Abstract
The ability of plants to adapt to changing light conditions depends on a protein kinase network in the chloroplast that leads to the reversible phosphorylation of key proteins in the photosynthetic membrane. Phosphorylation regulates, in a process called state transition, a profound reorganization of the electron transfer chain and remodeling of the thylakoid membranes. Phosphorylation governs the association of the mobile part of the light-harvesting antenna LHCII with either photosystem I or photosystem II. Recent work has identified the redox-regulated protein kinase STN7 as a major actor in state transitions, but the nature of the corresponding phosphatases remained unknown. Here we identify a phosphatase of Arabidopsis thaliana, called PPH1, which is specifically required for the dephosphorylation of light-harvesting complex II (LHCII). We show that this single phosphatase is largely responsible for the dephosphorylation of Lhcb1 and Lhcb2 but not of the photosystem II core proteins. PPH1, which belongs to the family of monomeric PP2C type phosphatases, is a chloroplast protein and is mainly associated with the stroma lamellae of the thylakoid membranes. We demonstrate that loss of PPH1 leads to an increase in the antenna size of photosystem I and to a strong impairment of state transitions. Thus phosphorylation and dephosphorylation of LHCII appear to be specifically mediated by the kinase/phosphatase pair STN7 and PPH1. These two proteins emerge as key players in the adaptation of the photosynthetic apparatus to changes in light quality and quantity.
Keywords: Photosynthesis, PP2C phosphatases, thylakoid, plastid
Plants are critically dependent on light as a source of energy to drive photosynthesis. However, in natural settings, both the intensity and the spectral quality of light vary extensively, sometimes within very short periods. Photosynthetic organisms posess an arsenal of mechanisms to adapt to such changes in their light environment, optimize photosynthesis, prevent photo-oxidation in excess light, and repair photo-damage (1, 2). These mechanisms operate on different time scales, ranging from seconds to days, and at all levels of organization, from the photosynthetic complexes in the thylakoid membranes to the morphology of the whole plant. Under low light intensity, light harvesting is maximized, but under excess light, acclimation responses lead to reduced light capture and enhanced energy dissipation.
Two photosystems, PSII and PSI, together with their associated light-harvesting antennae, function in series to drive linear electron flow in the thylakoid membranes, leading to the production of ATP and reductants such as reduced ferredoxin or NADPH. Cyclic electron flow around PSI allows synthesis of ATP without generating reducing power. Thus, the balance between linear and cyclic electron flow influences the ATP energy charge as well as the redox poise of the plant cell (2). The two photosystems have different light absorption characteristics; depending on the spectral composition of ambient light, a process called state transition regulates the relative cross-sections of their antennae to optimize linear electron flow (3 –5). In the green alga Chlamydomonas, state transitions also modulate cyclic electron flow and play a major regulatory role to respond to the metabolic requirements for ATP (6).
Two prominent features of state transitions are (i) the association of a mobile part of the LHCII antenna with either PSII or PSI, and (ii) changes in the structural organization of the thylakoid membranes. In state 1, the antenna is attached to PSII in grana stacks of the thylakoid membranes. In state 2, part of the antenna migrates and associates with PSI in stroma lamellae, grana margins, and grana ends, with a concomitant destacking of the thylakoid membranes (7, 8). State transitions are regulated by a protein kinase, called STN7 in Arabidopsis or Stt7 in Chlamydomonas, which is involved in the phosphorylation of some of the LHCII proteins (9 –11). The activity of the kinase is controlled by the redox state of the plastoquinone pool, or, more specifically, by binding of reduced plastoquinol to the Qo site of the b6f complex (12, 13). Thus, when light conditions favor the activity of PSII, reduction of the plastoquinone pool activates the STN7 kinase and causes a transition to state 2. The LHCII antenna is phosphorylated (5) and associates with PSI by binding to the PsaH subunit (14). The process is reversible, so that when PSI is more active and the plastoquinone pool is oxidized, the LHCII antenna is dephosphorylated and associates with PSII. Although the corresponding phosphatase activity has been assayed in thylakoid preparations, little is known on the molecular nature of the phosphatases involved in state transitions (15). Dephosphorylation of LHCII proteins was observed with isolated thylakoids, indicating that at least a portion of the phosphatase is membrane associated (16). It was further shown that thylakoid protein phosphatases are redox independent and kinetically heterogeneous (17). A 29-kDa stromal protein phosphatase was shown to act on LHCII in vitro (18). However, it is not clear whether this protein functions in the dephosphorylation of LHCII in vivo. Here we report the identification of a chloroplast protein phosphatase, PPH1, which is specifically required for efficient dephosphorylation of the LHCII antenna and transition from state 2 to state 1.
Results
Genetic Screen for Phosphatases Involved in State Transitions.
