One of the most profound and poorly understood processes in cell biology is compartmentalization – how the detailed atomic structures of proteins, lipids, carbohydrates, metabolites, and nucleic acids orchestrate the assembly of discrete organelles and cellular substructures. Compartmentalization demands the separation of immiscible phases into distinct domains. Understanding the forces and rules that govern biopolymer segregation will enrich our understanding of the chemistry that guides membrane and organelle biogenesis, and provide guiding principles for the burgeoning field of synthetic biology. Herein we take the fundamental first steps towards a β-peptide bundle that can be compartmentalized within a membrane environment.
We reported that certain β-peptides self-assemble into cooperatively folded bundles whose kinetic and thermodynamic metrics mirror those of natural α–helix bundle proteins.1-4 The structures of four such bundles are known in atomic detail.1,2 These structures reveal a solvent-sequestered, hydrophobic core stabilized by a unique arrangement of leucine side chains and backbone methylene groups. Here we report that this core can be re-engineered to contain a fluorous sub-domain while maintaining the characteristic β-peptide bundle fold. Like α-helical bundles possessing fluorous cores,5-15 fluorous β-peptide bundles are stabilized relative to hydrocarbon analogs and undergo cold-denaturation. β-peptide bundles with fluorous cores represent the essential first step in the synthesis of orthogonal protein assemblies that can sequester selectively in an interstitial membrane environment.
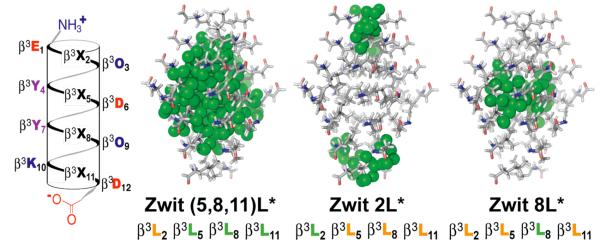
We initially synthesized Zwit-(5,8,11)L* (Figure 1), which contains hexafluoro-β3-leucine (L*) in place of three of four leucines in Zwit-YK, an analog of Zwit-1F. Zwit-(5,8,11)L* displayed no concentration-dependent increase in 14-helical structure as judged by circular dichroism (CD) analysis between 25 and 150 μM; even at high concentration the CD spectrum of Zwit-(5,8,11)L* was featureless (Figure S1A). In retrospect, the absence of bundle formation by Zwit-(5,8,11)L* was not surprising, as a single –CF3 substituent possesses a Van der Waals surface that is 2 to 3 times larger than a –CH group,16-18 and the octameric Zwit-(5,8,11)L* bundle would contain 48 such substitutions.
Figure 1.
Helical net diagram of β-peptide monomers and models of the octameric bundle cores that each might form. Hexafluoro-β3-leucine side chains are represented by green spheres.
Next, we interrogated the Zwit-1F structure1 to identify a sub-set of leucine side chains that would best establish a fluorous sub-core. Using Spartan,19 we individually substituted each β3-leucine residue of Zwit-1F (at positions 2, 5, 8, and 11) with hexafluoro-β3-leucine to generate models of octameric Zwit-2L*, Zwit-5L*, Zwit-8L*, and Zwit-11L* (Figure 1). Examination of these models suggested that only one–Zwit-8L*–would contain a single, solvent-excluded fluorous sub-core, shielding 8 CF3 groups from solvent water. All other models were predicted to contain multiple fluorous-rich regions, either buried within the core (Zwit-5L* and Zwit-11L*) or in solvent contact (Zwit-2L*).
