Abstract
Relatively little is understood about the dynamics of global host–pathogen transcriptome changes that occur during bacterial infection of mucosal surfaces. To test the hypothesis that group A Streptococcus (GAS) infection of the oropharynx provokes a distinct host transcriptome response, we performed genome-wide transcriptome analysis using a nonhuman primate model of experimental pharyngitis. We also identified host and pathogen biological processes and individual host and pathogen gene pairs with correlated patterns of expression, suggesting interaction. For this study, 509 host genes and seven biological pathways were differentially expressed throughout the entire 32-day infection cycle. GAS infection produced an initial widespread significant decrease in expression of many host genes, including those involved in cytokine production, vesicle formation, metabolism, and signal transduction. This repression lasted until day 4, at which time a large increase in expression of host genes was observed, including those involved in protein translation, antigen presentation, and GTP-mediated signaling. The interactome analysis identified 73 host and pathogen gene pairs with correlated expression levels. We discovered significant correlations between transcripts of GAS genes involved in hyaluronic capsule production and host endocytic vesicle formation, GAS GTPases and host fibrinolytic genes, and GAS response to interaction with neutrophils. We also identified a strong signal, suggesting interaction between host γδ T cells and genes in the GAS mevalonic acid synthesis pathway responsible for production of isopentenyl-pyrophosphate, a short-chain phospholipid that stimulates these T cells. Taken together, our results are unique in providing a comprehensive understanding of the host–pathogen interactome during mucosal infection by a bacterial pathogen.
Keywords: epithelial growth factor receptor, host pathogen, microarray, Streptococcus pyogenes, transcriptome
Identification of the specific molecular events that in the aggregate comprise host–pathogen interactions is important to our understanding of infectious disease pathogenesis. Advances in genome-wide analyses occurring in the last decade have facilitated the study of global transcriptome changes that occur during microbial infection. The vast majority of these studies, such as the recent investigation of typhoid fever (1), have addressed the pathogen or host transcriptome at a single or relatively few points in the infection cycle. Although much has been learned, a poorly understood area of infectious disease research revolves around the simultaneous changes in the host and pathogen transcriptomes occurring throughout the entire course of infection. This type of study, which we will refer to as longitudinal interactome analysis, has great promise to enhance our understanding of the molecular pathogenesis of infectious diseases. Indeed, one recent study provided new understanding of host–pathogen interactions that regulate early-stage replication of HIV-1 (2) and protein-protein interactome studies have been published for Epstein-Barr virus (3), varicella zoster virus (4), and Kaposi's sarcoma herpesvirus (4). Simultaneous analysis of the host and pathogen transcriptome during in vivo infections has been reported for malaria (5) and Escherichia coli (6) infections. However, these studies were limited to mouse models of infection and only inferred host–pathogen interactions based on known or putative biological roles of pathogen genes. In addition to their inherent basic science interest, global interactome analyses are critical to conduct because they promise to provide previously unexplored avenues for translation research, including development of unique diagnostics and therapeutics. Many factors account for the lack of longitudinal interactome analyses, several key points being complex experimental design and computational intensity.
Group A Streptococcus (GAS) is a Gram-positive, human bacterial pathogen responsible for 2 million cases of pharyngitis and 15,000 cases of invasive disease in the United States annually. GAS produces many actively secreted proteins that target the host cell, including numerous proteases, lipases, and immunomodulatory proteins. GAS alters the level of production of these virulence factors in response to changes in external stimuli, resulting in significant transcriptome remodeling (7, 8). Therefore, GAS is likely to have differing tissue-specific gene expression patterns that are influenced by the host microenvironment. Several studies have investigated GAS transcriptome changes that occur during culture in laboratory media, saliva, blood, and with host cells (7, 9, 10). However, these types of studies lack the ability to capture the complex host–pathogen interactions that take place during infection of an intact host. Therefore the use of a relevant animal infection model is critical to providing enhanced understanding of host–pathogen interactions (11, 12).
Far less is known about the host response to infection by this pathogen. Most studies have only analyzed the host transcriptional response in particular cell types, such as leukocytes or immortalized epithelial cell lines, generally at a single time-point. Although informative, the inherent limitation of these studies means that they do not adequately reflect the complexity of the host response to GAS in vivo. Moreover, they lack the ability to resolve temporal changes in the host transcriptome that likely occur during natural infection.
To test the hypothesis that GAS infection produces a distinctive host transcriptome response, we performed a longitudinal analysis of host and pathogen genome-wide expression levels during a 32-day acute pharyngeal infection by GAS in 20 cynomolgus macaques. Gene transcript levels were measured at nine time-points during GAS infection using host and custom pathogen-specific Affymetrix GeneChip arrays. We discovered that relative to mock-infected animals, the host transcriptome response is unique and differentially expressed at each time interval of the infection cycle. Unexpectedly, many host genes and gene categories were down-regulated within the first 24 and 48 h of infection. Interactome analysis provided unique insight into the interactions between host and pathogen biological process and provided many previously unexplored avenues for further molecular confirmation studies and pathogenesis research.