Comprehensive genomic surveys identified 159 genes that code for catalytic subunits of protein phosphatases in Arabidopsis (19 –21). We included all of these proteins, as well as others that are annotated in the Interpro database to contain domains of phosphatase regulatory subunits, in an initial candidate list. Their subcellular localization was predicted in silico using a panel of eight algorithms available through the Suba II web site (22). Those phosphatases that were predicted by at least one program to be targeted to the plastid were retained and were ordered according to the number of different algorithms that predicted plastid localization. Data from mass-spectrometry (MS) analysis of chloroplast proteins was also taken into account (23, 24). Coexpression of the putative chloroplast phosphatase genes with STN7 and STN8, based on an analysis of publicly available microarray data using the Genevestigator clustering tool (25) was used as a further criterion to rank the candidates. We obtained 84 homozygous T-DNA insertion lines disrupting the genes encoding 60 putative chloroplast phosphatases, and systematically analyzed seedlings for defects in protein dephosphorylation during a transition from state 2 to state 1, using the immunoblotting assay described in the next section. One of the high-ranking candidates, pph1-1, showed a strong defect in dephosphorylation of the LHCII antenna, and was selected for further analysis.
PPH1 Phosphatase Is Required for Dephosphorylation of LHCII Antenna.
When Arabidopsis seedlings are exposed to moderate levels of white light (50 μE m−2·s−1), their photosynthetic electron transfer chain is largely in state 2. Reduction of the plastoquinone pool leads to the association of the mobile part of the LHCII antenna with PSI and to the phosphorylation of the Lhcb proteins, which can be monitored by immunoblotting with antiphosphothreonine antibodies (Fig. 1A) (26). Other proteins of the thylakoid membrane are also phosphorylated, such as CP43, D1 and D2, which are subunits of the PSII core (27). Upon exposure to far-red light, excitation of PSI is favored over PSII, so that the plastoquinone pool is oxidized and the system shifts to state 1. The mobile LHCII antenna associates with PSII and the LHCII proteins are dephosphorylated (Fig. 1A). There is also a decrease in the phosphorylation of D1 and D2, and to a lesser degree of CP43. Phosphorylation of LHCII is strongly reduced after 20 min of exposure to far-red light and reaches a low level after 40 min in our experimental conditions. A similar effect is observed when seedlings are transferred from white light to the dark. Phosphorylation is rapidly restored if the seedlings are transferred from far-red light to blue light, which induces a transition to state 2.
Fig. 1.
Analysis of thylakoid protein dephosphorylation. (A) Dephosphorylation of thylakoid proteins. Twelve-day-old Col0 wild-type seedlings grown under white light were exposed to far-red light or transferred to the dark for the indicated times to induce a transition from state 2 to state 1. Seedlings treated with far red for 1 h were then exposed to blue light to induce the reverse transition from state 1 to state 2. Total protein was extracted and was analyzed by SDS/PAGE and immunoblotting with antiphosphothreonine antibodies. (B) The pph1 mutants are deficient in LHCII dephosphorylation under far-red light. Seedlings of the Col0 wild-type (WT), of pph1-1, and of pph1-2 were treated with far-red light and analyzed as in A. The same blot was decorated with antibody against the PsaA subunit of PSI as a control. (C) The pph1 mutants are deficient in dark-induced LHCII dephosphorylation. Four hours after the onset of the light period, mature leaves were collected and the plants were transferred to the dark for 5 h. Total protein was extracted and analyzed as in A. Note that the defect in dephosphorylation is more pronounced in pph1-3 than in pph1-1, which is a leaky allele containing residual amounts of PPH1 mRNA (Fig. S5).
In striking contrast, in pph1 mutants, LHCII remained strongly phosphorylated after 20 min of far-red light treatment and showed only a moderate decrease after 40 min (Fig. 1B). The effect appears to be specific for LHCII because the core subunits of PSII underwent dephosphorylation as in the wild type. Impaired dephosphorylation of LHCII proteins was similarly observed in pph1 mutants when a transition from state 2 to state 1 was induced by transferring adult plants from moderate white light to the dark (Fig. 1C). These observations identified the PPH1 phosphatase as an essential component required for the dephosphorylation of the LHCII antenna, but not of the PSII core, during a transition from state 2 to state 1. Although in pph1 mutants dephosphorylation was impaired during a transition to state 1, there was no apparent hyperphosphorylation of the Lhcb proteins under the conditions favoring state 2, which were used for growing the seedlings (moderate white light, 50 μE m−2·s−1).
Analysis of in Vivo Protein Phosphorylation in pph1-1 by MS.
Immunoblotting analysis of Arabidopsis seedlings exposed to far-red light showed that phosphorylation of LHCII proteins was significantly reduced in the wild-type but not in pph1-1 plants (Fig. 1B). The LHCII antenna comprises many isoforms of the Lhcb proteins. To determine more precisely which Lhcb polypeptides were not dephosphorylated in the mutant, we analyzed these proteins using differential stable isotope labeling and MS (SI Text). After exposure to far-red light, thylakoid membranes were isolated in parallel from the pph1-1 mutant and wild-type plants in the presence of NaF to inhibit dephosphorylation (28). The surface-exposed peptides from the wild-type and the mutant membranes were prepared by proteolytic shaving and were differentially labeled by esterification of carboxylic groups with hydrogen- or deuterium-containing methanol, respectively (29, 30). A 1:1 mixture of these two preparations was subjected to IMAC (immobilized metal ion affinity chromatography) so as to capture and enrich the phosphorylated peptide methyl esters. The phosphorylated peptides enriched by IMAC were then subjected to nano liquid chromatography and electrospray ionization MS (LC-MS), which allowed simultaneous measurements of light and heavy isotope–labeled phosphopeptide pairs. We also performed the reverse labeling of the wild-type and mutant peptides as an internal control and additional experiment for relative quantification of differentially labeled peptides. The difference in intensities of light and heavy phosphorylated peptides provided quantitative data for the phosphorylation differences between the mutant and wild-type after a transition from state 2 to state 1 induced with far-red light. (Fig. S1, Fig. S2, and Fig. S3).