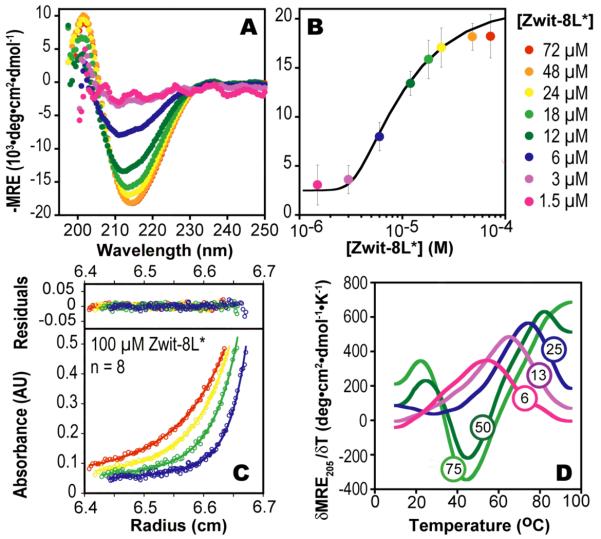
Zwit-2L*, and Zwit-8L* were characterized first using CD to determine if they exhibited the concentration-dependent change in 314-helical structure that characterizes all β-peptide bundles.1-4 Both do: In the case of Zwit-2L* the molar residue ellipticity at 214 nm (MRE214) decreases to a minimum of – 25,000 deg·cm2·dmol−1 at concentrations between 2 and 48 μM (Figure S1B) whereas in the case of Zwit-8L* the minimum decreases to −20,000 deg·cm2·dmol−1 between 1.5 and 76 μM (Figure 2A). Analysis of plots of MREmin as a function of [β-peptide] suggests that only Zwit-8L* formed an octamer, with ln Ka = 83.9 ± 0.6 (Figure 2B), whereas Zwit-2L* formed a tetramer, with ln Ka = 34.5 ± 0.12 (Figure S1C). The stability of the Zwit-2L* tetramer is comparable to that of the tetrameric Zwit-VY bundle reported by Goodman et al.4
Figure 2.
(A) Wavelength-dependent CD spectra of Zwit-8L*. (B) Plot of MREmin as a function of [Zwit 8L*] monomer. Curve shows the best fit to a monomer-octamer equilibrium.20 (C) AU data fit to a monomer-n-mer equilibrium (ln Ka = 82.0; n = 8.0). Residuals are displayed with a linear Y-axis scale. (D) First derivatives of the CD melting curves at the concentrations shown (in μM units).
Next we performed equilibrium sedimentation experiments to confirm the association states of Zwit-2L* and Zwit-8L* in solution and provide an independent measure of ln Ka. Sedimentation was monitored at four speeds (36, 42, 50 and 60 kRPM) at concentrations of 75, 100 and 150 μM for Zwit-8L* and 5, 25, and 50 μM for Zwit-2L*. For Zwit-8L* the AU data fit best to a monomer-n-mer equilibrium where n = 7.8 (n was allowed to vary) with an RMSD of 0.00697 (Figure 2C). Significantly poorer fits were observed when n was set to equal 6, 7 or 10, respectively, whereas comparable fits were found when n was set to equal 8 (RMSD = 0.00698). The ln Ka value calculated from the fits where n = 7.86 and n = 8 (81.1 ± 0.5 and 82.0 ± 0.2) agree well with the ln Ka value calculated from the CD data (83.9 ± 0.6), providing additional support for equilibration between monomeric and octameric Zwit-8L*. For Zwit-2L*, the AU data fit best to a monomertetramer equilibrium, as predicted by the CD data, with excellent agreement between the ln Ka values calculated from AU (34.1 ± 0.1) and CD (34.5 ± 0.12).
We also measured the temperature dependence of the 14-helix dependent CD signal for Zwit-8L*, and found it to undergo a cooperative melting transition (Figure 2D). At 50 μM, the T of the Zwit-8L* bundle is 82 °C, on par with that of the parental Zwit-YK (85 °C) and much higher than Zwit-1F (57 °C) and Zwit-2L* (52 °C). Examination of the Zwit-8L* melting data shows clear evidence for cold denaturation at concentrations where the octamer predominates (87.8% and 91.4% octamer at 50 and 75 μM, respectively). Cold denaturation results when the Gibbs free energy of hydrating non-polar side chains overcomes the Gibbs free energy of folding.21,22 Fluorocarbons are more hydrophobic than hydrocarbons,5,16,23 and thus proteins containing fluorous cores undergo cold denaturation at higher temperatures than those containing hydrocarbon cores.24 The observation of a cold denaturation transition is fully consistent with the presence of a discrete fluorous sub-core in the Zwit-8L* bundle.