Results and Discussion
Experimental Design.
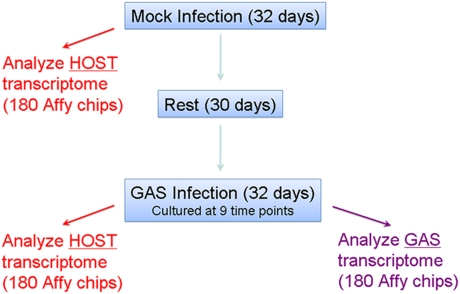
We used a three-phase experimental protocol consisting of mock-infection, rest, and GAS infection of 20 cynomolgus macaques (Fig. 1) (12). The unique experimental design allowed each animal to serve as its own experimental control, thereby reducing variability caused by interindividual differences in factors such as immune and genetic backgrounds. Genome-wide expression levels were assessed simultaneously in host and pathogen at nine time-points during both the mock and infection experimental phases. This longitudinal analysis of both host and pathogen transcriptome allowed us to probe the dynamic transcriptional events occurring during in vivo infection. Importantly, the protocol permitted us to identify individual host and pathogen gene pairs with correlated changes in gene expression, indicative of interaction between host and pathogen.
Fig. 1.
Overview of experimental design.
Long-Term Changes in the Host Transcriptional Response to GAS Infection.
To test the hypothesis that GAS produced significant changes in host-gene expression throughout the course of infection, a repeated measures statistical test with the Greenhouse-Geisser and Huhn-Feldt corrections was performed. For this test, 509 host genes were differentially expressed in the GAS-infected animals, compared with the mock-treatment group at a significance level of P < 0.005 (P < 0.05 for the Greenhouse-Geisser and Huhn-Feldt corrections) (Table S1). This genetic signature represents a comprehensive profile of the significant pharyngitis-specific host transcriptome response in the cynomolgus macaque for all time-points and all animals.
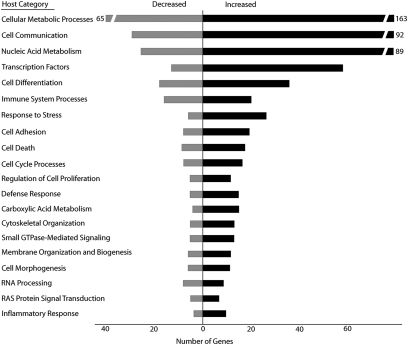
To probe the potential biological role that these differentially expressed genes play in the host, gene ontology (GO) categories were assessed using the DAVID application (13) (Fig. 2). We observed a significant increase in expression of genes involved in primary metabolic processes, such as ATP biosynthesis and carboxylic acid metabolism. We also discovered that genes involved in signaling categories, such as cell communication, transcription factors, Ras signaling, and GTPase-mediated signaling, were all significantly up-regulated. Additionally, we used Ingenuity Pathway Analysis software to determine the biological pathways that were differentially expressed. Seven canonical pathways were significantly overrepresented in the dataset at P < 0.05 (Fig S1). Among the differentially expressed pathways were several involved in the immune response to pathogens, such as acute phase response, eicosanoid signaling, Toll-like receptor signaling, and antigen presentation. Additionally, pathways involved in PPAR/RXR and apoptosis signaling were significantly overrepresented in our dataset, as assessed by Fisher's Exact test.
Fig. 2.
Host biological processes differentially expressed during GAS infection. GO categories were assigned using the DAVID application for each of the 509 host genes differentially expressed throughout the entire infection. Shown are the significantly overrepresented host biological processes (EASE score P < 0.05), ranked from top to bottom according to the number of differentially expressed genes in the category.
Short-Term Changes in the Host Transcriptional Response to GAS Infection.
To complement our repeated-measures analysis that identified host genes differentially expressed throughout the entire course of infection, we performed a serial analysis to track the short-term transcriptional response to GAS infection occurring between individual time-points. This process provides a high-resolution analysis of the temporal changes in gene expression and is not simply a breakdown of the repeated-measures analysis results by time. Therefore, it is important to note that the repeated-measures analysis is not simply a sum of these results. Two hundred fifty genes were differentially expressed (P < 0.01) during the first 24-h interval. The number of differentially expressed genes increased to a peak of 629 genes on day 4, a time corresponding to a very high pharyngitis score and GAS CFU level (11, 12). This peak was followed by a gradual decline to a low of 138 differentially expressed genes at day 16.
