Abstract
Bacterial pathogens deliver effector proteins with diverse biochemical activities into host cells, thereby modulating various host functions. Legionella pneumophila hijacks host vesicle trafficking to avoid phagosome–lysosome fusion, a mechanism that is dependent on the Legionella Dot/Icm type IV secretion system. SidM/DrrA, a Legionella type IV effector, is important for the interactions of Legionella-containing vacuoles with host endoplasmic reticulum–derived vesicles. SidM is the only known protein that catalyzes both the exchange of GDP for GTP and GDI displacement from small GTPase Rab1. We determined the crystal structures of SidM alone (residues 317–647) and SidM (residues 193–550) in complex with nucleotide-free WT Rab1. The SidM structure contains an N-terminal helical domain with a potential new function, a Rab1-activation domain, and a C-terminal phosphatidylinositol 4-phosphate–binding P4M domain. The Rab1-activation domain has extensive strong interactions mainly with Rab1 switch I and II regions that undergo substantial conformational changes on SidM binding. Mutations of switch-contacting residues in SidM attenuate both the nucleotide exchange and GDI displacement activities. Structural comparisons of Rab1 in the SidM complex with Rab1-GDP and Ypt1-GDP in the GDI complex identify key conformational changes that disrupt the nucleotide and GDI binding of Rab1. Further biochemical and structural analyses reveal a unique mechanism of coupled GDP release and GDI displacement likely triggered by the SidM-induced drastic displacement of switch I of Rab1.
Keywords: GDI-displacement factor, guanine nucleotide exchange factor, Legionella pneumophila, Rab GTPase, type IV secretion
Rab GTPases play important roles in eukaryotic vesicle trafficking through selective activation of downstream effector proteins (1, 2). Rabs cycle between a GTP-bound “on” state and a GDP-bound “off” state, which are structurally distinguished by the conformations of two flexible regions known as switch I and switch II. The switch regions, particularly switch II, are loosely packed in the GDP-bound state, and GTP binding renders these regions more compactly packed, due largely to structural contacts made by the GTP γ-phosphate. Conversion from one nucleotide-bound state to the other is catalyzed by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) that catalyze the exchange of GDP for GTP and promote hydrolysis of GTP into GDP, respectively. The C-terminal prenylated GDP-bound Rabs are extracted from the membrane into the cytosol by GDP-dissociation inhibitors (GDIs). For certain Rab-GDI complexes, GDI can be displaced by PRA1 (YIP3 in yeast). The GDI-displacement factor (GDF) activity of PRA1 allows Rab membrane insertion and GEF-mediated Rab activation (3, 4). Precise regulation of Rab activity with spatial and temporal resolution by GEFs/GAPs and GDIs/GDFs is critical for eukaryotic vesicle fusion and trafficking.
Manipulation of the Rab GTPase function is often used by intracellular pathogens to inhibit phagosome maturation and modulate the endocytic/lysosomal pathway (5, 6). Legionella pneumophila, the causative agent of Legionnaires’ disease, replicates in Legionella-containing vacuoles (LCVs) following uptake by alveolar macrophages. The bacteria hijack the trafficking of endoplasmic reticulum (ER)-derived vesicles and decorates LCVs into ER-like compartments that resist lysosomal fusion. Critical to this process is the large number of effectors translocated outside LCVs by the Legionella type IV secretion system (TFSS) (7, 8). LidA, LepB, and SidM (also known as DrrA) are three Legionella TFSS effectors that modulate the host Rab GTPase function. LidA specifically binds to Rab-GTP and collaborates with SidM to recruit early secretory vesicles (9). LepB and SidM directly subvert the function of Rab1, a key regulator of vesicle trafficking between the ER and the Golgi complex. Both effectors are colocalized with Rab1 on LCVs. Whereas LepB is a Rab1-specific GAP that promotes Rab1 inactivation and its removal from the membrane (10), SidM activates Rab1 through its Rab1-specific GEF activity (9, 11). SidM exhibits an additional GDF activity that activates Rab1 in the GDI complex (10, 12). The 647-aa bacterial GEF shows no sequence homologies to eukaryotic GEFs and is the only protein with dual GEF and GDF activities for Ras-like GTPases. Unlike the host GDF PRA1, a polytopic membrane protein (13) that binds to both Rab and GDI (14, 15), SidM interacts only with Rab1 (16) (Fig. S1), suggesting a distinct mechanism underlying SidM-catalyzed GDI displacement.
