J, et al. Poly(ADP-ribose) polymerase 1 regulates both the exonuclease and helicase activities of the Werner syndrome protein. Nucleic Acids Res. 2004;32:4003–4014. doi: 10.1093/nar/gkh721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, et al. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 1995;9:509–520. doi: 10.1101/gad.9.5.509. [DOI] [PubMed] [Google Scholar]
- Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, et al. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11:2347–2358. doi: 10.1101/gad.11.18.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe F, Fukazawa H, Masutani M, Suzuki H, Teraoka H, et al. Poly(ADP-ribose) polymerase-1 inhibits ATM kinase activity in DNA damage response. Biochem Biophys Res Commun. 2004;319:596–602. doi: 10.1016/j.bbrc.2004.05.031. [DOI] [PubMed] [Google Scholar]
- Watanabe K, Sakuraba Y, Inoue H. Genetic and molecular characterization of Neurospora crassa mus-23: a gene involved in recombinational repair. Mol Gen Genet. 1997;256:436–445. doi: 10.1007/s004380050587. [DOI] [PubMed] [Google Scholar]
- Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–1282. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]