Although lengthy lists of individual genes provide important detail and specificity, they produce relatively restricted biological clarity. To better understand the biological significance of these changes in host gene expression, we performed an analysis of the GO categories represented in our dataset using a computational method known as significance analysis of function and expression (SAFE). SAFE determines which GO categories are differentially expressed using a two-step approach that takes into account the entire gene-expression dataset, rather than restricting the analysis to a short list of differentially expressed genes (14). This approach allows the identification of categories with a large number of marginally significant genes that might have otherwise been excluded in a traditional gene list approach. Eight individual SAFE analyses were performed, representing each of the eight time intervals present in our analysis of the short-term changes in host gene expression. We found that 494 significant (P < 0.05) categories were discovered over the eight time intervals (Table S2). These categories were differentially expressed in infected relative to mock-treated animals. Unexpectedly, during the first 24 h of infection, none of the host-gene categories were up-regulated. Rather, we observed widespread down-regulation of genes involved in sensory stimulus, vacuole formation, tight junctions, cytokine biosynthesis, G protein signaling, cellular repair, angiogenesis, and inflammatory response. This initial period of down-regulation was followed by a dramatic increase in expression occurring during the 2 to 4 day interval, when numerous gene categories were up-regulated (Table S2). To summarize, host gene expression was greatly altered over the course of infection relative to mock treatment.
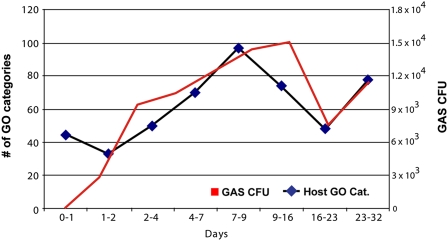
To test the hypothesis that increased bacterial growth correlated with increased activation of host biological processes, we examined the relationship between levels of GAS CFUs and expression of host-gene categories. Interestingly, when the quantity of GO categories per time interval was plotted against the average number of CFUs collected over time from the pharyngeal swabs obtained from all monkeys, we observed a striking, nearly superimposed relationship (Fig. 3). These data suggest that differential expression of GO category genes (relative to mock treatment) are related to the bacterial load at the infection site.
Fig. 3.
Comparison of biological process activity in host and pathogen during GAS infection. Graph showing the number of biological processes (GO categories) found to be differentially expressed in the host by SAFE analysis at each time interval is displayed along with GAS CFUs cultured from host. GAS values are shown in red; host values are in black.
Interactome Analysis.
One of our key goals was to gain enhanced understanding of the relationships between changes in the pathogen and host transcriptome over time: that is, the longitudinal interactome. Our experimental design permitted us to compare host and pathogen gene-expression profiles to identify individual host and the pathogen gene pairs that showed correlated levels of expression. In other words, we used a computationally intensive strategy to determine which host transcripts demonstrated expression parallel to pathogen transcripts and which transcripts demonstrated inverse-expression profiles. The goal was to determine which GAS transcripts influenced host-gene expression or vice versa.
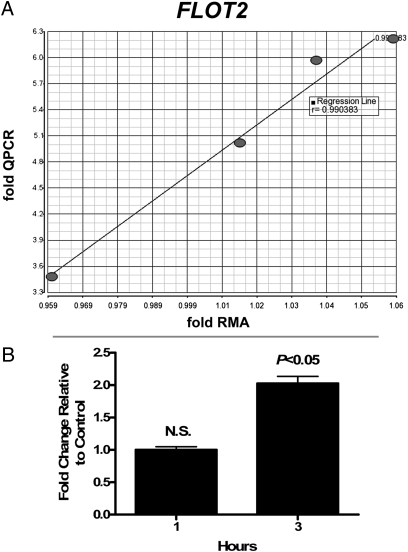
We analyzed several statistical algorithms for robustness and applicability to our datasets and experimental design, including Pearson's Correlation, Dynamic Programming, and Common Entropy. The SAFE method was extended for detection of host–pathogen gene expression coexpression analysis by using the Spearman Rank correlation for the local statistic and comparing all pair-wise combinations of individual host and pathogen gene expressions. The large number of individual comparisons (∼58 million) posed considerable multiple testing issues and produced a large list of correlations at P < 0.05; therefore, we refined this list by cross-referencing it with the significant results of our category-category analysis. By this method, we identified 73 unique host and pathogen gene pairs with correlated gene-expression levels. A summary of these results and individual correlation graphs are shown in Fig S2. Quantitative RT-PCR (qPCR) validation of two randomly selected genes from this dataset indicated a strong correlation between qPCR and expression microarray data (Fig. 4A and Fig S3).
Fig. 4.
TaqMan qPCR validation data. (A) Graph showing correlation between expression microarray and TaqMan qPCR validation data for flotillin 2 (FLOT2) gene. x axis represents RMA microarray values expressed as fold RMA values. y axis represents qPCR fold values (2-ddCT). The four data points are as follows, starting from Lower Left and moving to the Right: day 9, day 0, day 32, day 2. Each point represents the average of six animals. The regression line and r value of 0.99 are shown. (B) Increase in GAS metK expression following incubation with neutrophils. GAS metK transcript levels were measured by TaqMan after a 1- or 3-h incubation with human neutrophils. Changes in GAS metK mRNA levels after phagocytosis were determined by comparing normalized metK transcript levels in the presence and absence of human neutrophils. After 1 h, no significant changes in metK levels were detected. After 3 h, a significant (P < 0.05) 2-fold increase was seen in GAS incubated with neutrophils relative to bacteria alone.
