Abstract
Diffusion has been widely adopted in the clinical setting to study the microstructural tissue changes in conjunction with anatomic imaging and metabolic imaging to offer insights on the status of the tissue injury or lesion. However, geometric distortions caused by magnetic susceptibility effects, eddy currents, and gradient imperfections greatly affect the clinical utility of the diffusion images. Several diffusion methods have been proposed in the recent years to obtain diffusion parameters with increased accuracy. In most cases, the comparisons to the clinical standard EPI diffusion are done visually without quantitative measurements. In this study, we present three simple, complementary quantitative methods of nonrigid image registration and shape analyses for evaluating spatial distortions on MR images with application in comparing SSFSE and EPI based diffusion measurements. These methods have confirmed the SSFSE based diffusion method is less distorted than the EPI based one, which is generally accepted through visual inspection.
Keywords: distortion, diffusion imaging, ssfse, epi, brain
Introduction
Echo-planar (EPI) based diffusion has been widely adopted in the clinical setting to study microstructural tissue changes in conjunction with anatomic imaging and metabolic imaging to offer insights on the status of the tissue injury or lesion [1–5]. However, geometric distortions caused by magnetic susceptibility effects, eddy currents, and gradient imperfections greatly affect the clinical utility of the diffusion images [6]. These effects are particularly pronounced at air-tissue interfaces. The magnitude of these distortions increases with field strength; consequently, the increased use of 3 T and higher field magnets makes the problem of distortion more acute. Due to the difficulties that arise from EPI, efforts have been made to develop imaging techniques that minimize distortion artifacts as well as to develop post hoc correction schemes. Various acquisition techniques using RF refocusing, such as single-shot fast spin-echo (SSFSE) [7, 8], multi-shot FSE (PROPELLER) [9], and steady state free precession (SSFP) [10] pulse sequences in combination with parallel imaging have been utilized to reduce artifacts for diffusion imaging studies. Most often, comparisons are done qualitatively based on visual inspection. It is important to quantitatively measure the distortion effects when comparing these techniques as the difference in image quality becomes less and less apparent.
One class of post-processing methods applies nonrigid image registration to align the distorted image with an undistorted reference image; of particular interest to this study is the use of high-dimensional non-affine transformations [11–13]. It has been shown that deformation fields parameterized by cubic B-splines can be used to accurately relate a distorted EPI image to an undistorted anatomic image. These results have been verified by comparison of the computed deformation field with field map calculations [12]. Nonrigid image registrations have thus found an important use in the unwarping of distorted images [14]. Additionally, the information contained in the computed deformation field may be useful in characterizing and comparing the distortions from different acquisition schemes. Based on the existing literature on the use of nonrigid image registration for correction of EPI distortions and for quantitative analysis of tissue shifts, we have chosen to use nonrigid registration to characterize and compare the distortions occurring during different imaging sequences.
As distortions change the shape of the object, a second approach to distortion analysis is to consider the object shapes in an attempt to compare images from the various acquisitions. In this study, we chose to investigate the curvature of the outer edge of the brain, in part due to the expectation that magnetic susceptibility would distort the edges and cause sharp changes in the anterior portion of the brain near the sinuses and posteriorly near the skull base. The normals to the edges were analyzed. In addition, we compared distances from brain edges to brain centroids with the expectation that distortions would shift the location of centroid of the brain.
We used these methods to compare a standard spin-echo EPI sequence with a SSFSE sequence with a standard FSE sequence as reference.
Methods
Image Acquisition
A spin-echo EPI (SE-EPI) based diffusion sequence was compared with an SSFSE based diffusion sequence. An undistorted anatomical image was acquired using fast spin-echo (FSE) in addition to the two diffusion sets. The FSE images were acquired at the same location as the reference. All volunteer data were acquired on a 3 T GE Signa EXCITE scanner (GE Healthcare Technologies, Waukesha, WI). For each volunteer, all the images were acquired in the same examination. Informed consent was obtained from all subjects, as approved by the Committee on Human Research at our institution.
Six healthy volunteers were recruited and sixteen identically placed slices of the brain were acquired with each of three imaging sequences. The two 2D test sequences that were to be compared were an SSFSE (TR/TE = 16000/84 ms; necho = 72) and SE-EPI (TR/TE = 5000/84 ms). A fast spin-echo (FSE) sequence was acquired as the reference image (TR/TE = 800/84 ms). All images were acquired using a standard quadrature head coil with a FOV of 22 cm × 22 cm with a 256 × 128 encoding matrix (frequency × phase) and reconstructed on a 256 × 256 array. The nominal slice thickness of all scans was 5 mm. These imaging parameters are commonly used in the clinical setting, and optimized for SNR and coverage. All image analyses were performed on the b0 image of the diffusion sequences, in which no diffusion gradient was applied.