The LC-MS analyses (Table 1, and Fig. S1, Fig. S2, and Fig. S3) revealed very similar levels of phosphorylation for the photosystem II core proteins D1 and D2, but marked differences for the Lhcb proteins in the mutant compared with the wild type. In our analyses, we found two phosphorylated peptides from LHCII proteins. However, it should be noted that because the Lhcb isoforms have very similar sequences, a specific peptide sequence can originate from several gene products. Importantly, we found that each of these two phosphopeptides was present in significantly higher amounts in the mutant samples than in the wild type (Table 1). Thus, our data reveal that PPH1 deficiency resulted in significantly higher phosphorylation of LHCII peptides, which could correspond to up to seven different Lhcb gene products.
Table 1.
MS quantification of thylakoid protein phosphorylation in pph1-1 and wild type
| Phosphopeptide sequence | Protein name | At gene identifier | Mutant/wild type ratio | |
| LHCII | Ac-RKtVAKPK | LHCB1.1 | At1g29910 | 3.6 ± 0.2 |
| LHCB1.2 | At1g29920 | |||
| LHCB1.3 | At1g29930 | |||
| LHB1B2 (LHCB1.5) | At2g34420 | |||
| Ac-RRtVK | LHCB2.1 | At2g05100 | >13 (*) | |
| LHCB2.2 | At2g05070 | |||
| LHCB2.4 | At3g27690 | |||
| PSII core | Ac-tAILER | D1 | AtCg00020 | 1.1 ± 0.2 |
| Ac-tIALGK | D2 | AtCg00270 | 1.5 ± 0.3 |
Sequence of peptides is indicated with their modifications: Ac, N-terminal acetyl group; t, phosphorylated threonine residue. Mutant/wild-type ratios were determined in every LC-MS experiment from the areas of extracted ion chromatograms for each of the differentially labeled phosphorylated peptides corresponding to a particular protein. Data are the means (± SD) from four different experiments.
*Ac-RRtVK phosphopeptide signal was not detected in the wild-type sample in three experiments.
Alternative Splicing of PPH1 mRNA.
Three alternative transcript models are annotated for the PPH1 gene on the Arabidopsis Information Resource (TAIR) web site (31). The longest ORF corresponds to a fully spliced mRNA that contains 11 exons and is predicted to encode a 43-kDa polypeptide (AT4G27800.1; Fig. S4). Reverse transcription followed by PCR and sequencing confirmed the presence of this mature mRNA in rosette leaves (Fig. S5A). A longer alternatively spliced form of the mRNA was also observed (AT4G27800.2), which retains intron 9 and is predicted to encode a truncated 36 kDa polypeptide that lacks some of the highly conserved residues of PP2C phosphatases (Fig. S4). The longer predicted isoform of PPH1 has a single potential hydrophobic transmembrane helix, while the shorter predicted isoform may be a soluble protein (Fig. S4). We could not confirm the presence of a third form of the transcript, which would have contained a 4-bp insert at the end of exon 8. We obtained three alleles with T-DNA insertions in the PPH1 gene (Fig. S4). Two alleles, pph1-1 and pph1-2, have insertions in the 5′UTR of the gene. Another allele, pph1-3, has an insertion in exon 4.
To assess whether the mutations are null or hypomorphic, we examined the level of expression of the PPH1 gene. RNA was extracted from leaves of wild-type and pph1 mutant plants and was analyzed by RNA blotting with a PPH1 probe. The pph1-2 and pph1-3 plants had no detectable PPH1 mRNA. However the pph1-1 mutant showed residual levels of PPH1 mRNA, which accumulated to approximately one third of the amount in the wild-type (Fig. S5B). These data indicate that the pph1-1 mutant is leaky and that residual levels of the phosphatase might be present in the pph1-1 plants. This suggests that only partial defects should be expected in the phenotypic analysis of the pph1-1 mutant plants.
PPH1 Phosphatase Is a Chloroplast Protein.
To determine whether PPH1 is targeted to the chloroplast, 35S-labeled PPH1 preprotein was obtained by in vitro translation and presented to isolated chloroplasts in an in vitro import assay (Fig. 2A). After the incubation, the 44-kDa precursor was converted to a shorter form, which was protected from exogenous protease, showing that pre-PPH1 is imported and processed in the chloroplast to a mass corresponding to that of the predicted mature form. Moreover, the radio-labeled PPH1 precursor was also efficiently imported into isolated pea chloroplasts and processed to the mature form, which was protected from exogenous protease (Fig. 2B).
Fig. 2.