Hyaluronic Acid Biosynthesis and Endocytic Vesicle Formation.
The correlation analysis identified relationships between GAS genes involved in hyaluronic acid biosynthesis and host genes encoding proteins involved in the formation of endocytic vesicles and clathrin-coated pits. Specifically, expression of the GAS genes hasA, hasB, hasC, and hasC.2 was correlated with expression of host genes encoding the clathrin adapter proteins AP2A2 and AP2S1, epithelial growth factor receptor (EGFR), and the voltage-gated calcium channels CACNA1I and CACNA1G. These GAS genes catalyze the multistep biosynthesis of the hyaluronic acid capsule, which is antiphagocytic and a major bacterial virulence factor (15). Increased capsule production reduces internalization of GAS into host cells and enhances virulence (15, 16). The hyaluronic acid capsule facilitates adhesion of GAS to the surface of human epithelial cells by binding to CD44 protein, a hyaluronic acid receptor on host cells (17). Binding of GAS to CD44 increases epithelial cell motility and triggers formation of lamellipodia (16). These effects are mediated by signaling through the Rho-GTPase Rac1 (16). Importantly, EGFR is recruited to CD44 and forms a stable complex upon binding of native hyaluronic acid produced by the host to CD44 (18, 19). However, EGFR has not been previously implicated in the molecular events associated with binding of GAS hyaluronic capsule to host CD44, and thus our finding is unique. Furthermore, activation of EGFR induces changes in cell motility and morphology similar to those observed in GAS binding to host CD44. Activation of EGFR has been associated with the redistribution of tight junction proteins that maintain epithelial integrity (20). Moreover, inhibitors of EGFR disrupt changes in cell motility caused by binding of hyaluronic acid to CD44 (18). Together, our findings suggest that EGFR plays an integral role in mediating the downstream signaling effects of GAS binding to CD44, and implicates the voltage-gated calcium channels CACNA1I and CACNA1G in controlling flux of Ca2+ levels associated with increased motility (model shown in Fig S4).
We observed several additional correlations between has GAS gene expression and the expression of the clathrin adapter proteins AP2A2 and AP2S1. These two proteins form part of the AP2 adapter complex, which localizes to the cytosolic side of endocytic vesicles and links to clathrin. Specifically, we observed an inverse correlation between AP2A2 transcript and has gene transcripts, whereas AP2S1 was positively correlated with has expression. Although relatively little is known about the specific roles of AP2A2 and AP2S1 adapters, their involvement in sorting of protein cargo for internalization and receptor recycling in endocytic vesicles suggests they are involved in internalization of GAS or in alteration in levels of receptor proteins, such as EGFR or CD44 at the plasma membrane. Thus, the analysis provided an increased layer of complexity to the previously described interaction between hyaluronic acid and CD44.
An additional set of positive correlations was observed between expression of mipB and two host genes encoding the leukotriene B4 receptor (LTB4R) and cysteinyl leukotriene receptor 2 (CYSLTR2), membrane-bound G protein-coupled receptors that mediate the inflammatory response to eicosanoids.
GAS GTPase Signaling and Host Fibrinolytic Genes.
The GAS genome encodes several GTPase proteins that are widely conserved in bacteria, including many involved in ribosomal assembly and tRNA processing. Several GAS GTPases and host genes had correlated gene expression levels (Table S3). Intriguingly, expression of several of these GAS GTPases was correlated with expression of the host plasminogen (PLG) gene. For example, the GAS thdF gene had an inverse correlation with PLG expression. Although little is known about the function of many of these GTPases, thdF catalyzes the modification of certain tRNA species and is required for proper base-pairing during codon-anticodon recognition (21, 22). Inactivation of these genes induces frame-shifting and premature termination during translation (23), particularly affecting proteins with extensive regions of positive charge, such as DNA-binding transcription factors. Genetic inactivation of thdF results in decreased production of several virulence factors, with the notable exception that streptokinase expression increases (24). Although the exact mechanism is unknown, it has been hypothesized that decreased thdF results in decreased expression of a negative regulator of streptokinase (24). Thus, because streptokinase activates plasminogen cleavage, increased streptokinase levels would increase the rate of plasminogen activation to plasmin, leading to degradation of extracellular matrices and local fibrin deposits, thereby increasing GAS dissemination.
Several other host genes, including thrombin and macrophage stimulating protein (MSP), showed coregulation with multiple GAS GTPases. Plasminogen, thrombin, and MSP are each expressed as inactive zymogens that require cleavage for functional activation. All of these proteins have similar protein sequences, suggesting that a common factor, such as streptokinase, may act on all three (diagram shown in Fig S5). In this regard, we note that although traditionally considered components of blood, both plasminogen and MSP are present in the respiratory tract. Plasminogen is found in saliva and MSP is present in bronchoalveolar lavage fluid (25, 26). Additionally the MSP receptor, RON, is highly expressed in the upper respiratory tract where it regulates motility of ciliated epithelia (26). MSP activation of RON suppresses the inflammatory response by inhibiting nitric oxide production in macrophages and may represent a unique mechanism by which GAS evades the host immune response.
