Abstract
High levels of activated Stat3 are often found in human breast cancers and can correlate with poor patient outcome. We employed an activated-ErbB2 mouse model of breast cancer to investigate the in vivo role of Stat3 in mammary tumor progression and found that Stat3 does not alter mammary tumor initiation but dramatically affects metastatic progression. Four-fold fewer animals exhibited lung metastases in the absence of Stat3 and a 12-fold reduction in the number of lung lesions was observed in the Stat3-null tumors when compared to tumors from the wild type cohort. The decreased malignancy in Stat3-deficient tumors is attributed to a reduction in both angiogenic and inflammatory responses associated with a Stat3-dependent transcriptional cascade involving C/EBPδ.
Introduction
Constitutive activation of the transcription factor Stat3 is observed in 35 to 60% of human breast cancers (1, 2) and in a wide variety of other cancer types (3). In normal tissues Stat3 is involved in the direct transcriptional regulation of targets downstream of both cytokine and growth factor receptors. In tumors, Stat3 is activated downstream of oncogenes such as ErbB2/Neu, PyVMT, and Src (4-6). Overexpression of constitutively activated forms of Stat3 in fibroblast cells, either in isolation or in conjunction with oncogenes induces the formation of foci in vitro and tumors in orthotopic mouse models (6, 7). Moreover, loss of Stat3 function via RNA knockdown (8, 9), peptide inhibition (10), and expression of dominant negative forms (6, 11, 12) in cancer cells leads to a decrease in tumor cell growth and angiogenesis with a concomitant increase in apoptosis (9, 12, 13). Analyses of human tumor tissues have also shown that Stat3 expression and activation correlates with tumor grade, stage, or the presence of metastases (1, 14-16).
While studies suggest that activation of Stat3 is a critical event in the transformation of established cell lines in vitro, the in vivo role of Stat3 in mammary tumorigenesis is still unknown. To investigate the role of Stat3 in breast cancer, conditional Stat3 (Stat3flx) mice (17) were interbred with a novel transgenic strain (MMTV-NIC) where the expression of an activated form of ErbB2 is coupled to Cre recombinase via an internal ribosome entry site (IRES) (18). The resulting Stat3flx/flx/NIC mice exhibited a nearly 4-fold reduction in the incidence of tumor metastasis relative to the parental NIC strain, though tumor onset was not altered by mammary-specific, Cre-mediated ablation of Stat3. Using gene expression profiling, we observed that the expression of cebpd was downregulated in the Stat3-deficient tumors relative to their wild type counterparts. Consequently, Stat3flx/flx/NIC tumors lacked the ability to induced the expression of acute phase response genes downstream of both Stat3 and C/EBPδ (19). These results suggest that Stat3 may mediate a tumor inflammatory response through several downstream acute phase response genes and thus provide a pro-metastatic tumor environment.
Materials and Methods
Transgenic Mice
Mice harboring the conditional stat3 allele were generated in the Levy lab and characterized previously (17, 20). MMTV-NIC transgenic mice were generated as described (18). All mice were housed in the animal facility of the Royal Victoria Hospital and all experiments were performed in accordance with the animal care guidelines at the Animal Resource Centre of McGill. Mammary tumors were detected via biweekly physical palpation and animals were sacrificed 6 weeks following initial palpation. Material from necropsied mice was frozen in liquid nitrogen, in some cases tissues were set in an optimal cutting temperature media (OCT) prior to freezing, or was fixed in 10% neutral buffered formalin and embedded in paraffin wax. Fixed and embedded mammary tumors and lung lobes were sectioned at 4μm and either stained by hematoxylin and eosin (H&E) or processed further as indicated. Five H&E stained lung sections, taken at 50μm intervals, were examined by microscope for metastatic lesions. Experimental metastasis assays were performed by injecting 5×105 cells into the lateral tail vein of NCr mice (Taconic). Lungs were collected and processed as described above at 4 weeks post-injection.
Primary cell culture
Stat3wt/wt/ or Stat3flx/flx/ NIC mammary tumors were excised, finely chopped and dissociated in DMEM (Wisent) containing 2.4 mg/ml collagenase B (Roche), 2.4 mg/ml Dispase II (Roche) for 3 h at 37°C, with constant agitation. The cell suspensions were centrifuged at 1000RPM for 5 min, washed in a PBS/EDTA solution and respun at 1000RPM for 5 min. Pellets were resuspended in DMEM media containing 10% FBS (Wisent), MEGM SingleQuots (Clonetics), and 5% penicillin/streptomycin (Wisent). Cells were counted and plated on 10cm Nunc dishes.
Immunoblotting, ELISA, and Immunohistochemical Analyses
Frozen mammary tumors were lysed using a PLC-γ buffer and run on SDS-PAGE gels. Proteins were detected with antibodies against Stat3, Stat3-Y705-P (1:1000, 9132 and 9131, Cell Signaling), Neu (1:1000, sc 284, Santa Cruz), VEGF (1:1000, PC315, Calbiochem), β-actin (1:1000, A5441, Sigma), and Grb2 (1:1000, sc255, Santa Cruz). Tumor lysate VEGF was measured with a mouse Quantikine ELISA kit (MMV00, R&D Systems). Immunohistochemical analyses were performed on paraffin or OCT embedded sections as previously described (18). Antibodies used for immunohistochemical analyses include: Stat3 (1:100, 9139, Cell Signaling), Stat3-Y705-P (1:50, 9145, Cell Signaling), and Cre (1:600, PRB106C, Covance). Staining for CD31 (1:100, 550274, BD Biosciences), Ki67 (1:1000, ab15580, AbCam), and TUNEL (Apoptag Peroxidase Detection kit, Chemicon) was quantified using slides scanned with a Scanscope XT Digital Slide Scanner (Aperio) and corresponding positive pixel and nuclear IHC algorithms.
