Abstract
Protein kinases can adopt multiple protein conformations depending on their activation status. Recently, in drug discovery, a paradigm shift has been initiated, moving from inhibition of fully activated, phosphorylated kinases to targeting the inactive, unphosphorylated forms. For identification and characterization of putative inhibitors, also interacting with the latent kinase conformation outside of the kinase domain, highly purified and homogeneous protein preparations of unphosphorylated kinases are essential. The kinetic parameters of nonphosphorylated kinases cannot be assessed easily by standard kinase enzyme assays as a result of their intrinsic autophosphorylation activity. Kinetic binding rate constants of inhibitor-protein interactions can be measured by biophysical means upon protein immobilization on chips. Protein immobilization can be achieved under mild conditions by binding biotinylated proteins to streptavidin-coated chips, exploiting the strong and highly specific streptavidin–biotin interaction. In the work reported here, the cytoplasmic domains of insulin receptor and insulin-like growth factor-1 receptor fused to a biotin ligase recognition sequence were coexpressed individually with the phosphatase YopH and the biotin-protein ligase BirA upon triple infection in insect cells. Tandem affinity purification yielded pure cytoplasmic kinase domains as judged by gel electrophoresis and HPLC. Liquid chromatography-mass spectrometry analysis showed the absence of any protein phosphorylation. Coexpression of BirA led to quantitative and site-specific biotinylation of the kinases, which had no influence on the catalytic activity of the kinases, as demonstrated by the identical phosphorylation pattern upon autoactivation and by enzymatic assay. This coexpression approach should be applicable to other protein kinases as well and should greatly facilitate the production of protein kinases in their phosphorylated and unphosphorylated state suitable for enzymatic and biophysical studies.
Keywords: biotin, phosphatase, phosphorylation
INTRODUCTION
Protein kinases constitute a major fraction of the current drug targets,1 and numerous small molecule kinase inhibitors are currently in clinical trials for treatment of various human diseases, including cancer.2,3 Most protein kinases progress from their latent to their active state by phosphorylation through other kinases or by autophosphorylation and are able to adopt different conformations in each state.4,5
The majority of inhibitors on the market is directed against the active kinase conformation, and the inhibitors were identified and characterized using phosphorylated recombinant kinase domains. Recently, the emergence of kinase inhibitor resistance in the clinic and the hope for gaining better selectivity have shifted the drug discovery focus from targeting the active, fully phosphorylated protein to targeting the latent, nonphosphorylated protein and binding to allosteric sites outside of the conserved ATP-binding pocket, inhibiting the kinase through a non-ATP competitive mechanism.6–8 The protein production strategies to supply protein for initial inhibitor screening and secondary assays have to reflect this new development. When aiming at the identification of inhibitors binding to the latent protein, the challenge is to provide sufficient amounts of high-quality, nonphosphorylated, full cytoplasmic domains of kinases for screening and secondary assays. As a result of autophosphorylation, the kinetic parameters of nonphosphorylated kinases are not easy to measure, and thus, a repertoire of secondary assays comprising biophysical methods, such as surface plasmon resonance (SPR), can be helpful. For gentle binding to solid surfaces, the protein can be modified by an affinity tag, and amongst others, biotin labeling has often been used.
Using the insulin receptor (IR) family as model proteins, the purpose of the work reported here was to develop a system for the production of unphosphorylated and simultaneously, site-specifically, biotinylated receptor tyrosine kinases. IR and insulin-like growth factor-1 receptor (IGF1R) are usually considered as important regulators of carbohydrate metabolism but have also been implicated in neoplastic transformation of cells. In human cells, multiple tyrosine phosphorylation sites have each been reported for human IGF1R and IR.9–13 Screening for and characterization of inhibitors binding to the latent form of kinases require the production of unphosphorylated proteins. Although protein kinase domains differing in their phosphorylation status can be separated by conventional protein purification methods,14 this does not seem feasible for large quantities of full-length cytoplasmic domains with a complex phosphorylation pattern and limited amounts of each phosphorylated form. There are currently two major approaches to generating unphosphorylated protein kinases. One approach is to express kinases in Escherichia coli expression systems, which nevertheless, might in case of autophosphorylation, produce phosphorylated protein,15 and proteins are prone to aggregation, even in the presence of chaperones.16 Also, toxic effects of kinases have been observed in E. coli.17 An alternative approach is to express recombinant kinases in the baculovirus expression system and treat them with recombinant phosphatases during or after purification. However, this process is tedious and often needs additional purification steps. In addition, dephosphorylation is often incomplete, and protein yields might be low as a result of protein aggregation.18,19 Similar issues have been reported for in vitro biotinylation of proteins, as the level of biotinylation cannot be controlled easily, and nonspecific and over-biotinylation often causes loss of protein by precipitation or reduces protein activity.20,21 To overcome the limitations of in vitro protein modification, dephosphorylation and biotinylation have been achieved successfully in vivo by coexpression of phosphatases or biotin ligase, respectively.18,19,22
To produce large quantities of unphosphorylated and biotinylated protein in the work reported here, we expanded and optimized the expression approach for the simultaneous expression of phosphatase and biotin ligase, yielding homogeneous, completely dephosphorylated and site-specifically biotinylated kinases.