Nonrigid Image Registration
Because distortions within the brain are of the most interest and registration with segmented images has been found to be more robust, the scalp and skull of the SSFSE, FSE, and SE-EPI images were removed using an automated algorithm [15]. Fat suppression was performed during acquisition of the EPI images, and any incompletely suppressed scalp tissue was removed by manually deleting those areas from the images. Nonrigid registration was performed to align the test images to the reference image from the same patient. The registration was performed by optimizing the positions of an initially regularly spaced 3D lattice of cubic B-spline control points [16]. The optimization proceeded through a steepest ascent search algorithm to maximize a normalized mutual information cost function [17]. The initial control point spacing was 10 mm × 5 mm × 10 mm in the frequency, phase, and slice select directions, respectively. The comparisons were performed using Wilcoxon signed rank test.
Shape Analysis
Normal vectors were generated based on a Bezier spline fitted to the edge of the brain, and histogram similarity of their angles based on the χ2 test method, proposed by Puzicha et al. [18], was used for quantitative comparisons between the SE-EPI and SSFSE versus the FSE image as reference. In addition, distances from points on the borders to the centroids of the images were computed and results were analyzed in a similar way as for the curvature data. The comparisons between SSFSE and EPI diffusion images were performed using Wilcoxon ranked test.
Results
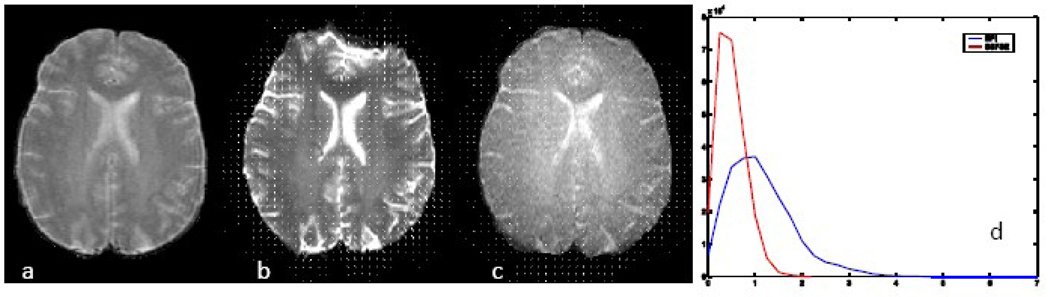
Shown in Figure 1 is an example of the nonrigid registration analysis, applied to the comparison between SE-EPI and SSFSE. The size and magnitude of the displacement vectors and displacement contours clearly reflect the large scale distortions of SE-EPI (as demonstrated in Table 1), and the general locations of the computed displacements agree well with visual analysis. The SSFSE image is essentially undistorted, and indeed, the displacement vectors and contours show very little effect. The Wilcoxon test demonstrated that the maximum distortion in the SE-EPI was significantly larger than the SSFSE on a subject-by-subject basis.
Figure 1.
Images of T2 FSE (a), EPI (b), SSFSE (c) with overlaid transformation vector. (d) is the histogram of the magnitude of the transformation vectors demonstrating larger deformation was needed for EPI compared to SSFSE, higher number of pixels with larger shifts.
Table 1.
Maximum voxel displacements in millimeters, using the FSE image as a reference, based on the whole brain.