Adaptive Response of GAS to Polymorphonuclear Neutrophils.
One of the major advantages of our in vivo primate model of infection is the ability to study the complex interplay between the host immune response and GAS gene expression. Although the pharyngeal swabs contained mainly epithelial cells, phagocytic leukocytes were also present. Polymorphonuclear neutrophils (PMNs) are actively recruited to the local site of GAS infection, and thus interaction between GAS and host PMNs has been an area of considerable interest for decades. However, relatively little is known about GAS–PMN interaction in the posterior pharynx. The correlation analysis identified several coregulated host and pathogen genes that provide further insight into the adaptive response of GAS to activated PMNs.
One of these correlations in gene expression was between the GAS metK gene, encoding an S-adenosylmethionine (SAM) synthetase, and host genes encoding CD64 (FCGR1A) and azurocidin 1 (AZU1). Azurocidin is a cationic antimicrobial peptide sequestered in neutrophil secretory vesicles and azurophilic granules. The molecule is released during PMN priming or activation by granule exocytosis and is chemotactic for monocytes and macrophages (27). CD64 is a leukocyte glycoprotein receptor that mediates phagocytosis of microbes opsonized by IgG (28), thereby eliciting phagocyte production of microbicidal reactive oxygen species (ROS). Recent studies by Soehnlein et al. demonstrated that macrophage CD64 is up-regulated by neutrophil azurocidin (29). Although resting neutrophils generally express low levels of CD64 at the plasma membrane, surface expression increases following activation by proinflammatory stimuli (28). Therefore, increased expression of AZU1 and FCGR1A during GAS pharyngitis is consistent with either increased PMN influx at the site of infection or indicative of the PMN response to infection.
Inasmuch as SAM is a precursor for biosynthesis of glutathione, a molecule important for moderating the effects of ROS, increased production of SAM by GAS might occur in response to interaction with PMNs. As a first step toward testing this hypothesis, we measured levels of GAS metK transcript following interaction of GAS with human PMNs. Compared with bacteria alone, GAS metK transcript levels increased 2.03-fold following interaction of GAS with human neutrophils for 3 h (Fig. 4B). These findings are compatible with the idea that increases in PMN FCGR1A and AZU1 are countered at least indirectly by up-regulation of GAS metK transcript.
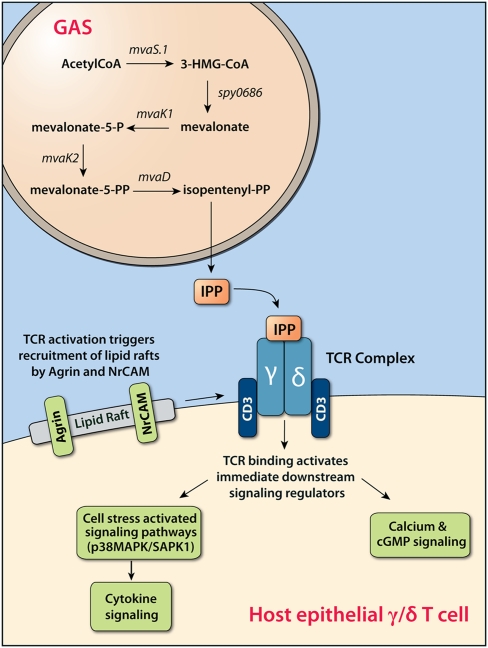
Signal for Activation of Host γδ T Cells by the GAS Mevalonic Acid Pathway.
One of the largest sets of host–pathogen interactions identified in the correlation analysis involved GAS genes in the mevalonic acid biosynthesis pathway. We identified correlations with four of the five GAS enzymes in the mevalonic acid pathway (mvaS.1, mvaK1, mvaK2, and mvaD) that convert acetyl-CoA to isopentenyl-pyrophosphate (IPP), with each step correlated to multiple overlapping host genes. IPP is short-chain phospholipid produced by GAS that is identical to IPP produced by host cells. In the host, accumulation of IPP is a marker for cellular stress and is detected by γδ T cells (30). These T cells recognize nonpeptide lipid antigens, with IPP being one of the primary host molecules. However, activation of host γδ T cells by IPP produced by GAS has largely been discounted because of low concentrations produced in vitro and the discovery that lipid intermediates produced by bacterial pathogens with a nonmevalonic IPP pathway (MEP) are over 10,000-fold more antigenic (31, 32).