Microarray Experiments
Total RNA was extracted from flash frozen mammary tumor samples using a Qiagen RNeasy Midi Kit and were labeled using an Amino Allyl MessageAmp II aRNA Amplification Kit (Applied Biosystems) and Cy3 and Cy5 dyes (Amersham Biosciences) in preparation for microarray hybridization. Dye-labeled RNA was hybridized onto a Whole Mouse Genome Oligo 4x44K Microarray platform (Agilent) against a universal mouse reference RNA (Stratagene). The resulting arrays were scanned using a Microarray Scanner (Model G2565BA, Agilent Technologies) and processed using Feature Extraction software (Agilent). Data processing, normalization and analysis were carried out using BIAS system (21). RMA background correction algorithm was used for correcting raw feature intensities (22). Resulting expression estimates were converted to log2-ratios. Spatial and intensity-dependent loess was performed to normalize within arrays (23). Median absolute deviation scale normalization was used to normalize between arrays (24). Differentially expressed genes were detected using rank product non-parametric statistic (p-value=0.01) (25). Microarray results have been deposited in the Gene Expression Omnibus (GEO) database under the accession number: GSE15632.
Results
Stat3 is not required for the initiation of mammary tumors but is important for tumor cell proliferation
To study Stat3 loss in a mouse model of tumorigenesis, we used Stat3 conditional mice harboring either one or two loxP-flanked stat3 alleles (stat3flx) (17). To circumvent the stochastic expression of Cre observed in many MMTV-Cre-based models (26), we mated the stat3flx animals with the transgenic NIC strain (18). In this model, mammary epithelial cells that express the activated ErbB2 oncogene will simultaneously express Cre recombinase resulting in the deletion of the conditional stat3 allele (18).
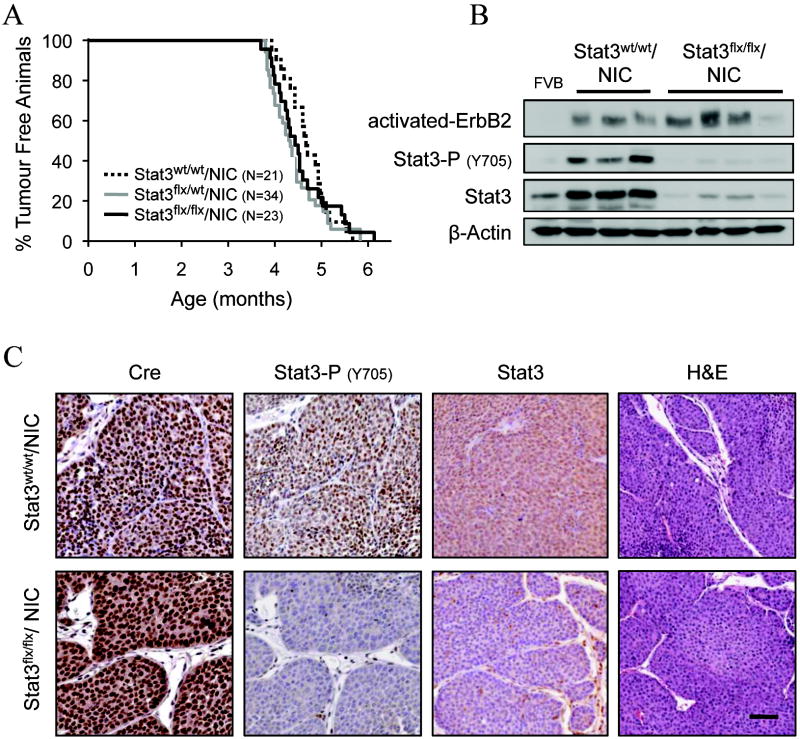
To investigate the role of Stat3 in the induction of mammary tumors, cohorts of virgin female Stat3wt/wt/NIC, Stat3flx/wt/NIC, and Stat3flx/flx/NIC mice were generated. All cohorts developed mammary tumors with statistically similar average onsets of 4.7±0.1, 4.4±0.1, and 4.6±0.1 months respectively (Fig. 1A). We verified the Cre recombinase-mediated stat3 deletion in the tumors from all three cohorts using PCR analyses (Supplementary Fig. S1). Ablation of Stat3 at the protein level was confirmed by both immunoblot and immunohistochemical analyses (Fig. 1B and 1C). Despite the fact that levels of Stat3 and its activated form, phospho-Y705-Stat3, were significantly reduced in Stat3flx/flx/NIC tumors (Fig. 1B and 1C), total activated-ErbB2 protein did not change across the samples (Fig. 1B). Tumors from NIC animals heterozygous for the stat3flx allele expressed total and activated Stat3 protein at levels comparable to the wild type NIC tumors (data not shown). The residual Stat3 protein observed in Stat3flx/flx/NIC mammary tumor lysates (Fig. 1B) is due to the retention of an intact stat3flx allele in the stroma as evidenced by a faint band by PCR (Supplementary Fig. S1) and by Stat3-positive stromal cells detected in the Stat3flx/flx/NIC tumor sections (Fig. 1C). Histological analysis of hematoxylin and eosin (H&E) -stained tumor sections showed that the Stat3flx/flx/NIC tumors displayed characteristics of solid adenocarcinomas consistent similar to wild-type activated ErbB2-driven tumors (Fig. 1C). Thus, Stat3flx/flx/NIC mice develop Stat3-null mammary epithelial tumors with the same onset as the wild type NIC group.
Figure 1. Stat3 activation is not required for the initiation of mammary tumorigenesis.
(A) Mammary tumor onset in Stat3wt/wt/NIC, Stat3flx/wt/NIC, and Stat3flx/flx/NIC cohorts (T50 = 4.7, 4.3, 4.5 months, respectively). (B) Immunoblot analysis of Stat3 and activated-ErbB2 expression in mammary tumor cell lysates from Stat3wt/wt/NIC and Stat3flx/flx/NIC animals. (C) Staining of paraffin-embedded sections of mammary tumors using antibodies against Cre (leftmost column), Stat3-Y705P and Stat3 (middle columns) or H&E (rightmost column). Scale bar: 50 μm.