A widely accepted diagnostic for a well-packed protein core is the inability to bind and increase the fluorescence of hydrophobic dyes such as 1-anilino-8-naphthalenesulfonate (ANS).25 Well-folded proteins, including previous β-peptide bundles,2,4 increase ANS fluorescence only minimally (<10-fold).26-28 Molten globules, by contrast, increase ANS fluorescence significantly (changes >100-fold).26 The relative fluorescence of ANS increased from a value of 1 at 1.56 μM Zwit-8L* (3% octamer) to a value of 15 at 200 μM Zwit-8L*, (98% octamer) (Figure S2). This increase is small relative to molten globules, although compared to the non-fluorinated β-bundles, it seems to suggest that the core of Zwit-8L* may be slightly more exposed. As the CD and AU data suggest that Zwit-8L* forms a well-folded octamer, we interpret this observation as further support for a distinct fluorous sub-core–the increased hydrophobicity and decreased polarizability of the fluorous core should induce greater dye emission due to even more unfavorable solvent relaxation pathways.29
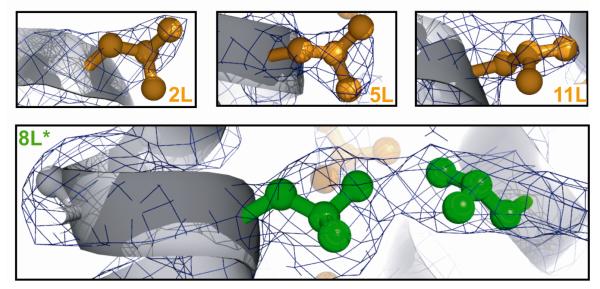
Finally, to validate formation of a central fluorous core in the Zwit-8L* bundle, we solved the structure to 2.75 Å. As expected, the electron density for the β3-homoleucine residues at positions 2, 5 and 11 closely outlines the leucine side chains with minimal unfilled density (Figure 3). Not surprisingly, the electron density at the location of the β3-hexafluoroleucine residue is larger than an unmodified leucine side chain. Furthermore, the electron density at 8L* is continuous with the electron density from 8L* in a neighboring peptide. Our previously solved structures1,2 do not show extended density or continuous electron density with neighboring leucine side chains. We attribute the unique features of the 8L* electron density in Zwit-8L* to the in-register alignment of the fluorinated residues and the formation of a protected discrete fluorous sub-core, analogous to the ones previously reported in fluorinated coiled coils.5,6,8,10,11,30
Figure 3.
X-ray structure of Zwit-8L*. Leucine residues at positions 2 (2L), 5 (5L), and 11 (11L) in Zwit-8L* are shown in orange with the corresponding electron density in dark blue.
Supplementary Material
Acknowledgement
This work was supported by the NIH (GM74756 to A.S. and GM65500 to K.K.), the NSF (CHE-0848098), and the National Foundation for Cancer Research.
Footnotes
Supporting Information Available: β-peptide synthesis and details of biophysical analyses. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Daniels DS, Petersson EJ, Qiu JX, Schepartz A. J. Am. Chem. Soc. 2007;129:1532–3. doi: 10.1021/ja068678n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodman JL, Petersson EJ, Daniels DS, Qiu JX, Schepartz A. J. Am. Chem. Soc. 2007;129:14746–51. doi: 10.1021/ja0754002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersson EJ, Craig CJ, Daniels DS, Qiu JX, Schepartz A. J. Am. Chem. Soc. 2007;129:5344–5. doi: 10.1021/ja070567g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goodman JL, Molski MA, Qiu J, Schepartz A. ChemBioChem. 2008 doi: 10.1002/cbic.200800039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilgiçer B, Fichera A, Kumar K. J. Am. Chem. Soc. 2001;123:4393–9. doi: 10.1021/ja002961j. [DOI] [PubMed] [Google Scholar]