Host genes correlated with the GAS mevalonic acid pathway fell into three distinct functional categories: receptors and receptor regulation, lipid raft formation/organization, and MAPK/SAPK signaling. Several of these genes are expressed only in neuronal tissue and T cells. These genes included agrin and the glutamate receptor GRIN1, both of which colocalize with the T-cell receptor (TCR) and are up-regulated in response to T cell activation (33, 34). Fig. 5 illustrates the effect of GAS IPP production on host γδ T cell signaling. In total, these correlated genes suggest that increased transcription of GAS mevalonic acid genes involved in production of IPP is correlated with a T cell-specific host transcriptional response characteristic of the molecular events triggered by antigen binding to the TCR. These events include the activation of the host cellular-stress signaling pathway. This contrasts with reports that GAS production of IPP does not specifically activate host T cells, including γδ T cells (31, 35). These conflicting data are likely explained by our use of an in vivo model where close physical proximity of GAS and host cells on the mucosal surface allows accumulation of IPP in the microenvironment. These GAS genes are positioned directly downstream of the sptR/sptS two-component system, an important regulator of GAS gene expression in saliva (10). We hypothesize that expression of these genes and consequent production of IPP enhances GAS survival in the oropharynx. Our findings suggest that study of the role of γδ T cells during GAS infection of the upper respiratory tract would be fruitful.
Fig. 5.
Relationship between increased production of IPP by GAS and signaling of γδ T cells. Up-regulation of mevalonic acid pathway genes (mvaS.1, spy0686, mvaK1, mvaK2, mvaD) results in increased production of IPP, which binds to the TCR of host γδ T cells present in the mucosal epithelium. Binding of IPP to the TCR stimulates recruitment of lipid rafts necessary for TCR signaling. Host genes involved in organization of lipid raft and receptor clustering, such as Agrin, Flotillin-2, and NrCAM, show increased expression levels to coordinate TCR and coreceptor signaling. The activated TCR initiates a signal transduction cascade leading to increased expression of host MAP3KIP1 and GADD45B, both members of the p38 MAP kinase pathway, and MAP4K1, a hematopoietic-specific signaling kinase that activates the cellular stress activated signaling pathway (SAPK1). Gamma-delta T cells are key defenders of the host mucosal surface and TCR signaling leading to increased proliferation or T-cell anergy is likely to influence the immune response to GAS.
Conclusion
This study provided unique insight into the host upper respiratory tract response to a common human bacterial pathogen, one result being many previously unexplored leads for pathogenesis research. Our strategy was distinct from previous investigations of host–pathogen interaction that relied on extensive in vitro yeast two-hybrid screens of protein–protein interaction or inferred interaction between host and pathogen transcriptomes based on previously described interactions reported in the literature. The unbiased strategy we used integrated paired host and pathogen transcriptomes during natural infection to identify interaction between individual host and pathogen genes and biological processes based on correlation of their gene expression profiles over time. Although it is important to interpret correlative microarray analyses with caution, our longitudinal interactome analysis yielded extensive data that can be exploited for subsequent hypothesis-driven research and translation research.
Materials and Methods
Bacterial Strain Cultivation.
GAS strain MGAS5005 (GenBank CP000017), genetically representative of contemporary serotype M1 strains, was grown as previously described (11, 12).
Infection Protocol.
Infection of cynomolgus macaques and sample preparation has been described extensively (11, 12). Briefly, animals were GAS-culture negative and had negligible antistreptolysin O titers, indicating no recent history of GAS exposure. Twenty animals were subjected to a mock-inoculation protocol (PBS only) for 5 weeks, rested for 4 weeks, and inoculated in the upper respiratory tract with 107 CFUs MGAS5005. Blood, saliva, and throat swabs were collected on days 0, 1, 2, 4, 7, 9, 16, 23, 32, 45, 58, 72, and 86. Only the first nine time-points were studied because specimens collected during days 0 to 32 had matching comparator specimens from the mock-infection protocol. Thirty-two clinical and laboratory parameters were measured by the same veterinarian during mock and infection periods. The study protocol was approved by the Animal Care and Use Committee, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases.
RNA Extraction, Microarray, and TaqMan Real-Time RT-PCR Protocols.
Details of these protocols are available in SI Materials and Methods.
Statistical Analysis of Host Gene Expression.
Analysis of the long-term host transcriptional response was determined by repeated-measures analysis of variance (RM-ANOVA) test. Partek Genomic Solution (Partek Inc.) was used to import Affymetrix CEL files, normalize with RMA and test by RM-ANOVA. The Greenhouse-Geisser and Huhn-Feldt corrections for nonsphericity were taken into account when determining significance. Geometric fold-changes ranged from −1.71 to +1.22. Although fold-changes appear low, this results from disease and gene expression changes varying across individual monkeys over the course of the experiment. Large and small gene-expression changes and clinical symptoms were observed within individual monkeys over time and in infected compared to mock. Fold-changes for individual subjects may be much higher. The large number of biological replicates (20) and sampling time-points (9) allowed for calculation of small fold-changes, demonstrating statistical significance. Short-term changes in the host transcriptional response were determined by comparing each pair of temporally adjacent time-points using an analysis of covariance (ANCOVA) test with corrections for variability because of time and kurtosis.