PPH1 is a chloroplast protein. (A) In vitro import into Arabidopsis chloroplasts. Radio-labeled PPH1 precursor obtained by in vitro translation was incubated for the indicated times with chloroplasts isolated from Arabidopsis leaves. After the incubation, an aliquot was treated with thermolysin (TLysin) as specified. The proteins were extracted and analyzed by SDS/PAGE and phosphorimaging. (B) In vitro import into pea chloroplasts. (C) Transient expression of PPH1::GFP in tobacco. Nicotiana benthamiana leaves were infiltrated with Agrobacterium containing the 35S::PPH1::GFP construct. After 2 days, protoplasts were isolated and observed under the confocal microscope. The four columns show chlorophyll autofluorescence, GFP fluorescence, merged images of the two, and transmission images. (D) Nontransformed protoplast as negative control.
To investigate the subcellular localization of the protein in vivo, a PPH1::GFP gene fusion under the CaMV 35S promoter was used to transform Nicotiana benthamia leaf cells by agroinfiltration. Confocal microscopy confirmed that the PPH1::GFP fusion localizes to the chloroplast (Fig. 2 C and D). The GFP signal formed distinct spots that suggested the fusion protein was localizing to a subcompartment of the chloroplast. A similar pattern was previously observed with STN7::GFP (9).
To determine where PPH1 is localized inside chloroplasts, we generated an antibody against a specific peptide sequence in the central part of the mature protein. This antibody recognized a band migrating at ≈40 kDa in immunoblots of wild-type chloroplast proteins (Fig. 3A). This corresponds to the size of the predicted mature form, and of the processed form observed in the in vitro import experiments. Comparison of chloroplasts from pph1-1 mutant and wild-type plants demonstrated a significant reduction of the 40-kDa band, which can thus be identified as PPH1 (Fig. 3A). These results are consistent with our finding that the pph1-1 mutant showed reduced levels of PPH1 mRNA (Fig. S5B). The nature of a cross-reacting band that migrated slightly faster is presently not known; it was detected only when the proteins were separated in PAGE gels containing urea (Fig. 3A), but not in those without urea (Fig. 3B). Immunoblots of membrane/thylakoid and soluble chloroplast fractions showed that a large portion of the PPH1 protein was present in the thylakoid membranes. Although it was also detected in the soluble fraction, its relative amount compared with the large subunit of Rubisco was much lower than in the total chloroplast sample, indicating that PPH1 is not enriched in the stroma. Further fractionation of thylakoid membrane domains by centrifugation after treatment with digitonin revealed that the phosphatase was highly enriched in stroma lamellae, whereas its signal in the grana fraction was significantly lower (Fig. 3B). These data indicate that, in chloroplasts, a large portion of PPH1 resides at the stroma lamellae of thylakoid membranes.
Fig. 3.
Localization of PPH1 in chloroplast subfractions. (A) Immunoblot analysis of proteins from intact chloroplasts, membrane fraction (thylakoids), and soluble fraction (stroma) of chloroplasts isolated from wild-type (WT) and pph1-1 plants with antibody against PPH1. Antibodies against reference proteins were used as controls for the stromal (Rubisco) and thylakoid (Lhcb1) fractions. The samples contained 1 μg of chlorophyll for the chloroplast and thylakoid samples, and 1.5 μg of protein for the stroma. (B) Immunoblot analysis of thylakoid membrane subfractions (Tk, thylakoids; G+M, grana + margins; SL, stroma lamellae) isolated from wild-type plants with antibody against PPH1. (Lower) Immunodetection with antibodies against D1 subunit of PSII and PsaF subunit of PSI. Loading of samples was on an equal chlorophyll basis (1 μg for PPH1; 0.2 μg for D1 and PsaF).
PPH1 Phosphatase Is Involved in State Transitions.
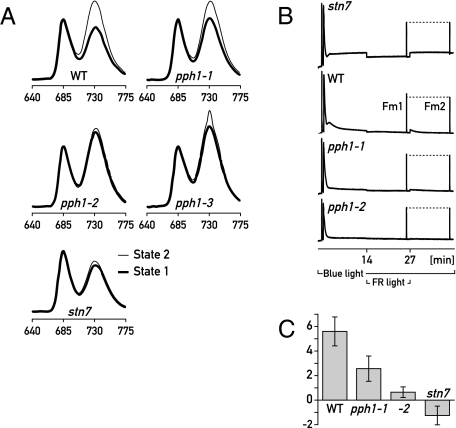
In state transitions, association of part of LHCII with either PSII or PSI causes changes in the relative cross-sections of their respective antennae. This can be monitored by determining the low-temperature fluorescence emission spectra of crude thylakoid preparations (Fig. 4A). For this analysis, leaves in state 1 and state 2 were obtained by exposure to far-red and blue light, respectively. As a control, the stn7 kinase mutant that is deficient in state transitions was also included in the analysis. A significant increase in PSI fluorescence was observed in wild-type seedlings in state 2 relative to state 1 (Fig. 4A). This difference was abolished in the stn7 kinase mutant, which is locked in state 1 and did not show an increased PSI fluorescence under state 2 conditions. In pph1-1, the differences in PSI fluorescence levels observed between state 1 and state 2 conditions were significantly lower compared with the wild type, and in pph1-2 and pph1-3 they were reduced even more strongly. Under state 1 conditions, the pph1 mutant seedlings showed higher relative PSI fluorescence than in the wild type. This suggests that the defect in dephosphorylation caused a bias toward state 2, with a tendency for the mobile antenna to be retained at PSI.