MATERIALS AND METHODS
Cloning
A polyhedrin promoter-GST fragment was PCR-amplified from pFastBacGST2 (Invitrogen, Basel, Switzerland) with upstream and downstream primers spanning Bst1107I and EcoRI restriction sites, respectively. The downstream primer introduced a PreScission protease recognition site. This fragment was subcloned via Bst1107I/EcoRI into pFastBac.
The cDNA encoding for the entire cytoplasmic domain of human IGF1R (Genbank NM_000875) from aa 960 to 1367 and the cDNA encoding for the entire cytoplasmic domain of human IR (Genbank NM_000208) from aa 980 to 1382 were amplified by PCR using existing, in-house expression plasmids as template.23 The downstream primers introduced a C-terminal 6× His-tag, and the products were subcloned via EcoRI/KpnI into the formerly generated pFastBacGST2-PreScission vector. For in vivo biotinylation, the recognition sequence (AviTag)24 for the BirA biotin-protein ligase was inserted in-frame as a double-stranded oligonucleotide into the unique EcoRI site.
Sequences of all plasmids were confirmed by sequencing (Solvias AG, Basel, Switzerland).
Recombinant baculovirus was generated with the Bac-to-Bac system (Invitrogen), following the manufacturer's instructions. The integrity of the gene of interest in the virus was confirmed by sequencing of viral genomic DNA (Solvias AG).
The baculovirus stock was amplified once and frozen following the Titerless Infected-Cells Preservation and Scale-up protocol (TIPS), as described.25 In brief, 1 × 108 Spodoptera frugiperda (Sf9) cells, in 100 ml ExCell 420 medium (JRH Biosciences, Lenexa, KS, USA) in an Erlenmeyer flask (300 ml), were infected with the virus of interest at a multiplicity of infection (MOI) = 3. After 24 h at 27°C on a shaker (130 rpm), the culture was transferred to two 50 ml tubes and centrifuged at 100 g for 10 min at room temperature. The cells were resuspended to 1 × 107 cells/ml in freezing medium [ExCell420 medium, 0.5×streptomycin/penicillin (Sigma-Aldrich, Switzerland), 10 g/l BSA, 10% DMSO]. Aliquots of 0.5 ml were transferred to 1.8 ml cryotubes and frozen in a Nalgene Cryo 1°C freezing container overnight at −80°C. An aliquot of frozen cells was diluted 1:100 and used for inoculation of expression cultures.
Protein Expression and Purification
Proteins were expressed in Sf9 cells in ExCell420 medium containing 0.5× streptomycin/penicillin solution, with 10 ml TIPS stock diluted 1:100 (1×106 cells, corresponding to MOI=0.1)/liter expression culture (1×109 cells) in 3–l shake flasks (expression volume, 1 liter; 120 rpm) or BioWave bioreactors (expression volume, 10 liters; 20 rpm; aeration 30 l/h; O2=30%) for 72 h at 27°C. Cells were harvested by centrifugation at 800 g and stored at −80°C.
A baculovirus vector containing the phosphatase YopH in pACMP3-GW and a baculovirus expressing biotin-protein ligase BirA were obtained from Janet Sim (Novartis Institutes for BioMedical Research, Emeryville, CA, USA) and Shari Caplan (Novartis Institutes for BioMedical Research, Cambridge, MA, USA), respectively. YopH and BirA were coexpressed with IGF1R and IR as TIPS using 10 ml/l and 2.5 ml/l diluted TIPS stock, respectively. For BirA coexpression, D-biotin (50 mM stock solution in 0.1 N NaOH) was added to 4 μM final concentration.
Cells from a 1-l expression aliquot were resuspended in 120 ml lysis buffer [50 mM Tris · HCl, pH 8.0 (25°C), 150 mM NaCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP), 10% glycerol, 10 μg/ml E-64 (Peptides International, Louisville, KY, USA; Buffer A)], supplemented with 1% Triton X-100, 4.8 ml protease inhibitor complete without EDTA (25×conc., Roche Diagnostics, Rotkreuz, Switzerland), and 60 μl Benzonase (25 U/μl, Merck Biosciences, Novagen, Darmstadt, Germany), and gently rocked for 30 min at 4°C. After sonication with a Branson sonifier 250 (2×30 s pulse, 1 min on ice, duty cycle 50%, output control 4–5), cell debris was precipitated by two consecutive centrifugation steps for 30 min at 4°C with 19,700 g and 47,800 g, respectively. The soluble fraction was filtered through 5 μM (PALL, Port Washington, NY, USA), 1.2 μM (PALL), and 0.45 μM Durapore filters (Millipore, Bedford, MA, USA) and incubated on a rotator with 4 ml glutathione sepharose 4B (GE Healthcare, Glattbrugg, Switzerland, Cat. No. 27-4574-01) for 4 h at 4°C. The glutathione sepharose was washed five times with 20 ml Buffer A, supplemented with 0.01% Triton X-100 at 4°C, and bound protein eluted four times for 10 min with Buffer A, supplemented with 0.01% Triton X-100 and 10 mM reduced L-glutathione. The pooled fractions were incubated with 200 units (2 U/μl) PreScission protease (GE Healthcare, Glattbrugg, Switzerland) during dialysis in 3 l Buffer A (0.01% Triton X-100) in Slide-A-Lyzer 3.5K dialysis cassettes (3500 MWCO, Thermo Scientific Pierce, Zug, Switzerland) overnight at 4°C.