In the present work, we examined the crystal structures of SidM317–647 alone and SidM193–550 in complex with nucleotide-free Rab1. In addition to the phosphatidylinositol 4-phosphate–binding P4M domain, the structure also reveals a central Rab1-activation domain bearing numerous strong interactions mainly with Rab1 switch regions. Structure-guided biochemical analyses identify several key switch-contacting residues in SidM that are required for both nucleotide exchange and GDI displacement activities. The complex structure further reveals the mechanism underlying SidM-catalyzed nucleotide and GDI release, which appears to be coupled and triggered by SidM-binding–induced dislocation of switch I in Rab1.
Results
Overall Structures of SidM Alone and in Complex with WT Rab1.
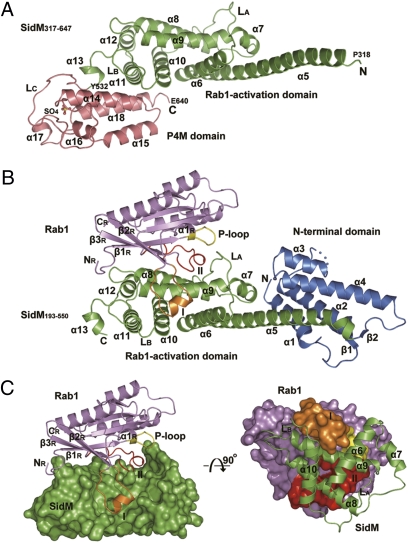
To elucidate the mechanism of SidM function, we solved the 3.45-Å structure of SidM317–647 (residues 317–647) alone (Fig. 1A) and the 2.85-Å structure of SidM193–550 in complex with nucleotide-free WT Rab1 (Fig. 1B). The majority of side-chain densities and the final model for the Rab1-free SidM317–647 structure are of relatively high quality (for details, see SI Materials and Methods), despite the fact that some side chains may not be accurately positioned due to the limited resolution. The Rab1-free structure misses the last 7 C-terminal residues, and the complex lacks the 16 N-terminal and 18 C-terminal residues of SidM193–550. SidM structure starting from Pro193, combined from the two structures, is well separated into three domains termed the N-terminal domain (residues 193–318), the Rab1-activation domain (residues 319–532), and the C-terminal P4M domain (residues 533–647) (Fig. 1 A and B). The N-terminal domain appearing in the complex structure consists of α1–α4 and a short two-stranded β-sheet (β1 and β2). Two helix pairs (α1/α2 and α3/α4), together with the N-terminal part of the long, rigid α5 helix from the Rab1-activation domain, constitutes a major helix bundle (Fig. 1B). The N-terminal domain does not contact Rab1, and its deletion has little effect on Rab1 binding and activation (16), suggesting a potential new function.
Fig. 1.
Crystal structures of SidM alone and the SidM-Rab1 complex. (A) Overall structure of SidM317–647 alone. Secondary structure elements are numbered from the N terminus to the C terminus starting with α5. The Rab1-activation domain and the P4M domain are shown in green and dark salmon, respectively. The starting and ending residues of each domain are labeled. The sulfate ion bound in the P4M domain is shown in a ball-and-stick presentation. (B) Overall structure of the SidM193–550–Rab1 complex. The N-terminal domain is in blue, and the Rab1-activation domain of SidM is in green. Rab1 is in dark purple, with switch I in orange, switch II in red, and the P-loop in yellow. The blue dashed line indicates the disordered loop. (C and D) Overview and the interface of Rab1 and the Rab1-activation domain of SidM. In C, the Rab1-activation domain is shown in surface presentation in green, and the ribbon structure of Rab1 is indicated as in B. In D, the presentation is rotated inward from that of C by 90 degrees, with Rab1 in the surface presentation and the Rab1-activation domain shown as a ribbon.