Fig. 4.
Analysis of state transitions. (A) Chlorophyll fluorescence emission spectra at low temperature. Rosette leaves from Col0 wild type, three pph1 alleles, and stn7 grown in short days under white light were treated for 30 min under far-red (state 1 conditions) or blue light (state 2 conditions). Crude extracts were frozen in liquid nitrogen, and fluorescence emission spectra were obtained with excitation at 480 nm. Curves were normalized for the PSII peak at 685 nm (average of four measurements). Fluorescence emission peak from PSI was at ≈732 nm. (B) Chlorophyll fluorescence analysis at room temperature. State transitions were measured with a pulse amplitude–modulated fluorimeter to monitor changes in the light-harvesting antenna associated with PSII. A dark-adapted leaf was exposed to blue light to induce state 2, then with additional far-red light to promote a transition to state 1, and was induced to return to state 2 after the far-red light was switched off. State transitions were estimated as the difference in maximal fluorescence in state 1 (Fm1) and in state 2 (Fm2), which was abolished in stn7, strongly reduced in pph1-1, and barely detectable in pph1-2. State transitions are also denoted by small changes in fluorescence when the lights are switched. These reflect changes in the redox state of the plastoquinone pool. In stn7, where the LHCII antenna stayed associated with PSII in blue light and the plastoquinone pool remained reduced, the decrease in fluorescence when the far-red light was turned on was more pronounced than in the wild type, where the state transition led to a gradual oxidation of the plastoquinone pool in blue light. As could be expected for a phosphatase mutant that favors state 2, only minor fluorescence changes were observed in pph1-1 and pph1-2 because the plastoquinone pool stayed predominantly oxidized. (C) Quantification of state transitions measured as in B: qT = (Fm1 − Fm2)/Fm1 × 100 (average of four different leaves, mean ± SD).
State transitions can also be measured using pulse amplitude–modulated chlorophyll fluorescence spectroscopy at room temperature, to monitor changes in the light harvesting antenna associated with PSII. In this analysis, state transitions were strongly impaired in the pph1-2 allele and were also affected in pph1-1 (Fig. 4 B and C). The data also suggested that in the pph1 mutants the plastoquinone pool tended to remain more oxidized than in the wild type, as might be expected if phosphatase deficiency favored the association of the mobile LHCII antenna with PSI.
Discussion
Reversible phosphorylation is a widespread protein modification involved in the regulation of metabolism and in many signal transduction pathways. State transitions are a particularly striking example because the phosphorylation of thylakoid proteins causes a profound reorganization of the electron transfer chain and remodeling of the thylakoid membrane system. Together with the b6f complex, the STN7 kinase acts as a sensor of the redox poise of the electron transfer chain, and relays this signal by phosphorylating numerous target proteins. This regulates both short term responses such as state transitions and long-term responses involving changes in the composition of the photosynthetic machinery (32). To allow acclimation to fluctuating light conditions and homeostasis, such responses must be reversible, and therefore require the activity of phosphatases. Here we identify the protein phosphatase PPH1, which is largely responsible for the dephosphorylation of the LHCII antenna.
PPH1 belongs to the PP2C type of protein phosphatases. This family of monomeric enzymes has been widely expanded in plants, and includes 76–80 genes in Arabidopsis or 78 in rice, compared with only 18 in humans (20, 21). Phylogenetic comparisons show that PPH1 belongs to a distinct clade together with similar proteins from other plants, but does not have close paralogs in Arabidopsis (20, 33) (Fig. S6). Members of this clade share a characteristic additional domain (residues 242–274), featuring many basic amino acids, which is absent in other PP2C phosphatases. Although it cannot be excluded that the role of PPH1 is indirect, the strong impairment of LHCII dephosphorylation in pph1 mutants suggests that PPH1 is the major phosphatase for the LHCII proteins and that other phosphatases may play only a minor role. This lack of redundancy is consistent with the absence of close PPH1 paralogs in Arabidopsis, but is surprising because protein phosphatases are generally thought to be rather unspecific. In turn, PPH1 appears to be specific for the Lhcb polypeptides, as the dephosphorylation of the core proteins D1, D2, and CP43 of PSII is not affected in the mutants (Fig. 1). In state 2, phosphorylated LHCII is associated with PSI in the stroma-exposed parts of the thylakoid membrane network. Thus, the localization of PPH1 at the thylakoid membranes and particularly at stroma lamellae is consistent with the role of PPH1 in LHCII dephosphorylation and transition from state 2 to state 1.
Using reverse transcription and PCR, we have detected an alternatively spliced form of the PPH1 mRNA in mature leaves. This form retains intron 9, which leads to a frame shift of the coding sequence and a premature stop codon. The predicted shorter truncated protein would lack residues that are highly conserved in PP2C phosphatases, as well as the putative hydrophobic transmembrane domain (Fig. S4). It will be of interest to determine whether the alternative forms are differentially regulated and have different functions.