After cleavage, the protein was loaded with an Aekta fast protein liquid chromatography (LC; GE Healthcare, Glattbrugg, Switzerland) onto a His-Trap HP 5-ml column (GE Healthcare, Cat. No. 17-5248-02), which was equilibrated with Buffer A (0.01% Triton X-100, 5 mM imidazole). After protein binding, the column was washed with 10 column volumes (CVs) Buffer A and then re-equilibrated with Buffer B (50 mM Hepes, 10% glycerol, 1 mM TCEP, 5 μg/ml E-64, supplemented with 5 mM imidazole), followed by another wash of 10 CVs. Bound protein was eluted with Buffer B, supplemented with 500 mM imidazole. Protein was aliquoted and snap-frozen in dry ice/ethanol.
Protein concentrations were determined by a modified Bradford method, as described,26 with a Bradford kit (BioRad, Reinach, Switzerland) in microtiter plates and albumin as a standard. For SDS-PAGE analysis, protein samples were heat-denatured in the presence of 1× lithium dodecyl sulfate sample buffer and reducing agent (Invitrogen) and loaded onto NuPAGE Bis-Tris 4–12% Novex gels (Invitrogen), which were stained with GelCode blue stain reagent (Thermo Scientific Pierce). Protein quality was assessed by HPLC and mass spectroscopy (MS).
Western Blotting
For Western blot analyses, the cells were lysed in 1× lysis buffer [50 mM Tris · HCl, pH 7.5 (Sigma-Aldrich), 1% Triton X-100 (Fluka, Buchs, Switzerland), 150 mM NaCl, 2 mM EDTA, 1 mM DTT, 1 mM activated Na3VO4 (Sigma-Aldrich), 10% glycerol, 1×protease inhibitor Complete (Roche Diagnostics), and Benzonase (2.5 μl/ml lysis buffer, VWR International, Dietikon, Switzerland)].
Cells from 1 ml expression culture were harvested by centrifugation (500 g), and the cell pellets were resuspended in 100 μl lysis buffer and incubated with gentle shaking for 30 min at room temperature (=total protein). Half of the sample was centrifuged for 5 min at 14,000 rpm to precipitate cell debris (=insoluble fraction) from supernatant (=soluble fraction). All samples were stored at −80°C until use.
For Western blotting, the proteins were transferred onto nitrocellulose membranes with an Xcell Blot module (Invitrogen). The proteins were detected with specific antibodies [Phospho-IGF1R (Tyr1161)/IR (Tyr1185; Cell Signaling Technologies, Danvers, MA, USA, Cat. No. 3021, 1:1000); IGF1Rβ (C-20; Santa Cruz Biotechnology, Heidelberg, Germany, Cat. No. Sc-713, 1:1000); and IRβ (C-19; Santa Cruz Biotechnology, Cat. No. Sc-711, 1:2000)]. The rabbit primary antibodies were detected by IRDye 700DX-conjugated, affinity-purified goat anti-rabbit IgG (Rockland Immunochemicals, Gilbertsville, PA, USA), diluted 1:20,000 (in Rockland blocking buffer), and membranes were scanned on an Odyssey fluorescence near infrared imager (Li-Cor, Bad Homburg, Germany).
HPLC-Tandem MS (MS/MS)
All chemicals were from Sigma-Aldrich, if not stated otherwise. Proteins were diluted at 2 μM in Hepes buffer, 50 mM, pH 7.2 (Gibco, Grand Island, NY, USA), 1 mM DTT, 1 mM MnCl2, and 10 mM MgCl2, in the presence or absence of 5 mM ATP (Sigma-Aldrich), in a final volume of 40 μl. Reactions were stopped with 40 μl 0.5 M EDTA and put in ice until LC-MS analysis. Separation of proteins was performed on a HP1100 HPLC system (Hewlett Packard, Palo Alto, CA, USA), using a 1-mm × 150-mm column packed with POROS R1/H (Perseptive Biosystems, Foster City, CA, USA). The column was kept at 80°C. Each sample (25 μl) was injected onto the column using a CTC PAL autosampler (CTC, Zwingen, Switzerland) fitted with a Valco model C6UW HPLC valve (Valco, Houston, TX, USA) and a 25-μl injection loop. HPLC was controlled by MassLynx software (Waters, Manchester, UK). UV detection was performed at λ = 214 nm. Eluent A was water containing 0.05% trifluoroacetic acid (TFA). Eluent B was a 1:9 mixture of water:acetonitrile containing 0.045% TFA. A gradient from 20% B to 90% B was run in 12 min. The flow rate was typically 80 μl/min. The total flow from the LC system was introduced into the UV detection cell prior to introduction in the electrospray ionization (ESI) source. MS was carried out using a quadrupole time-of-flight (Waters) hybrid MS/MS equipped with a Waters Z-type ESI source.