Although mammary epithelial disruption of Stat3 had no affect on the initiation of NIC tumors, Stat3 activation has been reported to enhance tumor growth in some in vitro studies (12, 27). Therefore, the total tumor burden in Stat3wt/wt/NIC, Stat3flx/wt/NIC, and Stat3flx/flx/NIC mice was measured at sacrifice (6 weeks post palpation). Wild type NIC animals exhibited an average total tumor volume of 3.5±0.4 cm3, whereas the total volume measured in Stat3flx/wt/NIC and Stat3flx/flx/NIC was 1.7±0.2 cm3 and 2.1±0.4 cm3, respectively (Supplementary Fig. S2A). Impaired tumor growth was attributed to a greater than 2-fold reduction in the number of proliferating cells in the Stat3-deficient mammary tumors as assessed by Ki67 immunohistochemical staining (Supplementary Fig. S2B), and not to a change in the apoptotic status of the same cells as determined by a TUNEL assay (Supplementary Fig. S2C). We also counted the number of tumors present in endstage animals. Multifocal tumors were observed in nearly all the tumor-bearing Stat3wt/wt/NIC and Stat3flx/wt/NIC mice: greater than 92.5% in both cases (Supplementary Fig. S3A). In contrast, only 62.5% of the Stat3flx/flx/NIC tumor-bearing animals developed multifocal tumors (Supplementary Fig. S3A). In addition, analysis of the adjacent tumor-free inguinal mammary glands from tumor-bearing mice at endstage revealed that animals lacking Stat3 in the mammary epithelium developed approximately 12 times fewer lesions than either wild type or heterozygous animals (Supplementary Fig. S3B and S3C). Taken together, these observations suggest that while Stat3 is dispensable for the initiation of activated ErbB2-induced mammary tumors, its loss causes a statistically significant reduction in total tumor burden attributed to both reduced tumor proliferation and penetrance.
Stat3 plays a critical role in the metastasis of ErbB2-driven mammary tumors
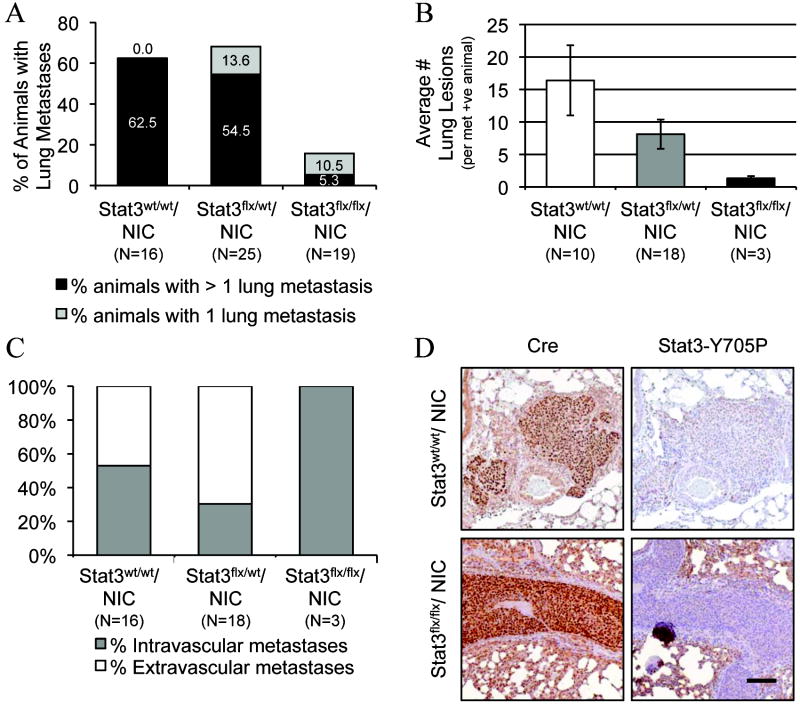
We next examined whether loss of Stat3 altered the metastatic capacity of ErbB2-induced tumors. Lungs from Stat3wt/wt/NIC, Stat3flx/wt/NIC, and Stat3flx/flx/NIC tumor-bearing mice were taken at 6 weeks post palpation and were scored for the presence of metastatic lesions. Only 15.8% of Stat3flx/flx/NIC animals exhibited lung lesions compared to 62.5% incidence in the wild type NIC cohort (Fig. 2A). Notably, mice heterozygous for the conditional stat3 allele, which presented with a statistically similar (p=0.47) tumor burden to the Stat3flx/flx/NIC mice (Supplementary Fig. S2A), exhibited comparable rates of metastasis to the wild type NIC animals (Fig. 2A). This precludes the possibility that the reduced tumor volume in the Stat3flx/flx/NIC cohort is responsible for the nearly 4-fold reduction in metastasis seen in the homozygous group (Fig. 2A). These results show that Stat3 is nearly indispensable for metastasis of activated-ErbB2 tumor cells to the lungs.
Figure 2. The presence of lung metastases is greatly reduced in mice bearing Stat3flx/flx/NIC mammary tumors.
Percentage of Stat3wt/wt/, Stat3flx/wt/, and Stat3flx/flx/ NIC animals harboring metastatic lesions in the lung (A) and quantification of the average number (±s.e.m.) of lesions per lung from animals positive for lung metastases (B). (C) Percentage of intra- versus extra-vascular lung lesions. (D) Immunohistochemical anti-Cre (right) and -Stat3Y705P (left) staining of paraffin-embedded lung sections from Stat3wt/wt/NIC, Stat3flx/flx/NIC animals. Scale bar: 100 μm. (p values were calculated using a two-tailed student’s t-test; *p=0.021).
We expanded this study to assess the malignancy of the Stat3wt/wt/NIC, Stat3flx/wt/NIC, and Stat3flx/flx/NIC tumors by counting the number of metastases present in the lungs and by characterizing the intra- or extra-vascular status of the lesions. The few Stat3flx/flx/NIC animals that developed metastases had 12-fold fewer lung lesions on average compared to their wild type counterparts (Fig. 2B). The rare lesions observed in the lungs of Stat3flx/flx/NIC mice were strictly confined within the pulmonary vasculature (Fig. 2C and 2D) while more than 45% of metastases present in the lungs of Stat3wt/wt/NIC and Stat3flx/wt/NIC animals were extravascular, demonstrating the ability of Stat3-proficient tumor cells to invade the surrounding lung parenchyma (Fig. 2C and 2D). To verify that the lesions observed in the Stat3flx/flx/NIC lungs did not express Stat3, the retention of Cre expression and the loss of active Stat3 in the metastatic lung lesions were confirmed by performing immunohistochemistry with antibodies directed to Cre recombinase and phospho-Y705-Stat3. Lesions from both Stat3wt/wt/NIC and Stat3flx/flx/NIC lungs revealed high expression of Cre recombinase while only the wild type-derived metastases retained expression of phospho-Stat3 (Fig. 2D). Collectively, these observations argue that Stat3 plays a critical role in the metastatic phase of activated-ErbB2 tumor progression.