It has been clearly demonstrated that the STN7 kinase is required for the phosphorylation of LHCII and for transitions from state 1 to state 2. Because LHCII is the major target of STN7, this provides evidence that phosphorylation of the antenna plays a key role in state transitions. Consistent with this model, we have found that the PPH1 phosphatase, which is required for dephosphorylation of LHCII, is required for efficient transitions from state 2 to state 1. This was apparent both from chlorophyll fluorescence emission spectra at low temperature and from chlorophyll fluorescence analysis at room temperature (Fig. 4). Thus, the STN7–PPH1 pair appears to be specific for the phosphorylation and dephosphorylation of the LHCII proteins respectively, and this correlates with their respective roles in state transitions.
After growth under conditions that favor state 2 (moderate white light), there is no apparent hyper-phosphorylation of LHCII in the pph1 mutants. This is perhaps not surprising, as the activity of the LHCII kinase STN7 is known to be regulated by the redox state of the plastoquinone pool. In a pph1 mutant, increased phosphorylation would result in enhanced association of the antenna with PSI. Increased PSI activity would in turn lead to oxidation of the plastoquinone pool, and therefore to reduced STN7 activity and diminished LHCII phosphorylation. Whether this negative feedback loop solely relies on the regulation of STN7 activity for homeostasis or whether the PPH1 phosphatase is also regulated remains to be determined.
In the pph1-1 mutant, the levels of the PPH1 mRNA are reduced approximately 3-fold compared with those of the wild type. The drastic effect of this mutation on the dephosphorylation of the LHCII antenna indicates that the reduced expression of PPH1 mRNA strongly limits the levels of phosphatase activity in the mutant, and thus that there is probably no excess of the mRNA in the wild type. This may be important in appropriately balancing the level of activity of the phosphatase and of the STN7 kinase. This was also reflected in the analysis of state transitions, where we found that the defect was more pronounced in the stronger alleles (pph1-2 and pph1-3) than in the leaky allele (pph1-1). Consistently with this observation, dephosphorylation of LHCII in dark-adapted leaves was also more affected in a strong allele than in the weaker allele (Fig. 1C).
The phosphatase activities in isolated thylakoids were previously found to be kinetically heterogeneous, with LHCII being dephosphorylated fastest, followed by D1 and D2, and then CP43 and PsbH (17). The specificity of PPH1 for LHCII proteins that we have observed supports the hypothesis that several phosphatases must be involved in dephosphorylation of thylakoid phosphoproteins. Thus, the kinetic differences could be due either to the different properties of distinct enzymes or to some different properties of the substrates. Although in vitro the phosphatase activities were reported not to be redox regulated (17), evidence has also been presented that they could be modulated by light or reducing agents (34). Our identification of PPH1 will facilitate future studies on the role and regulation of thylakoid protein phosphatases in light acclimation.
Materials and Methods
Plant Material and Growth Conditions.
For the genetic screen, Arabidopsis thaliana wild-type plants and T-DNA insertion mutants (all of Columbia-0 ecotype) were grown on Murashige-Skoog agar supplemented with 1.5% sucrose in a growth chamber (Percival CU36L5) in long days (16 h light, 8 h dark) at 22 °C under fluorescent white light (50 μE m−2·s−1). Plants on soil were cultivated in growth chambers at 24 °C under white light (100 μE m−2·s−1) in long days. Plants for chlorophyll fluorescence measurements were grown in short days (8 h light, 16 h dark). The T-DNA insertion lines (http://methylome.salk.edu/cgi-bin/homozygotes.cgi) (35) were obtained through the European Arabidopsis Stock Centre (NASC); pph1-1: SALK_025713C; pph1-2: SAIL_514-C03; pph1-3: GABI_232H12. The stn7 mutant was previously described (9). Primers for genotyping are described in SI Text.
Bioinformatic Tools.
Searches for protein domains were performed on the InterPro web site (http://www.ebi.ac.uk/interpro/).
Subcellular localization was predicted with Suba II (http://www.plantenergy.uwa.edu.au/applications/suba/index.php).
Genomic data for Arabidopsis were obtained from the Arabidopsis Information Resource web site (TAIR; http://arabidopsis.org), and T-DNA insertion lines were searched on the Salk web site (http://methylome.salk.edu/cgi-bin/homozygotes.cgi).
Assays of Thylakoid Protein Dephosphorylation.
For far-red light treatment, 12-day old seedlings grown on agar plates were placed under panels of infrared LEDs (L735; Epitex). Liquid nitrogen frozen samples were ground 4 × 10 s in a triturator (Silamat S5; Ivoclar Vivadent AG) using precooled glass beads. The frozen powder was thawed in 200 μL of lysis buffer containing 100 mM Tris-HCl, pH 7.7, 2% SDS, 50 mM NaF, and 2× Protease Inhibitor Mixture (Sigma-Aldrich) and incubated for 30 min at 37 °C. The lysate was centrifuged for 10 min at 14,000 rpm at room temperature. The samples (0.3 μg Chl/lane) were separated by Tris-glycine SDS/PAGE in 12% acrylamide gels containing 6 M urea. Immunoblotting was performed using antiphosphothreonine antibodies (Cell Signaling). The details of the quantitative MS analysis are provided in SI Text.