The C terminus of α5 forms a helix pair with α6, thereby linking the N-terminal domain and the Rab1-activation domain and stabilizing the overall structure. The Rab1-activation domain (α5–α13), corresponding to the minimal SidM region (residues 317–545) with both GDF and GEF activity (16), is roughly organized into two layers of helices (Fig. 1B). The α10/α11 pair extends the plane of the α5/α6 helix pair, which forms the bottom layer. α8 and α9, at the center of the SidM-Rab1 complex, are located on top of the bottom layer of helices. At the open end of the slightly V-shaped α8/α9 helix pair, the N terminus of α8 interacts vertically with α5/α6, and the C terminus of α9 is bound by α7, α5, and α6. The closed end of the α8/α9 pair is capped by α12 and loop LB, which connects α10 and α11. In the bottom layer, α10 contacts the underneath side of α8/α9 across the middle region of the helix pair. Away from the two layers of helices, α13 links the Rab-activation domain to the C-terminal P4M domain. When the structure of the Rab1-activation domain in the complex is superimposed on that of SidM alone, the only notable conformational change is in loop LA, which swings toward α8/α9 and Rab1 by 4.8 Å (Fig. 1 A and B and Fig. S2). This change is significant in that it is essential for formation of the switch II–binding pocket (see below). The overall structure of the Rab1-activation domain is not similar to that of any other known GEFs (Discussion).
The P4M Domain and Type IV Translocation Signal.
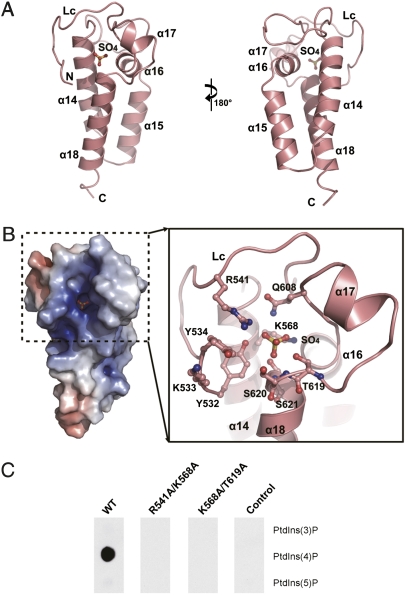
The C-terminal P4M domain (α14–α18) in the Rab1-free structure was recently defined as a PtdIns(4)P-binding domain that localizes SidM to LCVs (12). This helical domain is mushroom-shaped (Figs. 1A and 2A); a three-helix bundle (α14, α15, and α18) forms the stalk, and the cap is composed mainly by α16, α17, and a long loop connected to the Rab1-activation domain. The overall architecture of the P4M domain is not similar to that of any known phosphatidylinositol-binding domain. A large, positively charged pocket holding a phosphate-mimetic sulfate ion is seen at the surface of the mushroom cap (Fig. 2B). The pocket includes several arginine, tyrosine, and lysine residues (Fig. 2B) that often appear in the phosphatidylinositol-binding pockets of other phosphatidylinositol-binding domains. Substitution of R541/K568 or K568/T619 in this pocket into alanine consistently abolished the specific PtdIns(4)P-binding activity of SidM (Fig. 2C).
Fig. 2.
The C-terminal P4M domain. (A) Overview of the P4M domain, color-coded as in Fig. 1A. The bound sulfate is shown as a ball and stick. The presentation in the right panel is rotated by 180 degrees from that in the left panel. (B) The potential surface of the P4M domain (Left) and a magnified view of the sulfate-binding pocket (Right). The left panel has the same presentation as in the left panel in A, with the positive surface in blue and the negative surface in red. (C) Effects of mutations in the sulfate-binding pocket on PtdIns(4)P binding. Here 200 nM purified GST-SidM317–647 protein (WT or indicated mutants) was incubated with phosphatidylinositol phosphate (PIP) strips. The anti-GST Western blot of the overlaid membrane is shown. GST-SidM193–550 was used as the negative control.