The metastatic defect in Stat3-deficient tumors correlates with impaired tumor angiogenesis and cell autonomous defects in colonization
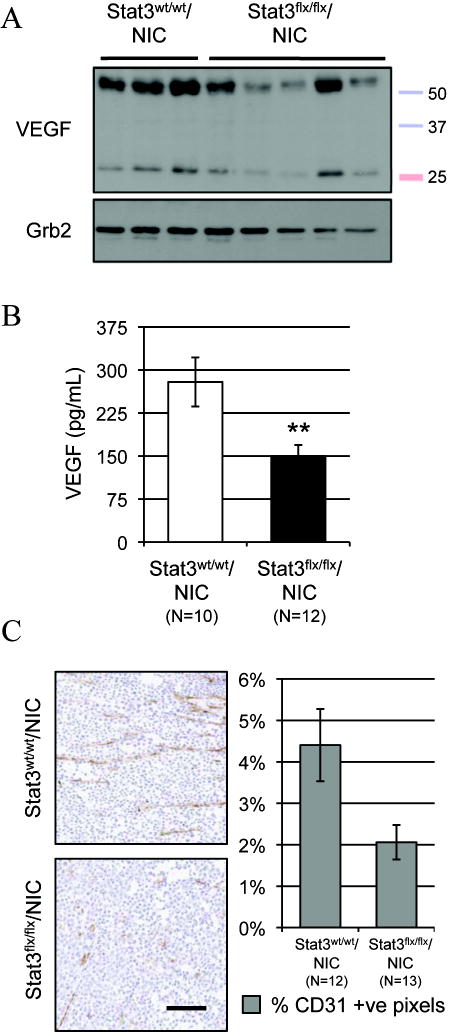
To further elucidate the molecular basis for the observed impairment of metastasis in the Stat3-deficient NIC tumors we assessed whether tumor angiogenesis was affected by ablation of Stat3. Tumor vascularization was quantified by measuring the endothelial content in Stat3wt/wt/NIC and Stat3flx/flx/NIC tumors stained with an antibody against CD31. Tumors from the Stat3flx/flx/NIC animals were less vascularized than tumors from the wild type cohort (Fig. 3C). In addition, a significant reduction of the pro-angiogenic factor, VEGF, (both 50 and 24 kDa isoforms) was observed in the Stat3flx/flx/NIC tumors by immunoblotting and ELISA (Fig. 3A and 3B). These results indicate that a lack of Stat3 impairs tumor angiogenesis which may, in turn, impede tumor cell metastasis.
Figure 3. Stat3 ablation leads to decreased VEGF production and angiogenesis.
Decreased VEGF protein levels in Stat3flx/flx/NIC tumors by immunoblotting (A) and ELISA (B) assays (average VEGF quantity ±s.e.m.). (C) Immunohistochemical staining using a CD31 antibody (left panel) and quantification of the average number (±s.e.m.) of CD31 positive pixels (right panel). Scale bar: 100 μm. (p values were calculated using a two-tailed student’s t-test; *p=0.017).
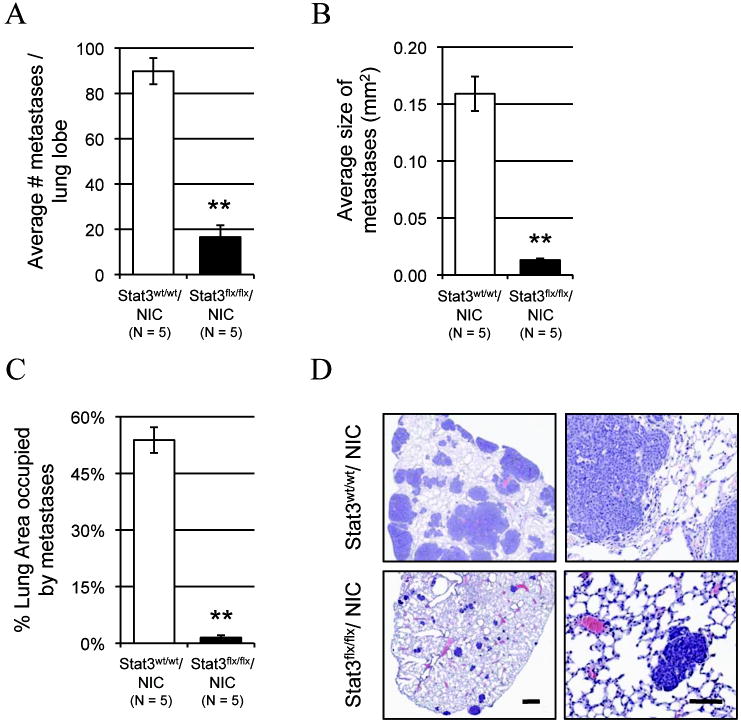
Whereas the above results suggest that defects in the tumor microenvironment may influence the metastatic capacity of Stat3-deficient tumor cells, it is also possible that the inability to efficiently metastasize to the lungs is due to a tumor cell intrinsic defect. To test this possibility, cells from both Stat3-proficient and -deficient NIC tumors were dissociated and established in cell culture. These primary tumor cells were injected into the lateral tail vein of immunodeficient mice and the number and size of the resulting lung lesions was scored. Injection of Stat3flx/flx/NIC tumor cells resulted in a 5-fold reduction in the number and a 12-fold decrease in the size of the resulting lung metastases compared to the Stat3wt/wt/NIC tumor cells (Fig. 4A and 4B). In addition, the total lung area occupied by metastatic lesions decreased 36-fold in the lungs of animals injected with Stat3flx/flx/NIC tumor cells compared to those injected with Stat3wt/wt/NIC tumor cells (Fig. 4C and 4D). Lesions from these lungs were subjected to immunohistochemical analysis which recapitulated the results observed in the lesions in the transgenic animals (Supplementary Fig. S4). Prior to injection, the expression of erbB2/neu, cre, and the excision of the stat3flx allele in the Stat3flx/flx/NIC tumor cells by PCR on DNA samples from the established mammary epithelial cultures (Supplementary Fig. S5A). As with the primary tumors, immunoblotting was performed on protein lysates from the tumor cell cultures (Supplementary Fig. S5B). In addition, the proliferative capacity of the primary tumor cells was monitored in vitro revealing that, in culture, Stat3-deficient and -proficient tumor cells grew at the same rate (Supplementary Fig. S5C). The fact that these primary Stat3-null tumor cells were impaired in their ability to metastasize even when introduced directly into the vasculature, suggests that the metastatic defect observed in the transgenic animals is due only in part to impaired angiogenesis. Indeed, this data suggests that Stat3 also mediates the ability of the tumor cells to colonize and grow in the lung in a cell autonomous manner.