| Sequence | Magnitude | Frequency encode | Phase encode | Slice select |
|---|---|---|---|---|
| SE-EPI | 3.3 ± 0.3 | 1.9 ± 0.3 | 2.9 ± 0.3 | 2.1 ± 0.4 |
| SSFSE | 1.4 ± 0.2 | 1.1 ± 0.2 | 0.9 ± 0.2 | 1.1 ± 0.2 |
| p value | <0.05 | <0.05 | <0.05 | <0.05 |
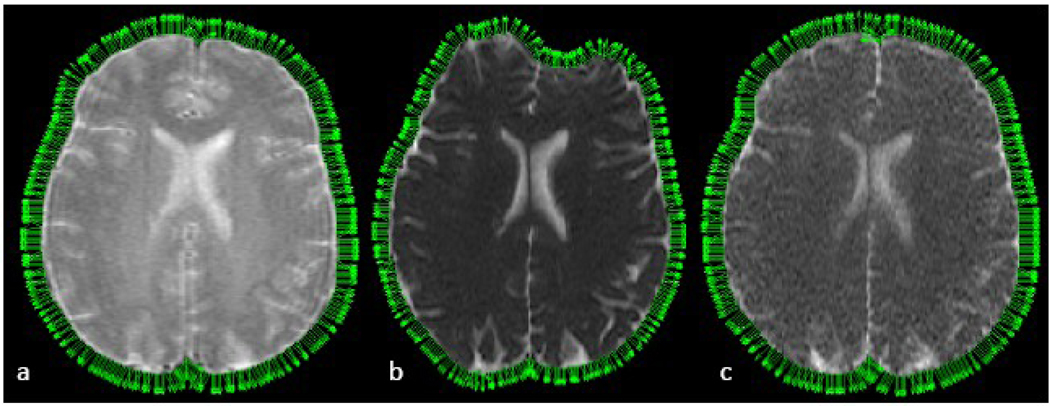
Figure 2 shows vectors normal to the edge of the brain and demonstrates that in and near regions of large field error due to magnetic susceptibility (anterior and posterior parts of the image), the vectors change direction as the distortions occur. The SE-EPI image showed large changes in the contour of the brain in comparison to that of the FSE and SSFSE images. This is confirmed by the quantitative results in Table 2. Both the vectoral direction change and distance to the centroid showed a significantly larger deviation in the SE-EPI compared with the SSFSE.
Figure 2.
Images of T2 FSE (a), EPI (b), SSFSE (c) with overlaid normal vectors, demonstrating larger change in the contour of the brain for EPI compared to SSFSE.
Table 2.
Curvature and distances to centroid comparisons between SE-EPI and SSFSE images with FSE as the reference.
| Sequence | Curvature | Centroid |
|---|---|---|
| SE-EPI | 0.45 ± 0.024 | 0.33 ± 0.088 |
| SSFSE | 0.24 ± 0.037 | 0.15 ± 0.030 |
| p value | <0.05 | <0.05 |
Discussion
In this work, we have demonstrated the use of nonrigid image registration and shape analysis in the objective experimental evaluation of image distortions. These methods can be applied to quantitatively compare spatially distorted images and therefore may be important in evaluating the variety of available and emerging methods for diffusion imaging. With improvement in parallel imaging (SENSE and GRAPPA) methods, it is increasingly difficult to visually assess the distortions in the images. For clinical diagnoses, it is critical to obtain the diffusion parameters that are from the exact location of the regions of interest, which are used in conjunction with traditional anatomic images that are minimally or not distorted.
This study provides quantitative confirmation that the SSFSE sequence yields images that are significantly less distorted than conventional SE-EPI sequences. The SSFSE distortions were found to be minimal throughout the image volume, with voxel displacements of less than 1.5 mm, while the SE-EPI image was significantly distorted throughout the entire volume. This confirms expectations based on visual assessment and on the physics of image distortions. Without rapid gradient switching, the SSFSE, which uses RF for refocusing, does not suffer from the same susceptibility artifacts as the SE-EPI. Similarly, the requirements for gradient performance are relaxed in the SSFSE compared with the SE-EPI, and so eddy currents and k-space alignment of the echoes are less problematic. The artifacts caused by susceptibility are in three dimensions as seen by results in Table 1. The largest difference came from the phase encode direction, which has the smallest effective bandwidth. The slice direction displacement may be due to additional requirement in the EPI diffusion sequence for subcutaneous lipid suppression. The SSFSE has similar characteristics in excitation and acquisition to the reference FSE, which has very small displacements in the all frequency, phase, and slice select directions. One also should note that the EPI based diffusion images have much higher SNR and CNR due to much shorter acquisition time in comparison to the SSFSE, which by the end of the spin echo train, the leftover signal is in the noise floor. However, the SNR and CNR do not have significant affect on the described non-rigid registration method for evaluating distortion for clinically used acquisitions, which are optimized to evaluate possible lesions. There should be enough SNR and contrast for non-rigid registration, as it is routinely performed in research imaging studies. They also should not strongly affect the quantitative evaluation using shape analysis, which is dependent on the overall shape and distribution of pixel values.
The described methods for the quantitative comparison of distortions can be applied to any imaging sequence. The major advantages of these approaches are that they are fully automated and therefore not subject to user bias, and the methods can be used to quantify arbitrarily complex spatially variant distortions. Moreover, the analysis is performed on experimental data rather than on purely theoretical considerations, and so may take into account more real-world acquisition factors. One should note that the reliability of the displacement vectors is entirely dependent on the robustness of the non-rigid deformation algorithm. Multiple algorithms will yield different results, but with a consistent method, the reproducibility of the analysis is high, which can be seen by the relatively small standard deviation of the displacement vectors. The two simple methods described above should provide the necessary quantitative information to evaluate various diffusion acquisition methods that are becoming available.