Significance Analysis of Function and Expression.
To assess the significant differential expression of gene categories, we used the permutation-based SAFE methodology (14) implemented in the statistical software R, and publicly available in the Bioconductor repository (http://www.bioconductor.org). Host categories were formed from GO using annotation packages for the array platform, and 61 functional categories in the GAS pathogen data were built as previously defined (36). The local statistics of gene-specific differential expression corresponded to t-statistics for the treatment effect from the ANCOVA model stated above. The Wilcoxon rank sum was used as the global statistic to measure increased amounts of differential expression in the gene categories. Empirical P values were computed from 10,000 permutations of the array assignment within each subject, along with the false-discovery rate for the sets significant findings (37). Data were grouped by pairs of temporally adjacent time points and analyzed to determine significant changes in gene expression from day to day. Analysis by both ANCOVA and SAFE was performed as noted above for each of the eight consecutive pairs of time-points.
Coexpression Correlation Analysis.
Spearman rank correlation was used to score all pairs of host and pathogen genes. P values for the total of 5.8 × 107 host–pathogen gene pairs were calculated for the correlation in the standard large-sample approximate manner. For each category and within each host–pathogen pair, the variance of the average across-species Spearman correlation was determined by repeatedly calculating the statistic against randomly generated expression profiles. P values were obtained in the large-sample approximate manner using the appropriately standardized forms of the observed Spearman correlations. Across-subject tests of the average correlation were performed under the assumption that host–pathogen pairs are independent. For comparing a host category to a pathogen category, no resampling-based procedure was available that appropriately accounts for the correlations within each organism. Thus, the most extreme average Spearman correlations are reported without providing a P value for the level of significance.
Supplementary Material
Acknowledgments
Quantitative PCR (TaqMan) validation and correlation analysis were performed by Hannah Wilder and Craig Martens. We thank K. Stockbauer for assistance with figures. This work was supported in part by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Raw .cel files for microarrays described in this manuscript have been deposited in the Gene Expression Omnibus (GEO) database (accession no. GSE2713 and GSE20262).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906384107/DCSupplemental.
References
- 1.Thompson LJ, et al. Transcriptional response in the peripheral blood of patients infected with Salmonella enterica serovar Typhi. Proc Natl Acad Sci USA. 2009;106:22433–22438. doi: 10.1073/pnas.0912386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.König R, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calderwood MA, et al. Epstein-Barr virus and virus human protein interaction maps. Proc Natl Acad Sci USA. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uetz P, et al. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 5.Lovegrove FE, et al. Simultaneous host and parasite expression profiling identifies tissue-specific transcriptional programs associated with susceptibility or resistance to experimental cerebral malaria. BMC Genomics. 2006;7:295. doi: 10.1186/1471-2164-7-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motley ST, et al. Simultaneous analysis of host and pathogen interactions during an in vivo infection reveals local induction of host acute phase response proteins, a novel bacterial stress response, and evidence of a host-imposed metal ion limited environment. Cell Microbiol. 2004;6:849–865. doi: 10.1111/j.1462-5822.2004.00407.x. [DOI] [PubMed] [Google Scholar]
- 7.Malke H, Steiner K, McShan WM, Ferretti JJ. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int J Med Microbiol. 2006;296:259–275. doi: 10.1016/j.ijmm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Shelburne SA, Davenport MT, Keith DB, Musser JM. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 2008;16:318–325. doi: 10.1016/j.tim.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham MR, et al. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am J Pathol. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shelburne SA, 3rd, et al. Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci USA. 2005;102:16037–16042. doi: 10.1073/pnas.0505839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Virtaneva K, et al. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 2003;71:2199–2207. doi: 10.1128/IAI.71.4.2199-2207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Virtaneva K, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dennis G, Jr, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 14.Barry WT, Nobel AB, Wright FA. Significance analysis of functional categories in gene expression studies: a structured permutation approach. Bioinformatics. 2005;21:1943–1949. doi: 10.1093/bioinformatics/bti260. [DOI] [PubMed] [Google Scholar]