Figure 4. Stat3flx/flx/NIC tumor cells show impaired lung colonization in an experimental metastasis assay.
Metastatic burden as measured by the average number of lesions per lung lobe (A), the average size of the metastases (B), and the average percentage of total lung area covered by metastatic lesions (C) (±s.e.m.) based on lesions resulting from experimental metastasis assays using 5×105 wild type or Stat3-null primary tumor cells. (D) Representative H&E stained sections of lungs from experimental metastasis assays. Scale bars: left column, 0.5 mm; right column, 100 μm. (p values were calculated using a two-tailed student’s t-test; a. **p=1.42E-5, b. **p=5.83E-4, c. **p=7.01E-5).
Gene expression profiling of Stat3-deficient tumors reveals a Stat3-dependent transcription network involved in the regulation of inflammation and angiogenesis
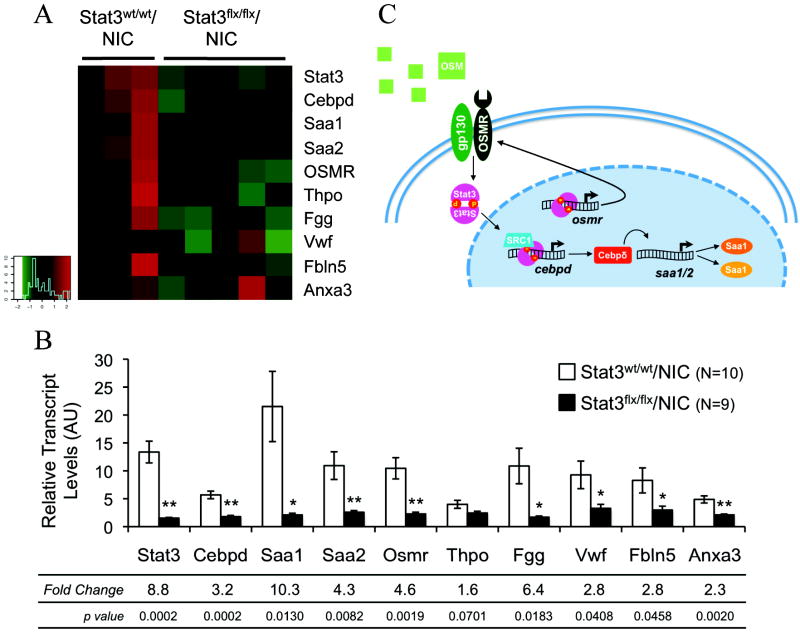
To further evaluate the impaired metastatic phenotype observed in Stat3flx/flx/NIC mice and to identify molecular profiles that would explain the decrease in malignancy, we compared the gene expression profile of Stat3wt/wt/NIC and Stat3flx/flx/NIC tumors using Agilent oligonucleotide microarrays. We found that Stat3flx/flx/NIC tumors expressed lower levels of both cebpd and osmr, two genes that are induced by Stat3 (28, 29) These genes code for the transcription factor CCAAT enhancer/binding protein δ(C/EBPδ) and the oncostatin M receptor (OSMR), and are known to potentiate the acute phase response (APR), an early stage of inflammation. While the OSMR acts upstream of Stat3 in the APR, C/EBPδ is a downstream target that promotes the expression of serum amyloid genes including saa1 and saa2 (29, 30). The expression levels of the serum amyloid genes were significantly reduced by more than 10-fold in the Stat3flx/flx/NIC tumors (Fig. 5A). We found additional targets involved in the APR and in tumor angiogenesis that were significantly downregulated in the Stat3flx/flx/NIC tumors compared to the wild type NIC tumors including genes encoding for von willebrand factor, thrombopoeitin, fibrinogen gamma, fibulin 5, and annexin a3 (Fig. 5A). Thus, loss of Stat3 from NIC tumors leads to the downregulation of many APR and pro-angiogenic genes which may, in turn, lead to reduced tumor inflammation and angiogenesis.
Figure 5. Stat3 expression causes upregulation of targets involved in angiogenesis and inflammation in NIC tumors.
(A) Heatmap of selected genes differentially expressed between Stat3wt/wt/NIC and Stat3flx/flx/NIC tumors. (B) Average transcript levels (±s.e.m.) of selected genes relative to GAPDH as calculated by quantitative RT-PCR. Fold change calculated by dividing the relative transcript level of the Stat3wt/wt/NIC samples by the value for the Stat3flx/flx/NIC samples. (C) Schematic of Stat3-dependent transcriptional network controlling inflammation. (p values were calculated using a student’s t-test).