The 20 C-terminal residues in several Dot/Icm substrates, such as RalF, contain a sufficient secretion signal thought to be recognized by the Dot/Icm machinery (17 –19). The role of SidM C-terminal residues in secretion is not known; however, residues 444–647 in the SidM C terminus bear the minimal region capable of translocating a reporter (12). The 7 invisible C-terminal residues in SidM likely are disordered, similar to the 20 C-terminal residues in RalF (20). Unlike RalF, in which the 20 residues are projected away from the RalF molecule and fully accessible for interactions with other proteins (20), residues −20 to −12 in SidM are structurally integral to the P4M domain. The last 11 flexible residues are projected toward α6 along the surface of the Rab1-activation domain and likely are only partially exposed (Figs. 1A and 2A).
Binding Interface Between the SidM Rab1-Activation Domain and the Rab1 Switch Regions, and SidM Selectivity for Rab1.
In the SidM-Rab1 complex, Rab1 sits on the top layer of helices of the Rab1-activation domain, and the two molecules contact each other with an extensive surface of 1,779.7 Å2 (Fig. 1 C and D). Structural elements of Rab1 involved in interactions with SidM include the interswitch region (β2R and β3R, R, on Rab1) and, more intensively, switch I and II (Fig. 1C), which form a concave binding surface for the SidM α8/α9 helix pair (Fig. 1D). Interactions between the interswitch region and α8/α9 involve several hydrogen bonds, along with another four hydrogen bonds between the N terminus of Rab1 and the loop linking α8 and α9 in SidM (Fig. S3). Switch I and switch II of Rab1 clamp the α8/α9 helix pair from both sides of α9 (Fig. 1 C and D). On one side, an open pocket formed by α6, α9, and α10 accommodates switch I. Loop LA, which undergoes significant conformational changes on Rab1 binding, packs together with α8/α9, generating a large cleft to hold switch II on the other side of α9.
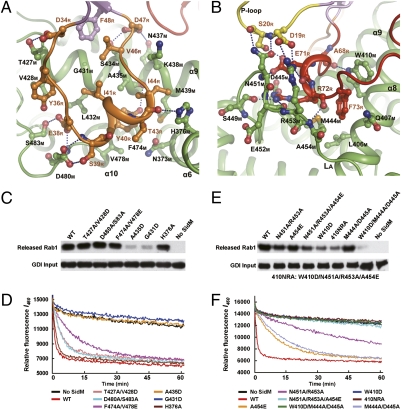
Switch I and adjacent residues (residues from D34R to D47R) loop around the switch I–binding pocket and make extensive contact with the SidM Rab1-activation domain (Fig. 3A). Consequently, the middle region of switch I forms a new helix. This helix bears three hydrophobic interaction cores surrounding Y40R, I41R, and I44R. At the bottom of the pocket, the side chains of A435M, F474M, V478M, and N373M hydrophobically interact with the aromatic ring of Y40R, and the phenolic oxygen of Y40R has van der Waals and hydrogen-bond interactions with M439M and the carbonyl oxygen of A435M, respectively. G431M, L432M, and A435M form the second hydrophobic interaction core with I41R and V46R clustered together with another hydrophobic interaction between F48R from β2R and V46R. H376M and the aliphatic side chain of K438M form a hydrophobic cleft that binds I44R, extending the two aforementioned hydrophobic interaction cores. Notably, G431M, located at the center of the hydrophobic cluster, provides the space needed for I41R and V46R to form hydrophobic interactions. These hydrophobic interactions are strengthened by two groups of hydro-gen bonds that stabilize the switch I helix from its two ends. Four hydrogen bonds are formed between S483M/D480M and E38R/S39R at the N-terminal end of the helix. At the C terminus, H376M captures T43R through another hydrogen bond. Moreover, the N terminus of switch I adheres to the pocket through a hydrophobic stack between V428M and Y36R, as well as a hydrogen bond between T427M and D34R. S434M and N437M each make a hydrogen-bond contact with the side chain of D47R at the C terminus of switch I.