Enzymatic Activity Assay
The catalytic activity of purified IGF1R and IR was determined by a radiometric assay as described.27 In brief, the AviTag-IGF1R, AviTag-IR, IGF1R, and IR (1.3 mM) proteins were incubated for 15 min in reaction buffer (50 mM Hepes, pH 7.4, 1 mM MnCl2, 10 mM MgCl2, 1 mM DTT, 0.01% Brij35) in the presence of 5 mM ATP. The autophosphorylation reactions were diluted into reaction buffer containing DMSO and incubated for 30 min. The peptide (Ac-EQEDEPEGDYFEWLE-NH2, 15 μM) and 33γP-ATP (0.003 μCi/μl) were added, and the reactions were run for 10 min [final concentrations: (ATP)=6 μM; (enzyme)=2 nM; (DMSO)=3%]. The reactions were stopped by the addition of stop solution (200 mM phosphoric acid) and transferred onto preactivated filter plates (96-well MultiScreen plates, Millipore). The plates were washed twice with 0.5% phosphoric acid and dried at room temperature. Microsint 40 (Perkin Elmer, Wellesley, MA, USA) was dispensed, and the bound radioactivity was counted in a TopCount NXT (Hewlett Packard).27
SPR Assay
Neutravidin (Thermo Scientific Pierce) was immobilized on the surface of a carboxymethylated dextran matrix (CM5) sensor chip (GE Healthcare) by the standard amine-coupling procedure. All BIAcore experiments were done at a flow rate of 10 μl/min on a BIAcore T100 machine. Buffer of this reaction was: PBS 1× pH 7.5, TCEP 1 mM, and Tween 20 0.05%. Reagents were injected in the following order: 0.05 M N-hydroxysuccinimide/0.2 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide mixture and 1 M ethanolamine (pH 8.5; all GE Healthcare) and neutravidin (45 μg/ml), diluted in buffer (1×). The amount of immobilized neutravidin was approximately 20,000 resonance units. The biotinylated AviTag-IGF1R and the untagged IGF1R were diluted in buffer 1× at a final concentration of 300 nM (15 and 14 μg/ml) and injected over the neutravidin surface during 60 s at a flow rate of 10 μl/min.
RESULTS
Small molecular weight protein kinase inhibitors are a well-established treatment regimen for several cancers. Recently, the emergence of resistance in the clinic and the lack of strong selectivity causing undesired side-effects have triggered the development of new classes of kinase inhibitors targeting the latent conformation or binding in a non-ATP competitive manner to allosteric sites outside of the ATP-binding pocket. The enzymatic and biophysical characterization of the new inhibitor classes sets high standards in terms of protein quantity and quality. Protein kinases isolated from eukaryotic expression systems are usually phosphorylated and as such, in their active conformation, not suitable for the characterization of compounds binding to the latent conformation. Consequently, they have to be dephosphorylated prior to use. In addition, labeling with an affinity tag, e.g., biotin, would facilitate SPR studies. Although dephosphorylation and biotinylation can be achieved in vitro, both methods have some disadvantages. In vitro dephosphorylation is often incomplete, the protein might aggregate, leading to reduced yields, and additional purification steps are required.18,19 Similar issues are known for in vitro biotinylation reactions, as the level of biotinylation cannot be controlled, and nonspecific and over-biotinylation can cause loss of protein by precipitation or reduces protein activity.20,21
To overcome the limitations of the in vitro reactions, we aimed at achieving both protein modifications in a more physiological way by enzymatic reactions in vivo in the baculovirus expression system. The model proteins IGF1R and IR, respectively, were expressed simultaneously with the phosphatase YopH and the biotin-protein ligase BirA, aiming at modifying the target proteins concomitantly during expression.
The full-length cytoplasmic domains of IGF1R (960–1367) and IR (980–1382) were cloned into pFastBac vectors and expressed in the baculovirus system following the TIPS protocol.25 Both proteins were expressed as fusion proteins with a C-terminal His-tag and an N-terminal GST-tag with an adjacent PreScission protease cleavage site. The double-tagging allowed for an efficient tandem-affinity purification strategy via glutathione sepharose, removal of the GST-tag by PreScission protease cleavage, and Ni-affinity purification, which yielded the full-length cytoplasmic domains of IGF1R and IR.