To validate the results of these microarray-based results, we performed quantitative real-time PCR analysis of these candidate genes on total RNA extracted from Stat3wt/wt/ and Stat3flx/flx/NIC tumors. First, we established that stat3 transcript levels were significantly reduced in the Stat3flx/flx/NIC tumors compared to the wild type tumors (Fig. 5B). Next we determined that the expression levels of direct Stat3 transcriptional targets such as C/EBPδ and the OSMR were also reduced by 3.2- and 4.6-fold, respectively. The expression of the direct transcriptional targets of C/EBPδ, serum amyloid a1 and a2, was reduced in the Stat3flx/flx/NIC tumors by 10.3- and 4.3-fold, respectively (Fig. 5B). Given the importance of the Stat3 and C/EBPδ transcriptional network in regulation of the acute phase response and inflammation during normal mammary gland involution (31), these observations suggest that the impaired metastatic phenotype in Stat3-deficient tumors results from a dramatic reduction in tumor inflammation and angiogenesis.
Discussion
The dysregulated activation of Stat3 observed in many human breast cancers (1, 2, 32) suggests that it is of central importance to tumorigenesis and that loss of Stat3 should impair the progression or malignancy of the disease. This is supported by in vitro models, where the inhibition of Stat3 alters tumor growth, invasiveness, and, in a few cases, prevents tumor initiation (11, 12, 33). Since previous studies were largely based on established tumor cells, the role Stat3 plays in the induction or progression of mammary tumors in vivo is still poorly understood. Using a spontaneous transgenic mouse model of tumor progression, we have shown that loss of Stat3 in an activated-ErbB2 model of mammary tumorigenesis does not affect the initiation or survival of NIC tumors but does hinder tumor cell proliferation and angiogenesis. The Stat3flx/flx/NIC tumors are less metastatic and mammary epithelial cells from these cultures are unable to colonize or grow in the lung. Stat3 ablation also significantly reduced the gene expression of a number of acute phase response genes including cebpd, osmr, saa1 and saa2. These results implicate Stat3 in the transcriptional control of tumor inflammation and angiogenesis and suggest a major role for Stat3 expression and activation in the metastatic potential of ErbB2-induced tumors.
While Stat3 promotes cell death during normal mammary gland involution (20), it enhances proliferation and prevents apoptosis in a variety of tumorigenic cells (13, 34). In our model of ErbB2-induced tumorigenesis, the Stat3flx/flx/NIC tumors exhibited a 2-fold reduction in the number of proliferating cells, but showed no change in apoptotic index compared to wild type NIC tumors. The fact that the Stat3flx/flx/NIC tumors did not exhibit increased apoptosis may reflect the fact that Stat3 ablation occurs prior to ErbB2-driven tumor induction, thus allowing for the selection of cells expressing Stat3-independent survival pathways. These observations suggest that tumor cells can be selected to survive in the absence of Stat3 but that activated-ErbB2 tumors are at a proliferative disadvantage when lacking activated Stat3 expression.
Although several studies attempt to correlate the presence of activated Stat3 in human breast tumors with prognostic factors such as tumor stage, tumor size, or patient survival, few reports correlate Stat3 expression with breast cancer metastasis (1). Therefore, one of the most striking aspects of this study is the observation that the Stat3-deficient tumors exhibit a dramatic impairment in their capacity to metastasize to the lung. While Stat3 can regulate the transcription of pro-invasive matrix metalloproteinases (33, 35), neither MMP2 nor MMP9 protein levels changed in NIC tumors in the absence of Stat3 expression (data not shown). However, we did observe that Stat3flx/flx/NIC tumor cells were impaired in their ability to colonize and grow in the lung. Surprisingly, our microarray data did not reveal the differential expression of any genes directly implicated in metastasis. The absence of these genes further supports the hypothesis that Stat3 is involved in promoting metastasis via the upregulation of a number of genes to illicit a pro-metastatic response (eg; through the upregulation of angiogenic and inflammatory responses) rather than direct transcriptional regulation of any one specific gene involved in invasion or metastasis. Thus, the impaired metastasis seen in both the Stat3flx/flx/NIC mice and in the primary Stat3-null tumor cells may indicate a general requirement for Stat3 in tumor cell dissemination, establishment, and/or growth at a secondary site.
In this model, the metastatic defect was also correlated with impaired tumor angiogenesis. Solid tumors that are impaired in vascularization are often limited in their ability to efficiently metastasize (36). The Stat3-deficient tumors exhibited reduced expression of VEGF, which is considered a major mediator of the angiogenic process (36). Consistent with these observations, Stat3 expression corresponds with VEGF protein levels in a variety of transformed cell lines (37, 38). Additionally, downregulating Stat3 activity by siRNA or through expression of a dominant negative form causes a decrease in VEGF downstream of IL6 in cervical cancer cells, and Src and ErbB2 in human melanoma and breast cancer cells (39). More recently, expression of the activated mutant of Stat3, Stat3-Y705F, was positively correlated with microvessel density in human hepatocellular carcinoma tissue sections (14). Thus, the presence of Stat3 serves to promote the expression of VEGF and allow for tumor angiogenesis.
Whereas the above studies suggest that tumor cells are the principle source of VEGF, it has recently been reported that infiltrating tumor-associated macrophages can also contribute to VEGF production (40). It is thus interesting to note that gene expression profiling revealed a profound reduction in the expression of genes involved in both the inflammatory and angiogenic responses in the Stat3-deficient tumors. Given that Stat3, also known as the acute phase response factor (APRF), is a master regulator of this inflammation-based response (41), it is conceivable that impairment of inflammation through the downregulation of this network may compromise the metastatic capacity of these tumors. In the normal mammary gland, activation of Stat3 and C/EBPδ during involution leads to an inflammatory cascade that is characterized by the recruitment and activation of leukocytes, partly via the expression of the serum amyloid a genes (19, 42), and by the production of various growth factors (20, 31, 43, 44).
In tumors, the recruitment of inflammatory cells, such as macrophages, and release of growth factors, such as VEGF, can potentiate angiogenesis and metastasis (31, 40, 45). Indeed, the recruitment of macrophages was demonstrated to be a critical in promoting metastatic progression in the PyVmT mammary tumor transgenic model (40). Recently, mammary specific ablation of steroid receptor coactivator 1 (SRC-1) in the PyVmT tumor model also resulted in a block in tumor metastasis (46). Significantly, the SRC-1/Stat3 complex is involved in the transcriptional up-regulation of cebpd (47) (Fig. 5C). The direct transcriptional targets of C/EBPd include saa1 and saa2 both of which are involved in the mobilization of immune cells during normal mammary gland involution and may serve a similar role in the tumor microenvironment, effectively engaging pro-metastatic immune cells such as macrophages (42). Thus, this transcription regulatory network is strongly implicated as a critical modulator of the metastatic process via the dysregulation of the acute phase response, inflammation, and angiogenesis. The future development of therapeutics directed at suppressing this Stat3-dependent inflammatory cascade may be a promising treatment regime for metastatic breast cancers.