Fig. 3.
Interface between SidM and the switch regions of Rab1 and mutation analyses. (A) The detailed interactions between SidM (green) and switch I of Rab1 (orange). The interacting residues of Rab1 and SidM are shown in sticks with orange and black labels, respectively. Secondary structures of SidM involved in the interaction are marked as well. (B) The detailed interactions between SidM (green) and switch II (red) and the P-loop (yellow) of Rab1. The interacting residues shown in sticks are marked as in A. (C–F) Mutation analyses of the switch-contacting residues in SidM. C and E show GDF assays measuring Rab1 release from the Rab1-GDI complex purified from yeast. The upper panels are immunoblots of released Rab1 in the supernatant using the Rab1 antibody, and the lower panels are immunoblots of GDI on beads as input. D and F show the GEF assay of mant-GDP release from bacterially purified Rab1. Residues of SidM mutated in C and D are the residues contacting switch I of Rab1 identified in A, whereas those mutated in E and F are the residues binding to switch II or to the P-loop identified in B. All of the GDF and GEF assays were repeated at least three times, and representative results are shown.
Various strong interactions also occur around the switch II region of (Fig. 3B). W410M is inserted into switch II and interacts with A68R through a side chain–main chain hydrogen bond. W410M also has an aromatic π-stacking interaction with F73R, which is held in a hydrophobic pocket formed by W410M and four other residues (M444M, A454M, L406M, and Q407M). D445M, S449M, E452M, and A454M make a cleft into which R72R is inserted through formation of a hydrogen bond with each of the four residues. The Rab1 binding–induced conformational change of loop LA provides two residues, N451M and R453M, that are hydrogen-bound to E71R. Notably, the side chains of N451M and R453M protrude toward and have four hydrogen-bond contacts with S20R and D19R in the P-loop. SidM binding creates another hydrogen bond between E71R in switch II and S20R in the P-loop.
SidM contacts a number of residues in Rab1 concentrated in the interswitch and two switch regions (Fig. S4). The majority of SidM-contacting residues in switch I (from D34R to I41R) are poorly conserved in Rab GTPases, and these residues are part of the Rab subfamily–specific sequence region (RabSF2) (21). This provides a structural explanation for the SidM substrate selectivity for Rab1 reported previously (9).
Mutation Analyses of the Binding Interface.
To assess the role of switch I/II–contacting residues of SidM in mediating Rab1 activation, we purified a series of SidM mutants and performed GDF assays with the Rab1-GDI complex purified from yeast. We also performed GEF assays with bacterially purified Rab1 and measured both the release of fluorescent mant-GDP and the loading of mant-GTP; the results were similar to those for GDP release. All of the mutants appeared to behave well in terms of solubility and stability (Fig. S5). Among mutations of switch I–binding residues, G431D and A435D mutations, which likely disrupt the hydrophobic patch accommodating I41R and V46R, resulted in the most deleterious effect, loss of both GDF and GEF activity (Fig. 3C and D). Single or double mutations of other switch I–contacting residues in SidM had little effect on Rab1 activation, consistent with the large interface and extensive interactions observed in the complex structure (Fig. 3A). Among mutations of switch II–contacting residues in SidM, the W410D mutant was largely deprived of both GDF and GEF activity (Fig. 3E and F), highlighting the key role of the W410M–F73R π-stacking interaction. The N451A/R453A double mutant also exhibited attenuated GDF and GEF activity; the mutation appeared to have a more severe effect on GEF activity, consistent with the fact that N451/R453 are on the conformationally changed loop LA and are in direct contact with the P-loop. The three inactive mutants (G431D, A435D, and W410D) can still bind to Rab1 with a weaker affinity than WT SidM. The foregoing analyses also demonstrate that mutations affecting GEF activity also interfere with SidM GDI displacement activity.