- 6.Bilgiçer B, Xing X, Kumar K. J. Am. Chem. Soc. 2001;123:11815–6. doi: 10.1021/ja016767o. [DOI] [PubMed] [Google Scholar]
- 7.Niemz A, Tirrell DA. J. Am. Chem. Soc. 2001;123:7407–7413. doi: 10.1021/ja004351p. [DOI] [PubMed] [Google Scholar]
- 8.Tang Y, Ghirlanda G, Vaidehi N, Kua J, Mainz DT, Goddard IW, DeGrado WF, Tirrell DA. Biochemistry. 2001;40:2790–6. doi: 10.1021/bi0022588. [DOI] [PubMed] [Google Scholar]
- 9.Tang Y, Tirrell DA. J. Am. Chem. Soc. 2001;123:11089–90. doi: 10.1021/ja016652k. [DOI] [PubMed] [Google Scholar]
- 10.Tang Y, Ghirlanda G, Petka WA, Nakajima T, DeGrado WF, Tirrell DA. Angew. Chem, Int. Ed. 2001;40:1494–1496. [PubMed] [Google Scholar]
- 11.Bilgiçer B, Kumar K. Tetrahedron. 2002;58:4105–4112. [Google Scholar]
- 12.Bilgiçer B, Kumar K. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15324–9. doi: 10.1073/pnas.0403314101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Naarmann N, Bilgiçer B, Meng H, Kumar K, Steinem C. Angew. Chem, Int. Ed. 2006;45:2588–91. doi: 10.1002/anie.200503567. [DOI] [PubMed] [Google Scholar]
- 14.Chiu HP, Suzuki Y, Gullickson D, Ahmad R, Kokona B, Fairman R, Cheng RP. J. Am. Chem. Soc. 2006;128:15556–7. doi: 10.1021/ja0640445. [DOI] [PubMed] [Google Scholar]
- 15.Son S, Tanrikulu IC, Tirrell DA. ChemBioChem. 2006;7:1251–7. doi: 10.1002/cbic.200500420. [DOI] [PubMed] [Google Scholar]
- 16.Seebach D. Angew. Chem, Int. Ed. 1990;29:1320–1367. [Google Scholar]
- 17.OHagan D, Rzepa HS. Chem. Commun. 1997:645–652. [Google Scholar]
- 18.Budisa N, Pipitone O, Siwanowicz I, Rubini M, Pal PP, Holak TA, Gelmi ML. Chem. Biodiversity. 2004;1:1465–75. doi: 10.1002/cbdv.200490107. [DOI] [PubMed] [Google Scholar]
- 19.Spartan '04. Wavefunction, Inc.; Irvine, CA: 2004. [Google Scholar]
- 20.Degrado WF, Lear JD. J. Am. Chem. Soc. 1985;107:7684–7689. [Google Scholar]
- 21.Privalov PL. Crit. Rev. Biochem. Mol. Biol. 1990;25:281–305. doi: 10.3109/10409239009090612. [DOI] [PubMed] [Google Scholar]
- 22.Dias CL, Ala-Nissila T, Karttunen M, Vattulainen I, Grant M. Phys. Rev. Lett. 2008;100:118101. doi: 10.1103/PhysRevLett.100.118101. [DOI] [PubMed] [Google Scholar]
- 23.Resnati G. Tetrahedron. 1993;49:9385–9445. [Google Scholar]
- 24.Graziano G, Catanzano F, Riccio A, Barone G. J. Biochem. 1997;122:395–401. doi: 10.1093/oxfordjournals.jbchem.a021766. [DOI] [PubMed] [Google Scholar]
- 25.Matulis D, Baumann CG, Bloomfield VA, Lovrien RE. Biopolymers. 1999;49:451–8. doi: 10.1002/(SICI)1097-0282(199905)49:6<451::AID-BIP3>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 26.Lumb KJ, Kim PS. Biochemistry. 1995;34:8642–8648. doi: 10.1021/bi00027a013. [DOI] [PubMed] [Google Scholar]
- 27.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas AF, Gilmanshin RI. Biopolymers. 1991;31:119–28. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 28.Bruckner AM, Chakraborty P, Gellman SH, Diederichsen U. Angew. Chem, Int. Ed. 2003;42:4395–9. doi: 10.1002/anie.200351871. [DOI] [PubMed] [Google Scholar]
- 29.Hawe A, Sutter M, Jiskoot W. Pharm. Res. 2008;25:1487–1499. doi: 10.1007/s11095-007-9516-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu JC, O'Shea EK, Kim PS, Sauer RT. Science. 1990;250:1400–3. doi: 10.1126/science.2147779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.