Supplementary Material
Acknowledgments
We would like to thank H. Lahlou and G. Ursini-Siegel for advice and discussions, T. Rao, R. Annan, and D. Dankort for critical reading of the manuscript, and B. Schade for help with microarray experiments. JJR and WJM conceived and designed the experiments and wrote the paper. JJR performed the experiments and analyzed the data. SS and MH processed and analyzed the microarray data. DL provided the Stat3flx mice. This work was supported by an NCIC/Terry Fox Foundation New Frontiers Program Project Team Grant # 017003. JJR is funded by a CIHR Canada Graduate Scholarships Doctoral Award. WJM is the recipient of a Canadian Research Chair in Molecular Oncology, McGill University.
References
- 1.Hsieh FC, Cheng G, Lin J. Evaluation of potential Stat3-regulated genes in human breast cancer. Biochemical and biophysical research communications. 2005;335(2):292–9. doi: 10.1016/j.bbrc.2005.07.075. [DOI] [PubMed] [Google Scholar]
- 2.Yeh YT, Ou-Yang F, Chen IF, et al. STAT3 ser727 phosphorylation and its association with negative estrogen receptor status in breast infiltrating ductal carcinoma. Int J Cancer. 2006;118(12):2943–7. doi: 10.1002/ijc.21771. [DOI] [PubMed] [Google Scholar]
- 3.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4(2):97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 4.Ren Z, Schaefer TS. ErbB-2 activates Stat3 alpha in a Src- and JAK2-dependent manner. The Journal of biological chemistry. 2002;277(41):38486–93. doi: 10.1074/jbc.M112438200. [DOI] [PubMed] [Google Scholar]
- 5.Garcia R, Yu CL, Hudnall A, et al. Constitutive activation of Stat3 in fibroblasts transformed by diverse oncoproteins and in breast carcinoma cells. Cell Growth Differ. 1997;8(12):1267–76. [PubMed] [Google Scholar]
- 6.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18(5):2553–8. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bromberg JF, Wrzeszczynska MH, Devgan G, et al. Stat3 as an oncogene. Cell. 1999;98(3):295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 8.Grandis JR, Drenning SD, Zeng Q, et al. Constitutive activation of Stat3 signaling abrogates apoptosis in squamous cell carcinogenesis in vivo. Proc Natl Acad Sci U S A. 2000;97(8):4227–32. doi: 10.1073/pnas.97.8.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sumita N, Bito T, Nakajima K, Nishigori C. Stat3 activation is required for cell proliferation and tumorigenesis but not for cell viability in cutaneous squamous cell carcinoma cell lines. Exp Dermatol. 2006;15(4):291–9. doi: 10.1111/j.0906-6705.2006.00407.x. [DOI] [PubMed] [Google Scholar]
- 10.Turkson J, Ryan D, Kim JS, et al. Phosphotyrosyl peptides block Stat3-mediated DNA binding activity, gene regulation, and cell transformation. J Biol Chem. 2001;276(48):45443–55. doi: 10.1074/jbc.M107527200. [DOI] [PubMed] [Google Scholar]
- 11.Zhang YW, Wang LM, Jove R, Vande Woude GF. Requirement of Stat3 signaling for HGF/SF-Met mediated tumorigenesis. Oncogene. 2002;21(2):217–26. doi: 10.1038/sj.onc.1205004. [DOI] [PubMed] [Google Scholar]
- 12.Burke WM, Jin X, Lin HJ, et al. Inhibition of constitutively active Stat3 suppresses growth of human ovarian and breast cancer cells. Oncogene. 2001;20(55):7925–34. doi: 10.1038/sj.onc.1204990. [DOI] [PubMed] [Google Scholar]
- 13.Al Zaid Siddiquee K, Turkson J. STAT3 as a target for inducing apoptosis in solid and hematological tumors. Cell Res. 2008;18(2):254–67. doi: 10.1038/cr.2008.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang SF, Wang SN, Wu CF, et al. Altered p-STAT3 (tyr705) expression is associated with histological grading and intratumour microvessel density in hepatocellular carcinoma. J Clin Pathol. 2007;60(6):642–8. doi: 10.1136/jcp.2006.036970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma XT, Wang S, Ye YJ, Du RY, Cui ZR, Somsouk M. Constitutive activation of Stat3 signaling pathway in human colorectal carcinoma. World J Gastroenterol. 2004;10(11):1569–73. doi: 10.3748/wjg.v10.i11.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdulghani J, Gu L, Dagvadorj A, et al. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol. 2008;172(6):1717–28. doi: 10.2353/ajpath.2008.071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raz R, Lee CK, Cannizzaro LA, d’Eustachio P, Levy DE. Essential role of STAT3 for embryonic stem cell pluripotency. Proc Natl Acad Sci U S A. 1999;96(6):2846–51. doi: 10.1073/pnas.96.6.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ursini-Siegel J, Hardy WR, Zuo D, et al. ShcA signalling is essential for tumour progression in mouse models of human breast cancer. The EMBO journal. 2008;27(6):910–20. doi: 10.1038/emboj.2008.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suffredini AF, Fantuzzi G, Badolato R, Oppenheim JJ, O’Grady NP. New insights into the biology of the acute phase response. Journal of clinical immunology. 1999;19(4):203–14. doi: 10.1023/a:1020563913045. [DOI] [PubMed] [Google Scholar]
- 20.Humphreys RC, Bierie B, Zhao L, Raz R, Levy D, Hennighausen L. Deletion of Stat3 blocks mammary gland involution and extends functional competence of the secretory epithelium in the absence of lactogenic stimuli. Endocrinology. 2002;143(9):3641–50. doi: 10.1210/en.2002-220224. [DOI] [PubMed] [Google Scholar]
- 21.Finak G, Godin N, Hallett M, et al. BIAS: Bioinformatics Integrated Application Software. Bioinformatics (Oxford, England) 2005;21(8):1745–6. doi: 10.1093/bioinformatics/bti170. [DOI] [PubMed] [Google Scholar]