Structural Mechanism for SidM-Catalyzed Nucleotide Exchange of Rab1.
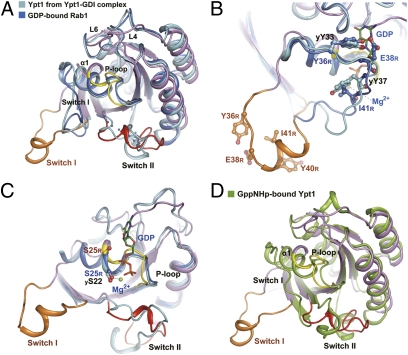
To gain insight into SidM GEF activity, we first compared the structure of Rab1 in the SidM complex with the structures of Rab1-GDP and Ypt1-GDP (yeast Rab1 homolog) in the Ypt1-GDI complex (22). Rab1-GDP and Ypt1-GDP have similar structures, and their nucleotide-binding regions adopt a similar conformation (22) (Fig. 4A). Superimposition of the three structures revealed drastic conformational changes of the switch regions and the P-loop on SidM binding (Fig. 4A). Most notably, switch I, which adopts a closed conformation to bind GDP and magnesium in Rab1-GDP and Ypt1-GDI, is dislocated from the main body of Rab1/Ypt1 and rotated almost 180 degrees in the SidM complex. Y36R (yY33, y, on Ypt1), the residue known as the G1 site, which is essential for nucleotide binding through interactions with the guanosine base (23), moves away from its position in Rab1-GDP and Ypt1 by almost 21 Å (Fig. 4B). yY37 (Y40R), contacting sugar and phosphate groups of GDP in Ypt1-GDP, shows a similar movement of 22.6 Å (Fig. 4B). E38R binds to the sugar group of GDP in both Rab1-GDP and Ypt1-GDI, and I41R is close to the magnesium in Rab1-GDP. In the SidM complex, E38R and I41R are displaced by about 25 Å and 15 Å, respectively. As a result of the drastic switch I displacement, the C terminus of α1R is disassembled, and its N terminus together with the preceding P-loop is twisted and adopts a distorted conformation (Fig. 4C). This further drives the N terminus of the P-loop to move toward the nucleotide-binding pocket. The P-loop harbors a conserved phosphate- and magnesium-binding PM2 motif (GxxxxGK[S, T]) (23). The magnesium-binding S25R (yS22) in the PM2 motif is displaced by about 3.5 Å on SidM binding (Fig. 4C). Thus, the dramatic SidM-induced conformational changes of switch I and the P-loop, particularly the displacement of Y36R and S25R, are expected to abolish the interaction between Rab1 and GDP. Other notable structural changes between Rab1-GDP/Ypt1-GDP and Rab1 in the SidM complex occur on loops L4R and L6R, which harbor the guanosine-binding G2 and G3 sites, respectively (Fig. 4A). These structural changes generate an enlarged and more open guanosine-binding pocket, and cooperate with switch I and the P-loop to facilitate GDP release.
Fig. 4.
Structural basis for SidM-catalyzed nucleotide exchange. (A) Superimposition of Rab1 from the SidM complex with Rab1-GDP and Ypt1-GDP from the Ypt1-GDI complex. Rab1 from the SidM complex is color-coded as in Fig. 1B. Rab1-GDP (PDB ID code 2FOL) is in blue and Ypt1-GDP (PDB ID code 1U.K.V) is in cyan. Regions with considerable conformational changes in the SidM-Rab1 complex are denoted by black or orange labels. (B) The dislocated switch I residues involved in nucleotide binding. The switch I regions from Rab1 in the SidM complex and Rab1-GDP and Ypt1-GDP in the Ypt1-GDI complex are color-coded as in A. The residues binding to nucleotide or magnesium in Rab1-GDP and/or Ypt1-GDP are denoted by blue and black labels, respectively, and the corresponding residues in Rab1 from the SidM complex are in orange. All residues are shown as ball-and-stick representations. GDP and magnesium are from Rab1-GDP. (C) Conformational changes of the P-loop and the displacement of magnesium-binding residue S25 in Rab1. The structural presentation is similar to that in A, except for omission of switch I of Rab1-GDP and Ypt1. S25 in Rab1 (S25R) and S22 in Ypt1 (yS22) are shown as ball-and-stick representations and labeled as in B. GDP and magnesium are from Rab1-GDP. (D) Superposition of Rab1 from the SidM complex with Ypt1 bound with the GTP analog GppNHp (PDB ID code 1YZN) (green). Switch regions, the P-loop, and α1 are labeled and colored as shown.