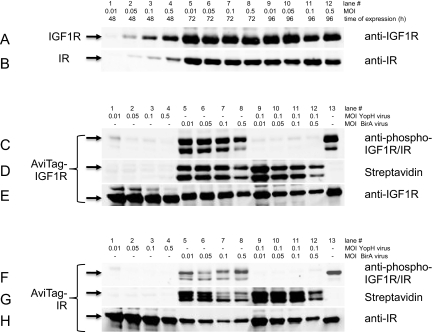
Optimization of MOI and time of expression revealed good expression levels for both proteins after 72 h at all virus levels tested, as judged by Western blotting with anti-IGF1R and IR-specific antibodies. The MOI = 0.01 corresponds to 0.1 ml baculovirus-infected insect cells (BIIC)/100 ml expression volume. Expression levels of IGF1R and IR did not improve with longer expression time (96 h), and thus, the parameters for all future protein expression experiments were set to MOI = 0.1 (1 ml BIIC from TIPS/100 ml expression volume) and 72 h (Fig. 1A and B).
FIGURE 1.
Expression optimization: Western blotting of cell extracts from Sf9 BIIC. Time course and virus titration for expression of IGF1R (A) and IR (B). For all coexpression experiments, the MOI IGF1R and IR was kept at MOI = 0.1. (C) Phospho-AviTag-IGF1R Western blot detection upon YopH and BirA virus titration. (D) Visualization of biotin-labeled AviTag-IGF1R by fluorescence-labeled streptavidin after virus titration as in C. (E) AviTag-IGF1R detection by Western blotting as in C and D. (F) Phospho-AviTag-IR detection as in C. (G) Detection of biotinylation of AviTag-IR as in D. (H) Western blotting of AviTag-IR.
The strong signal with phospho-specific antibodies of cell lysates indicated that the untreated, full-length cytoplasmic domains expressed in insect cells are phosphorylated (Fig. 1C and F, lane 13). Aiming at producing unphosphorylated proteins, the multi-specific tyrosine phosphatase YopH28 was coexpressed from a second virus. YopH from the pathogen Yersinia pseudotuberculosis29,30 has been coexpressed successfully in bacteria for dephosphorylation of Abl and Src31 and in the baculovirus-expression system to act on Bruton's tyrosine kinase, Janus Kinase 3, and EPH receptor A2.18 As shown in Figure 1 for the AviTag-IGF1R and IR constructs, coexpression of YopH from a second virus added at a very low MOI already reduced the strong tyrosine phosphorylation of both proteins to levels almost nondetectable by Western blotting with a phospho-IGF1R (Y1161)/phospho-IR (Y1185) dual-specific antibody (Fig. 1C and F, lanes 1–4 and 13).
The intensity differences of the phospho-specific signal for IGF1R (Fig. 1C, lanes 5–8 and 13) and IR (Fig. 1F, lanes 5–8 and 13) in the absence of phosphatase might stem from the dual-specific antibody having a different affinity to each of the two proteins. The absence of a phospho-specific signal upon YopH coexpression demonstrates an efficient dephosphorylation in the cell. There was also no difference in terms of dephosphorylation efficiency between double and triple infection (Fig. 1C and F, lanes 1–4 and 9–12).
A MOI = 0.1 was chosen for all future YopH coexpression experiments. The good dephosphorylation efficiency during coexpression, as demonstrated here, may not only be a result of the dephosphorylation as such but might also be attributed to the prevention of autophosphorylation by keeping the activation status of the kinases below a critical threshold, rendering them inactive.
Ideally, for the purpose of characterization by SPR, not only dephosphorylation but also biotinylation, enabling gentle immobilization of the target protein to solid surfaces, should be achieved. Thus, we asked the question of whether it would be feasible to dephosphorylate and site-specifically biotinylate the proteins at the same time, and for this purpose, we have expanded the concomitant protein expression and dephosphorylation by an enzymatic biotinylation step in vivo, which makes the purified proteins readily suitable for assays using biotin for immobilization or labeling.
The coexpression of BirA with IGF1R and IR led to a strong biotinylation of both receptor tyrosine kinases, as shown by Western blotting with a fluorescent-labeled streptavidin (Fig. 1D and G, lanes 5–8). Best results were obtained with a MOI = 0.01 and 0.05, and a MOI = 0.025 was selected for future expressions. The same strong level of biotinylation could be achieved when YopH was coexpressed with IGF1R/IR and BirA (Fig. 1D and G, lanes 9–12). Although biotin-dependent enzymes are ubiquitous in nature, biotinylation is a relatively rare modification, and only a few biotinylated protein species are found in different organisms.32 Accordingly, the biotin-protein ligase BirA used here belongs to an enzyme family that catalyzes a reaction of high specificity, usually yielding protein with a single biotin moiety attached.33 As the degree of in vivo biotinylation by coexpression of BirA depends on the concentration of D-biotin, and exogenous D-biotin can drive the enzymatic reaction,34,35 D-biotin was added to the expression medium at a final concentration of 4 μM.