In Vitro Import into Isolated Chloroplasts and Localization of PPH1::GFP in Tobacco.
In vitro import into isolated chloroplasts was performed as described for pea and Arabidopsis (36, 37). Leaves of Nicotiana benthamiana were infiltrated with Agrobacterium (35S::PPH1::GFP). After 48 h, protoplasts were isolated and fluorescence was monitored using a laser scanning confocal microscope as described in SI Text.
Cell Fractionation.
Chloroplasts and thylakoids were isolated essentially as previously described (38). The soluble stromal fraction was obtained as the supernatant after lysis of the chloroplasts. Fractionation of thylakoids into grana and stroma lamellae was performed by the use of digitonin. Thylakoids at a concentration of 0.6 mg of chlorophyll/mL were incubated with 1% digitonin (Sigma-Aldrich) for 5 min and then centrifuged at 1,000 × g. The supernatant was collected and centrifuged at 40,000 × g for 40 min to pellet grana membranes and grana margins. The remaining supernatant was centrifuged at 140,000 × g to collect stroma lamellae.
Immunoblotting was performed essentially as described previously (39) by SDS/PAGE using 14% acrylamide gels. Polyclonal antibody to PPH1 was raised against a synthetic peptide corresponding to residues 186–199 (Innovagen). Antibodies toward D1, PsaF, Rubisco large subunit, and Lhcb1 were purchased from Agrisera.
Chlorophyll Fluorescence Measurements.
Detached rosette leaves from short day-grown plants floating on distilled water in Petri dishes were placed under panels of far-red or blue LEDs (L735 and L470; Epitex) for 30 min to induce state 1 or state 2. Leaves were then frozen in liquid nitrogen and membrane extracts were prepared (SI Text). Chlorophyll fluorescence at 77 K was recorded on a Jasco FP-750 spectrofluorometer. Excitation was at 480 nm (slit width, 5 nm) and emission was recorded in the 600- to 800-nm range (slit width, 5 nm). Room temperature chlorophyll fluorescence was recorded using an FMS1 Pulse Modulated Fluorimeter (Hansatech) as detailed in SI Text.
Supplementary Material
Acknowledgments
This work was supported by grants from SystemsX.ch (RTD project “Plant Growth in a Changing Environment”; to M.G.-C., J.-D.R., and F.K.); by grants from the Swedish Research Council, the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (to A.V.V.), and from the NCCR Plant Survival and the Swiss National Foundation (3100AO-117712; to J.-D.R. and M.G.-C.). We thank Cyril Montandon and Birgit Agne for help with the in vitro import assays, Etienne Bucher for an actin probe, and Nicolas Roggli for help with the figures.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913810107/DCSupplemental.
References
- 1.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 2.Eberhard S, Finazzi G, Wollman FA. The dynamics of photosynthesis. Annu Rev Genet. 2008;42:463–515. doi: 10.1146/annurev.genet.42.110807.091452. [DOI] [PubMed] [Google Scholar]
- 3.Wollman FA. State transitions reveal the dynamics and flexibility of the photosynthetic apparatus. EMBO J. 2001;20:3623–3630. doi: 10.1093/emboj/20.14.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rochaix JD. Role of thylakoid protein kinases in photosynthetic acclimation. FEBS Lett. 2007;581:2768–2775. doi: 10.1016/j.febslet.2007.04.038. [DOI] [PubMed] [Google Scholar]
- 5.Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim Biophys Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- 6.Finazzi G, et al. Involvement of state transitions in the switch between linear and cyclic electron flow in Chlamydomonas reinhardtii. EMBO Rep. 2002;3:280–285. doi: 10.1093/embo-reports/kvf047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tikkanen M, et al. Phosphorylation-dependent regulation of excitation energy distribution between the two photosystems in higher plants. Biochim Biophys Acta. 2008;1777:425–432. doi: 10.1016/j.bbabio.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Chuartzman SG, et al. Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell. 2008;20:1029–1039. doi: 10.1105/tpc.107.055830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellafiore S, Barneche F, Peltier G, Rochaix JD. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- 10.Depege N, Bellafiore S, Rochaix JD. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- 11.Bonardi V, et al. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- 12.Vener AV, van Kan PJ, Rich PR, Ohad I, Andersson B. Plastoquinol at the quinol oxidation site of reduced cytochrome bf mediates signal transduction between light and protein phosphorylation: Thylakoid protein kinase deactivation by a single-turnover flash. Proc Natl Acad Sci USA. 1997;94:1585–1590. doi: 10.1073/pnas.94.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zito F, et al. The Qo site of cytochrome b6f complexes controls the activation of the LHCII kinase. EMBO J. 1999;18:2961–2969. doi: 10.1093/emboj/18.11.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lunde C, Jensen PE, Haldrup A, Knoetzel J, Scheller HV. The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature. 2000;408:613–615. doi: 10.1038/35046121. [DOI] [PubMed] [Google Scholar]