Switch II is largely disordered in Rab1-GDP. Except for D66R, which bonds with and stabilizes magnesium through a water molecule, most of the residues in or around the switch II region do not directly participate in interactions with GDP in Rab1-GDP and Ypt1-GDP in the GDI complex. D66R does not undergo significant positional changes despite the fact that switch II in the SidM-Rab1 complex adopts an ordered conformation different from that in the Ypt1-GDI complex. Thus, conformational changes of switch II likely do not directly release GDP, but instead may cooperate with SidM-induced repositioning of switch I and the P-loop given that mutation of switch II–contacting residues in SidM reduces its GEF activities (Fig. 3 E and F). Notably, comparison of the structure of Rab1 in the SidM complex with that of Ypt1 bound with a GTP analog showed that the switch II C-terminal region was organized similarly (Fig. 4D). The N terminus of switch II in the SidM complex moves outward and, together with the conformationally changed P-loop, generates a pocket that should be amenable for subsequent GTP loading following GDP release.
Structural Mechanism for GDI Displacement.
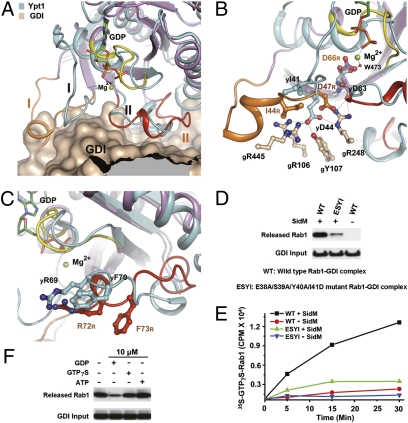
Along with catalyzing the nucleotide exchange, SidM also displaces GDI from the Rab1-GDI complex. The C terminus of switch I and the entire switch II region in Ypt1 interact with the Rab-binding platform in GDI, which is important for maintaining the YptI-GDI complex (22). SidM binding of switch I residues that are exposed in the YptI-GDI complex (see below) triggers substantial conformational or positional changes of those residues that contact the Rab-binding platform in GDI (Fig. 5A). In the switch I region, the side chain of yI41 (I44R) loses its strong hydrophobic interactions with GDI and is placed into the hydrophobic cleft formed by K438M and H376M in the SidM complex (Fig. 5B). yD44 (D47R) forms a hydrogen-bond network with gR106, gY107, gR445, and gR248 (g, on GDI). D47R moves by 1.4 Å, and its side chain rotates 108 degrees in the SidM complex (Fig. 5B). Disruption of this hydrogen-bond network by SidM binding further affects the interaction between yD63 (D66R) in switch II and gR248 in GDI. The well-ordered switch II interacts strongly with GDI in the Ypt1-GDI complex. In the SidM-Rab1 complex, the N terminus of switch II moves slightly outward, and the C terminus is lifted up from its position in the GDI complex (Fig. 5A). yF70 and yR69 are two important residues for binding to GDI (22). In the SidM complex, F73R (yF70) rotates clockwise by ∼70 degrees and is displaced by 4.6 Å into a hydrophobic pocket formed by W410M (Fig. 5C). R72R (yR69) moves downward by 3.7 Å and is also bound into a cleft in SidM. Thus, SidM targeting of some switch I residues induces the aforementioned conformational changes of GDI-binding regions in Rab1 and disrupts their interactions with GDI, resulting in “peeling” of GDI from Rab1.
Fig. 5.