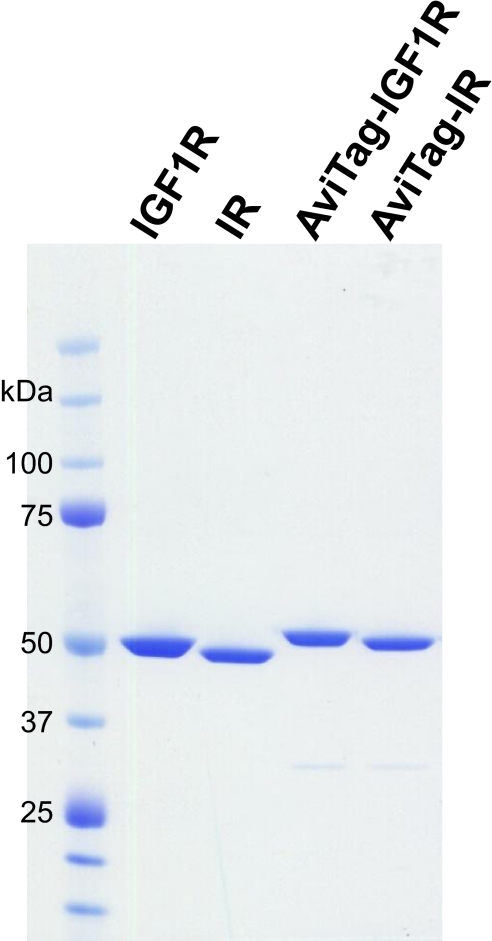
The triple-infection approach yielded up to a 30-mg electrophoretically homogeneous protein/liter baculovirus expression (Fig. 2) at a concentration of 2 mg/ml. With some batch-to-batch variability, the yield of pure protein was about 2% of the total protein lysate, which corresponds to approximately 8 pg-purified protein/cell. HPLC analysis of the tandem affinity-purified proteins showed that the amount of impurities for all proteins was below the detection limit of the method, and thus, the protein purity was determined as >95% (Fig. 3 insets). As expected, too high virus levels impaired the yield of recombinant protein (Fig. 1E and H, lanes 5–8 and 9–12).
FIGURE 2.
Stained gel of purified proteins. The four purified proteins used in this study were loaded (2 μg each) onto a NuPAGE Bis-Tris 4–12% Novex gel (Invitrogen) and stained with GelCode blue stain reagent (Thermo Scientific Pierce).
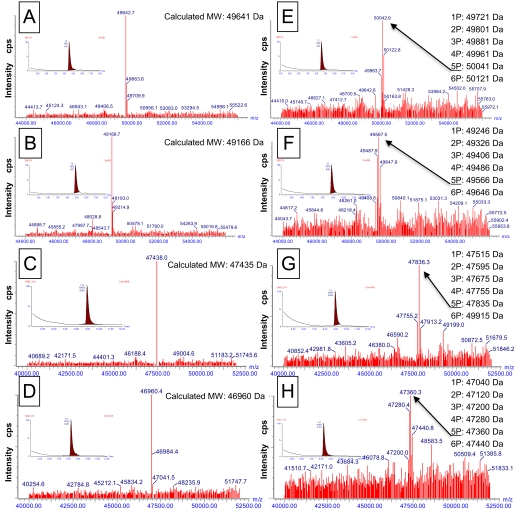
FIGURE 3.
HPLC and MS analysis of purified IGF1R and IR cytoplasmic domains. Unphosphorylated and biotinylated or nonbiotinylated proteins were analyzed prior to and after activation. Arrows indicate proteins with five phosphates. (A) Biotin-IGF1R; (B) biotin-IR; (C) IGF1R; (D) IR; (E) biotin-IGF1R-autophosphorylated; (F) biotin-IR-autophosphorylated; (G) IGF1R-autophosphorylated; (H) IR-autophosphorylated. (Insets) HPLC chromatogram.
The biotinylation as well as the phosphorylation status of the proteins were determined by LC-MS. As shown in Figure 3, the presence of a single peak in the LC-MS spectra of dephosphorylated IGF1R and IR from all constructs confirmed the quantitative dephosphorylation of the proteins by coexpression of YopH (Fig. 3A–D). The single peak of AviTagged but nonactivated IGF1R and IR showed that the proteins were quantitatively biotinylated. The molecular weight shift of 225 Da of the AviTagged IGF1R and IR confirms the presence of a single biotin molecule/protein, which was attached specifically by the biotin-protein ligase BirA (Fig. 3C and D). After activation, the LC-MS spectra revealed a step-wise molecular weight increase with a multiple of 80 Da, corresponding to addition of phosphates to the proteins, and the majority of molecules has four, five, and six phosphates attached (Fig. 3B, D, F, and H).