Acknowledgements
The authors would like to thank Dr. Cornelius von Morze for insightful discussions and ICURP UC Discovery ITLBIO04-10148 for funding support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Field AS, Alexander AL. Diffusion tensor imaging in cerebral tumor diagnosis and therapy. Top Magn Reson Imaging. 2004;15(5):315–324. doi: 10.1097/00002142-200410000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Kurhanewicz J, et al. Multiparametric magnetic resonance imaging in prostate cancer: present and future. Curr Opin Urol. 2008;18(1):71–77. doi: 10.1097/MOU.0b013e3282f19d01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukherjee P. Diffusion tensor imaging and fiber tractography in acute stroke. Neuroimaging Clin N Am. 2005;15(3):655–665. xii. doi: 10.1016/j.nic.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Sotak CH. The role of diffusion tensor imaging in the evaluation of ischemic brain injury - a review. NMR Biomed. 2002;15(7–8):561–569. doi: 10.1002/nbm.786. [DOI] [PubMed] [Google Scholar]
- 5.Ge Y, Law M, Grossman RI. Applications of diffusion tensor MR imaging in multiple sclerosis. Ann N Y Acad Sci. 2005;1064:202–219. doi: 10.1196/annals.1340.039. [DOI] [PubMed] [Google Scholar]
- 6.Le Bihan D, et al. Artifacts and pitfalls in diffusion MRI. Journal of Magnetic Resonance Imaging. 2006;24(3):478–488. doi: 10.1002/jmri.20683. [DOI] [PubMed] [Google Scholar]
- 7.Alsop DC. Phase insensitive preparation of single-shot RARE: application to diffusion imaging in humans. Magn Reson Med. 1997;38(4):527–533. doi: 10.1002/mrm.1910380404. [DOI] [PubMed] [Google Scholar]
- 8.Xu D, et al. Single-shot fast spin-echo diffusion tensor imaging of the brain and spine with head and phased array coils at 1.5 T and 3.0 T. Magn Reson Imaging. 2004;22(6):751–759. doi: 10.1016/j.mri.2004.01.075. [DOI] [PubMed] [Google Scholar]
- 9.Pipe JG, Farthing VG, Forbes KP. Multishot diffusion-weighted FSE using PROPELLER MRI. Magn Reson Med. 2002;47(1):42–52. doi: 10.1002/mrm.10014. [DOI] [PubMed] [Google Scholar]
- 10.Bosak E, Harvey PR. Navigator motion correction of diffusion weighted 3D SSFP imaging. Magma. 2001;12(2–3):167–176. doi: 10.1007/BF02668098. [DOI] [PubMed] [Google Scholar]
- 11.Rohde GK, et al. Comprehensive approach for correction of motion and distortion in diffusion-weighted MRI. Magnetic Resonance in Medicine. 2004;51(1):103–114. doi: 10.1002/mrm.10677. [DOI] [PubMed] [Google Scholar]
- 12.Studholme C, et al. Estimating tissue deformation between functional images induced by intracranial electrode implantation using anatomical MRI. Neuroimage. 2001;13(4):561–576. doi: 10.1006/nimg.2000.0692. [DOI] [PubMed] [Google Scholar]
- 13.Kybic J, et al. Unwarping of unidirectionally distorted EPI images. IEEE Trans Med Imaging. 2000;19(2):80–93. doi: 10.1109/42.836368. [DOI] [PubMed] [Google Scholar]
- 14.Lupo JM, et al. Feasibility of dynamic susceptibility contrast perfusion MR imaging at 3T using a standard quadrature head coil and eight-channel phased-array coil with and without SENSE reconstruction. Journal of Magnetic Resonance Imaging. 2006;24(3):520–529. doi: 10.1002/jmri.20673. [DOI] [PubMed] [Google Scholar]
- 15.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17(3):143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rueckert D, et al. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18(8):712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- 17.Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recognition. 1999;32(1):71–86. [Google Scholar]
- 18.Puzicha J, Hoffman T, Buhmann J. Non-parametric similarity measures for unsupervised texture segmentation and image retrieval; Proceedings of IEEE Conference Computer Vision and Pattern Recognition; 1997. [Google Scholar]