- 22.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford, England) 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 23.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods (San Diego, Calif) 2003;31(4):265–73. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 24.Yang YH, Buckley MJ, Speed TP. Analysis of cDNA microarray images. Briefings in bioinformatics. 2001;2(4):341–9. doi: 10.1093/bib/2.4.341. [DOI] [PubMed] [Google Scholar]
- 25.Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics (Oxford, England) 2006;22(22):2825–7. doi: 10.1093/bioinformatics/btl476. [DOI] [PubMed] [Google Scholar]
- 26.White DE, Kurpios NA, Zuo D, et al. Targeted disruption of beta1-integrin in a transgenic mouse model of human breast cancer reveals an essential role in mammary tumor induction. Cancer cell. 2004;6(2):159–70. doi: 10.1016/j.ccr.2004.06.025. [DOI] [PubMed] [Google Scholar]
- 27.Kijima T, Niwa H, Steinman RA, et al. STAT3 activation abrogates growth factor dependence and contributes to head and neck squamous cell carcinoma tumor growth in vivo. Cell Growth Differ. 2002;13(8):355–62. [PubMed] [Google Scholar]
- 28.Cantwell CA, Sterneck E, Johnson PF. Interleukin-6-specific activation of the C/EBPdelta gene in hepatocytes is mediated by Stat3 and Sp1. Mol Cell Biol. 1998;18(4):2108–17. doi: 10.1128/mcb.18.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clarkson RW, Boland MP, Kritikou EA, et al. The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development. Molecular endocrinology (Baltimore, Md) 2006;20(3):675–85. doi: 10.1210/me.2005-0392. [DOI] [PubMed] [Google Scholar]
- 30.Ray A, Ray BK. Serum amyloid A gene expression under acute-phase conditions involves participation of inducible C/EBP-beta and C/EBP-delta and their activation by phosphorylation. Mol Cell Biol. 1994;14(6):4324–32. doi: 10.1128/mcb.14.6.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stein T, Morris JS, Davies CR, et al. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6(2):R75–91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diaz N, Minton S, Cox C, et al. Activation of stat3 in primary tumors from high-risk breast cancer patients is associated with elevated levels of activated SRC and survivin expression. Clin Cancer Res. 2006;12(1):20–8. doi: 10.1158/1078-0432.CCR-04-1749. [DOI] [PubMed] [Google Scholar]
- 33.Xie TX, Wei D, Liu M, et al. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23(20):3550–60. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 34.Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251(2):199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 35.Dechow TN, Pedranzini L, Leitch A, et al. Requirement of matrix metalloproteinase-9 for the transformation of human mammary epithelial cells by Stat3-C. Proc Natl Acad Sci U S A. 2004;101(29):10602–7. doi: 10.1073/pnas.0404100101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carmeliet P. VEGF as a key mediator of angiogenesis in cancer. Oncology. 2005;69(Suppl 3):4–10. doi: 10.1159/000088478. [DOI] [PubMed] [Google Scholar]
- 37.Niu G, Wright KL, Huang M, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21(13):2000–8. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 38.Xu Q, Briggs J, Park S, et al. Targeting Stat3 blocks both HIF-1 and VEGF expression induced by multiple oncogenic growth signaling pathways. Oncogene. 2005;24(36):5552–60. doi: 10.1038/sj.onc.1208719. [DOI] [PubMed] [Google Scholar]
- 39.Wei LH, Kuo ML, Chen CA, et al. Interleukin-6 promotes cervical tumor growth by VEGF-dependent angiogenesis via a STAT3 pathway. Oncogene. 2003;22(10):1517–27. doi: 10.1038/sj.onc.1206226. [DOI] [PubMed] [Google Scholar]
- 40.Lin EY, Li JF, Gnatovskiy L, et al. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 2006;66(23):11238–46. doi: 10.1158/0008-5472.CAN-06-1278. [DOI] [PubMed] [Google Scholar]
- 41.Alonzi T, Maritano D, Gorgoni B, Rizzuto G, Libert C, Poli V. Essential role of STAT3 in the control of the acute-phase response as revealed by inducible gene inactivation [correction of activation] in the liver. Mol Cell Biol. 2001;21(5):1621–32. doi: 10.1128/MCB.21.5.1621-1632.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pensa S, Watson C, Poli V. Stat3 and the Inflammation/Acute Phase Response in Involution and Breast Cancer. J Mammary Gland Biol Neoplasia. 2009:1–9. doi: 10.1007/s10911-009-9124-x. [DOI] [PubMed] [Google Scholar]
- 43.Thangaraju M, Rudelius M, Bierie B, et al. C/EBPdelta is a crucial regulator of pro-apoptotic gene expression during mammary gland involution. Development (Cambridge, England) 2005;132(21):4675–85. doi: 10.1242/dev.02050. [DOI] [PubMed] [Google Scholar]
- 44.Watson CJ. Immune cell regulators in mouse mammary development and involution. Journal of animal science. 2008 doi: 10.2527/jas.2008-1333. [DOI] [PubMed] [Google Scholar]
- 45.McDaniel SM, Rumer KK, Biroc SL, et al. Remodeling of the mammary microenvironment after lactation promotes breast tumor cell metastasis. Am J Pathol. 2006;168(2):608–20. doi: 10.2353/ajpath.2006.050677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang S, Yuan Y, Liao L, et al. Disruption of the SRC-1 gene in mice suppresses breast cancer metastasis without affecting primary tumor formation. Proc Natl Acad Sci U S A. 2009;106(1):151–6. doi: 10.1073/pnas.0808703105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Sif S, DeWille J. The mouse C/EBPdelta gene promoter is regulated by STAT3 and Sp1 transcriptional activators, chromatin remodeling and c-Myc repression. Journal of cellular biochemistry. 2007;102(5):1256–70. doi: 10.1002/jcb.21356. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.