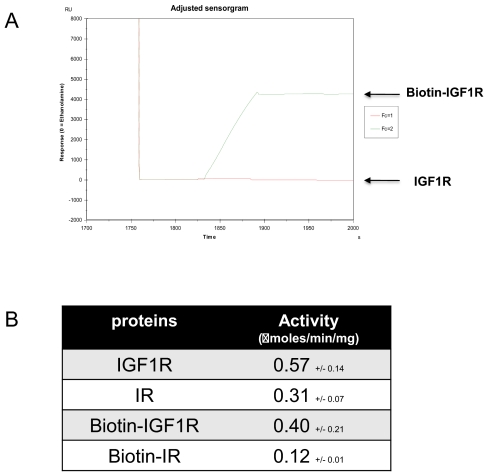
Nontagged, nonbiotinylated and AviTagged, biotinylated proteins could be activated equally well by ATP and showed consistently the addition of four, five, and six phosphates upon activation (Fig. 3B, D, F, and H). Although the autoactivation and phosphorylation pattern indicated that the purified proteins were active, the catalytic activity of phosphorylated IGF1R and IR was assessed by an enzyme assay with an exogenous peptidic substrate (Ac-EQEDEPEGDYFEWLE-NH2) prior to and after activation by incubation with 5 mM ATP for 15 min.27 Biotinylation had no or very little impact on enzyme activity, as the catalytic activity of biotinylated IGF1R and IR was 0.4 (nonbiotinylated 0.57) and 0.12 (nonbiotinylated 0.31) μmoles/min/mg, respectively (Fig. 4B). This indicated that neither the presence of the AviTag nor the biotin attached to it had any negative effect on the catalytic activity of the enzymes, and the modification of the enzymes, as applied here, renders them suitable for in-depth characterization of their enzymatic properties.
FIGURE 4.
Functional analysis of purified IGF1R and IR cytoplasmic domains. (A) The accessibility of the biotin for immobilization was assessed by binding of untagged and biotinylated AviTag-IGF1R to neutravidin-coated CM5 chips on a Biacore T100. (B) The enzyme activity in μmoles/min/mg protein was determined as described.
The accessibility of the attached biotin for binding to a neutravidin-coated solid surface was proven by a SPR assay of biotinylated AviTag-IGF1R and nontagged IGF1R as control (Fig. 4A). The functional characterization by SPR experiments revealed that the biotin attached to the AviTag fused to both target proteins was accessible and well suited for high-affinity binding to neutravidin-coated chips.
DISCUSSION
In conclusion, not only complete dephosphorylation but also quantitative biotinylation could be achieved, and the triple infection approach is a convenient and fast means to obtain unphosphorylated and quantitatively biotinylated protein kinase cytoplasmic domains as described here, and initial results in our laboratory indicate that it might also be applicable to other protein families that are then suitable for a whole variety of biochemical assays.
ACKNOWLEDGMENTS
We are grateful to J. Sim (Novartis Institutes for BioMedical Research, Emeryville, CA, USA) and S. Caplan (Novartis Institutes for BioMedical Research, Cambridge, MA, USA) for providing plasmids and baculoviruses of YopH and BirA, respectively. We thank S. Rieffel for BioWave fermentation and F. Bitsch and P. Brunet Lefeuvre (Novartis Institutes for BioMedical Research, Basel, Switzerland) for help with LC-MS.
REFERENCES
- 1. Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science 2002;298:1912–1934 [DOI] [PubMed] [Google Scholar]
- 2. Dancey J, Sausville EA. Issues and progress with protein kinase inhibitors for cancer treatment. Nat Rev Drug Discov 2003;2:296–313 [DOI] [PubMed] [Google Scholar]
- 3. Cohen P. Protein kinases—the major drug targets of the twenty-first century? Nat Rev Drug Discov 2002;1:309–315 [DOI] [PubMed] [Google Scholar]
- 4. Chene P. Challenges in design of biochemical assays for the identification of small molecules to target multiple conformations of protein kinases. Drug Discov Today 2008;13:522–529 [DOI] [PubMed] [Google Scholar]
- 5. Liao JJ, Andrews RC. Targeting protein multiple conformations: a structure-based strategy for kinase drug design. Curr Top Med Chem 2007;7:1394–1407 [DOI] [PubMed] [Google Scholar]
- 6. Adrian FJ, Ding Q, Sim T, et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol 2006;2:95–102 [DOI] [PubMed] [Google Scholar]
- 7. Bogoyevitch MA, Fairlie DP. A new paradigm for protein kinase inhibition: blocking phosphorylation without directly targeting ATP binding. Drug Discov Today 2007;12:622–633 [DOI] [PubMed] [Google Scholar]
- 8. Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer 2009;9:28–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Phospho.ELM Database Version 8.2, April 2009. http://phospho.elm.eu.org/, 2009.