- 15.Aro EM, Ohad I. Redox regulation of thylakoid protein phosphorylation. Antioxid Redox Signal. 2003;5:55–67. doi: 10.1089/152308603321223540. [DOI] [PubMed] [Google Scholar]
- 16.Bennett J. Chloroplast phosphoproteins. Evidence for a thylakoid-bound phosphoprotein phosphatase. Eur J Biochem. 1980;104:85–89. doi: 10.1111/j.1432-1033.1980.tb04403.x. [DOI] [PubMed] [Google Scholar]
- 17.Silverstein T, Cheng L, Allen JF. Chloroplast thylakoid protein phosphatase reactions are redox-independent and kinetically heterogeneous. FEBS Lett. 1993;334:101–105. doi: 10.1016/0014-5793(93)81690-2. [DOI] [PubMed] [Google Scholar]
- 18.Hammer MF, Markwell J, Sarath G. Purification of a protein phosphatase from chloroplast stroma capable of dephosphorylating the light-harvesting complex-II. Plant Physiol. 1997;113:227–233. doi: 10.1104/pp.113.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerk D, et al. The complement of protein phosphatase catalytic subunits encoded in the genome of Arabidopsis. Plant Physiol. 2002;129:908–925. doi: 10.1104/pp.004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerk D, Templeton G, Moorhead GB. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008;146:351–367. doi: 10.1104/pp.107.111393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue T, et al. Genome-wide and expression analysis of protein phosphatase 2C in rice and Arabidopsis. BMC Genomics. 2008;9:550. doi: 10.1186/1471-2164-9-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heazlewood JL, Verboom RE, Tonti-Filippini J, Small I, Millar AH. SUBA: The Arabidopsis Subcellular Database. Nucleic Acids Res. 2007;35:D213–D218. doi: 10.1093/nar/gkl863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Wijk KJ. Plastid proteomics. Plant Physiol Biochem. 2004;42:963–977. doi: 10.1016/j.plaphy.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Baerenfaller K, et al. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320:938–941. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann P, Hennig L, Gruissem W. Gene-expression analysis and network discovery using Genevestigator. Trends Plant Sci. 2005;10:407–409. doi: 10.1016/j.tplants.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Rintamaki E, et al. Phosphorylation of light-harvesting complex II and photosystem II core proteins shows different irradiance-dependent regulation in vivo. Application of phosphothreonine antibodies to analysis of thylakoid phosphoproteins. J Biol Chem. 1997;272:30476–30482. doi: 10.1074/jbc.272.48.30476. [DOI] [PubMed] [Google Scholar]
- 27.Vener AV. Environmentally modulated phosphorylation and dynamics of proteins in photosynthetic membranes. Biochim Biophys Acta. 2007;1767:449–457. doi: 10.1016/j.bbabio.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Vener AV, Rokka A, Fulgosi H, Andersson B, Herrmann RG. A cyclophilin-regulated PP2A-like protein phosphatase in thylakoid membranes of plant chloroplasts. Biochemistry. 1999;38:14955–14965. doi: 10.1021/bi990971v. [DOI] [PubMed] [Google Scholar]
- 29.Vener AV, Harms A, Sussman MR, Vierstra RD. Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J Biol Chem. 2001;276:6959–6966. doi: 10.1074/jbc.M009394200. [DOI] [PubMed] [Google Scholar]
- 30.Ficarro SB, et al. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 31.Swarbreck D, et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesaresi P, et al. Arabidopsis STN7 kinase provides a link between short- and long-term photosynthetic acclimation. Plant Cell. 2009;21:2402–2423. doi: 10.1105/tpc.108.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Hammer MF, Gautam S, Osterman JC, Markwell J. Assessing modulation of stromal and thylakoid light-harvesting complex-II phosphatase activities with phosphopeptide substrates. Photosynth Res. 1995;44:107–115. doi: 10.1007/BF00018301. [DOI] [PubMed] [Google Scholar]
- 35.Alonso JM, Ecker JR. Moving forward in reverse: Genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet. 2006;7:524–536. doi: 10.1038/nrg1893. [DOI] [PubMed] [Google Scholar]
- 36.Agne B, et al. A toc159 import receptor mutant, defective in hydrolysis of GTP, supports preprotein import into chloroplasts. J Biol Chem. 2009;284:8670–8679. doi: 10.1074/jbc.M804235200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith MD, Fitzpatrick L, Keegstra K, Schell DJ. In vitro analysis of chloroplast protein import. Current Protocols in Cell Biology. 2002:pp 11.16.11–11.16.21. doi: 10.1002/0471143030.cb1116s17. [DOI] [PubMed] [Google Scholar]
- 38.Schubert M, et al. Proteome map of the chloroplast lumen of Arabidopsis thaliana. J Biol Chem. 2002;277:8354–8365. doi: 10.1074/jbc.M108575200. [DOI] [PubMed] [Google Scholar]
- 39.Ingelsson B, Shapiguzov A, Kieselbach T, Vener AV. Peptidyl-prolyl isomerase activity in chloroplast thylakoid lumen is a dispensable function of immunophilins in Arabidopsis thaliana. Plant Cell Physiol. 2009;50:1801–1814. doi: 10.1093/pcp/pcp122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.