- 15.Wessels MR, Moses AE, Goldberg JB, DiCesare TJ. Hyaluronic acid capsule is a virulence factor for mucoid group A streptococci. Proc Natl Acad Sci USA. 1991;88:8317–8321. doi: 10.1073/pnas.88.19.8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cywes C, Wessels MR. Group A Streptococcus tissue invasion by CD44-mediated cell signalling. Nature. 2001;414:648–652. doi: 10.1038/414648a. [DOI] [PubMed] [Google Scholar]
- 17.Cywes C, Stamenkovic I, Wessels MR. CD44 as a receptor for colonization of the pharynx by group A Streptococcus . J Clin Invest. 2000;106:995–1002. doi: 10.1172/JCI10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y, et al. CD44-epidermal growth factor receptor interaction mediates hyaluronic acid-promoted cell motility by activating protein kinase C signaling involving Akt, Rac1, Phox, reactive oxygen species, focal adhesion kinase, and MMP-2. J Biol Chem. 2008;283:22513–22528. doi: 10.1074/jbc.M708319200. [DOI] [PubMed] [Google Scholar]
- 19.Bourguignon LY, Gilad E, Brightman A, Diedrich F, Singleton P. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase C epsilon-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J Biol Chem. 2006;281:14026–14040. doi: 10.1074/jbc.M507734200. [DOI] [PubMed] [Google Scholar]
- 20.Takai E, et al. Correlation of translocation of tight junction protein Zonula occludens-1 and activation of epidermal growth factor receptor in the regulation of invasion of pancreatic cancer cells. Int J Oncol. 2005;27:645–651. [PubMed] [Google Scholar]
- 21.Martínez-Vicente M, et al. Effects of mutagenesis in the switch I region and conserved arginines of Escherichia coli MnmE protein, a GTPase involved in tRNA modification. J Biol Chem. 2005;280:30660–30670. doi: 10.1074/jbc.M503223200. [DOI] [PubMed] [Google Scholar]
- 22.Yim L, Moukadiri I, Björk GR, Armengod ME. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli . Nucleic Acids Res. 2006;34:5892–5905. doi: 10.1093/nar/gkl752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brégeon D, Colot V, Radman M, Taddei F. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 2001;15:2295–2306. doi: 10.1101/gad.207701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho KH, Caparon MG. tRNA modification by GidA/MnmE is necessary for Streptococcus pyogenes virulence: a new strategy to make live attenuated strains. Infect Immun. 2008;76:3176–3186. doi: 10.1128/IAI.01721-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moody GH. Plasminogen in human saliva. Int J Oral Surg. 1982;11:110–114. doi: 10.1016/s0300-9785(82)80019-7. [DOI] [PubMed] [Google Scholar]
- 26.Sakamoto O, et al. Role of macrophage-stimulating protein and its receptor, RON tyrosine kinase, in ciliary motility. J Clin Invest. 1997;99:701–709. doi: 10.1172/JCI119214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira HA, Shafer WM, Pohl J, Martin LE, Spitznagel JK. CAP37, a human neutrophil-derived chemotactic factor with monocyte specific activity. J Clin Invest. 1990;85:1468–1476. doi: 10.1172/JCI114593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fanger MW, Shen L, Graziano RF, Guyre PM. Cytotoxicity mediated by human Fc receptors for IgG. Immunol Today. 1989;10:92–99. doi: 10.1016/0167-5699(89)90234-X. [DOI] [PubMed] [Google Scholar]
- 29.Soehnlein O, et al. Neutrophil primary granule proteins HBP and HNP1-3 boost bacterial phagocytosis by human and murine macrophages. J Clin Invest. 2008;118:3491–3502. doi: 10.1172/JCI35740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka Y, et al. Natural and synthetic non-peptide antigens recognized by human gamma delta T cells. Nature. 1995;375:155–158. doi: 10.1038/375155a0. [DOI] [PubMed] [Google Scholar]
- 31.Jomaa H, et al. Vgamma9/Vdelta2 T cell activation induced by bacterial low molecular mass compounds depends on the 1-deoxy-D-xylulose 5-phosphate pathway of isoprenoid biosynthesis. FEMS Immunol Med Microbiol. 1999;25:371–378. doi: 10.1111/j.1574-695X.1999.tb01362.x. [DOI] [PubMed] [Google Scholar]
- 32.Puan KJ, et al. Preferential recognition of a microbial metabolite by human Vgamma2Vdelta2 T cells. Int Immunol. 2007;19:657–673. doi: 10.1093/intimm/dxm031. [DOI] [PubMed] [Google Scholar]
- 33.Jury EC, Eldridge J, Isenberg DA, Kabouridis PS. Agrin signalling contributes to cell activation and is overexpressed in T lymphocytes from lupus patients. J Immunol. 2007;179:7975–7983. doi: 10.4049/jimmunol.179.11.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miglio G, Varsaldi F, Lombardi G. Human T lymphocytes express N-methyl-D-aspartate receptors functionally active in controlling T cell activation. Biochem Biophys Res Commun. 2005;338:1875–1883. doi: 10.1016/j.bbrc.2005.10.164. [DOI] [PubMed] [Google Scholar]
- 35.Eberl M, et al. Microbial isoprenoid biosynthesis and human gammadelta T cell activation. FEBS Lett. 2003;544:4–10. doi: 10.1016/s0014-5793(03)00483-6. [DOI] [PubMed] [Google Scholar]
- 36.Graham MR, et al. Analysis of the transcriptome of group A Streptococcus in mouse soft tissue infection. Am J Pathol. 2006;169:927–942. doi: 10.2353/ajpath.2006.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yekutieli DBY. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J Statist Plann Inference. 1999;82:171–196. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.