- 10. Peterson JE, Kulik G, Jelinek T, Reuter CW, Shannon JA, Weber MJ. Src phosphorylates the insulin-like growth factor type I receptor on the autophosphorylation sites. Requirement for transformation by src. J Biol Chem 1996;271:31562–31571 [DOI] [PubMed] [Google Scholar]
- 11. White MF, Kahn CR. The insulin signaling system. J Biol Chem 1994;269:1–4 [PubMed] [Google Scholar]
- 12. Jiang Y, Chan JL, Zong CS, Wang LH. Effect of tyrosine mutations on the kinase activity and transforming potential of an oncogenic human insulin-like growth factor I receptor. J Biol Chem 1996;271:160–167 [DOI] [PubMed] [Google Scholar]
- 13. Ouwens DM, van der Zon GC, Pronk GJ, et al. A mutant insulin receptor induces formation of a Shc-growth factor receptor bound protein 2 (Grb2) complex and p21ras-GTP without detectable interaction of insulin receptor substrate 1 (IRS1) with Grb2. Evidence for IRS1-independent p21ras-GTP formation. J Biol Chem 1994;269:33116–33122 [PubMed] [Google Scholar]
- 14. Favelyukis S, Till JH, Hubbard SR, Miller WT. Structure and autoregulation of the insulin-like growth factor 1 receptor kinase. Nat Struct Biol 2001;8:1058–1063 [DOI] [PubMed] [Google Scholar]
- 15. Du P, Loulakis P, Luo C, et al. Phosphorylation of serine residues in histidine-tag sequences attached to recombinant protein kinases: a cause of heterogeneity in mass and complications in function. Protein Expr Purif 2005;44:121–129 [DOI] [PubMed] [Google Scholar]
- 16. Haacke A, Fendrich G, Ramage P, Geiser M. Chaperone over-expression in Escherichia coli: apparent increased yields of soluble recombinant protein kinases are due mainly to soluble aggregates. Protein Expr Purif 2009;64:185–193 [DOI] [PubMed] [Google Scholar]
- 17. Elling RA, Tangonan BT, Penny DM, et al. Mouse Aurora A: expression in Escherichia coli and purification. Protein Expr Purif 2007;54:139–146 [DOI] [PubMed] [Google Scholar]
- 18. Wang L, Foster M, Zhang Y, et al. High yield expression of non-phosphorylated protein tyrosine kinases in insect cells. Protein Expr Purif 2008;61:204–211 [DOI] [PubMed] [Google Scholar]
- 19. Smith CK, Carr D, Mayhood TW, Jin W, Gray K, Windsor WT. Expression and purification of phosphorylated and non-phosphorylated human MEK1. Protein Expr Purif 2007;52:446–456 [DOI] [PubMed] [Google Scholar]
- 20. Høyer-Hansen G, Hamers MJ, Pedersen AN, et al. Loss of ELISA specificity due to biotinylation of monoclonal antibodies. J Immunol Methods 2000;235:91–99 [DOI] [PubMed] [Google Scholar]
- 21. Santala V, Lamminmaki U. Production of a biotinylated single-chain antibody fragment in the cytoplasm of Escherichia coli. J Immunol Methods 2004;284:165–175 [DOI] [PubMed] [Google Scholar]
- 22. Chapman-Smith A, Cronan JE., Jr. In vivo enzymatic protein biotinylation. Biomol Eng 1999;16:119–125 [DOI] [PubMed] [Google Scholar]
- 23. Garcia-Echeverria C, Pearson MA, Marti A, et al. In vivo antitumor activity of NVP-AEW541—a novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell 2004;5:231–239 [DOI] [PubMed] [Google Scholar]
- 24. Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (NY) 1993;11:1138–1143 [DOI] [PubMed] [Google Scholar]
- 25. Wasilko DJ, Lee SE. TIPS: Titerless Infected-Cells Preservation and Scale-up. BioProcess J 2006;5:29–32 [Google Scholar]
- 26. Zor T, Selinger Z. Linearization of the Bradford protein assay increases its sensitivity: theoretical and experimental studies. Anal Biochem 1996;236:302–308 [DOI] [PubMed] [Google Scholar]
- 27. Chene P. Study of the selectivity of type I insulin-like growth factor receptor (IGF1R) inhibitors, in press, Open Enz Lnh J. [Google Scholar]
- 28. Bliska JB, Clemens JC, Dixon JE, Falkow S. The Yersinia tyrosine phosphatase: specificity of a bacterial virulence determinant for phosphoproteins in the J774A.1 macrophage. J Exp Med 1992;176:1625–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guan KL, Dixon JE. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science 1990;249:553–556 [DOI] [PubMed] [Google Scholar]
- 30. Zhang ZY, Clemens JC, Schubert HL, et al. Expression, purification, and physicochemical characterization of a recombinant Yersinia protein tyrosine phosphatase. J Biol Chem 1992;267:23759–23766 [PubMed] [Google Scholar]
- 31. Seeliger MA, Young M, Henderson MN, et al. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci 2005;14:3135–3139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cronan JE., Jr. Biotination of proteins in vivo. A post-translational modification to label, purify, and study proteins. J Biol Chem 1990;265:10327–10333 [PubMed] [Google Scholar]
- 33. Chapman-Smith A, Cronan JE., Jr. The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem Sci 1999;24:359–363 [DOI] [PubMed] [Google Scholar]
- 34. Smith PA, Tripp BC, DiBlasio-Smith EA, Lu Z, LaVallie ER, McCoy JM. A plasmid expression system for quantitative in vivo biotinylation of thioredoxin fusion proteins in Escherichia coli. Nucleic Acids Res 1998;26:1414–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J, Jaramillo A, Shi R, Kwok WW, Mohanakumar T. In vivo biotinylation of the major histocompatibility complex (MHC) class II/peptide complex by coexpression of BirA enzyme for the generation of MHC class II/tetramers. Hum Immunol 2004;65:692–699 [DOI] [PubMed] [Google Scholar]