Structural basis for GDI displacement coupled with nucleotide release. (A) Overview of SidM-induced conformational changes of GDI-binding regions in Rab1. Rab1 from the SidM complex, color-coded as in Fig. 4A, is superimposed on Ypt1 (cyan) in the Ypt1-GDI complex. The partial surface of GDI involved in binding to Ypt1 switch regions is in gray. Switch I and switch II are in black (Ypt1) and orange (Rab1), respectively. GDP and magnesium bound in Ypt1 are represented as sticks and spheres, respectively. (B) SidM-induced positional changes of GDI-binding residues in switch I. yI41 (I44R), yD44 (D47R), and yD63 (D66R) interacting with GDI, are shown as ball-and-stick representations. The hydrogen-bond network formed by yD44 with the indicated GDI residues is denoted by a dashed line. Residues from the Ypt1-GDI complex and the SidM-Rab1 complex are denoted by black and orange labels, respectively. (C) SidM-induced conformational changes of switch II and positional movements of yR69 and yF70. yR69 (R72R) and yF70 (R72R), represented as sticks, are two important GDI-binding residues that are partially exposed in the Ypt1-GDI complex. (D) GDF activities of SidM on WT and mutant Rab1-GDI complexes. E38A/S39A/Y40A/I41D is the quadruple mutant of Rab1. Experiments were performed and data are presented similarly as in Fig. 3C. (E) GEF activities of SidM on WT and the quadruple mutant Rab1-GDI complexes. Incorporation of 35S-labled GTPγS on Rab1 was determined as the increase of radioactivity over time. (F) Effects of excess GDP and GTPγS on SidM-catalyzed GDI displacement. 10 μM of indicated nucleotides was added into the GDF reaction with 0.33 μM of Rab1-GDI complex.
Coupled Nucleotide Release and GDI Displacement Triggered by Switch I Dislocation.
In the Ypt1-GDI complex, switch II residues are largely buried or are not spatially available for SidM binding (Fig. S6). In contrast, SidM-contacting residues in switch I, including E38R/S39R/Y40R/I41R, do not contact GDI and are mostly exposed. These findings indicate that SidM-induced conformational change of switch II follows and requires that of switch I, and further suggest that recognition and dislocation of switch I by SidM may play a determining role in triggering the structural changes leading to nucleotide liberation and GDI release. A mutant Rab1-GDI complex containing a quadruple Rab1 mutant (E38A/S39A/Y40A/I41D) was largely resistant to SidM-catalyzed GDI displacement (Fig. 5D), as well as to GTP loading (Fig. 5E). Moreover, mutations of switch II–contacting residues in SidM interfered with the GDF activity of SidM (Fig. 3E), suggesting that the switch II conformational change following switch I displacement is also important for complete GDI displacement from Rab1.
GDP release is generally more difficult in the Rab-GDI complex due to the inhibitory effect of the bound GDI. Disruption of Rab1 interactions with GDI in the switch I/II regions should help relieve the structural constraints for GDP release imposed by GDI binding. This analysis predicts that nucleotide release is coupled with GDI displacement on the action of SidM. This hypothesis is supported by the finding that almost all of the examined mutations in SidM affecting GDP release also attenuated GDI displacement (Fig. 3). Moreover, excess GDP (but not GTPγS or ATP), which would be expected to inhibit GDP release, significantly inhibited the GDF activity of SidM (Fig. 5F). Meanwhile, 100 μM GTPγS could not promote SidM-catalyzed GDI displacement (Fig. S7). Thus, GDP release and the accompanying conformational reorganization of Rab1 are required for complete displacement of GDI from Rab1, and the two reactions likely are coupled, at least to some extent.
Discussion
Four classes of eukaryotic proteins (Mss4, VPS9, Sec2p, and the TRAPP complex) are thought to have RabGEF activities, and their structural mechanisms have been revealed. The structure of SidM from L. pneumophila does not resemble that of any of these proteins (Fig. S8 B–F). Mss4, with low GEF activity, primarily recognizes β1 and β3 of Rab and forms an intermolecular β-sheet zipper with β2, thereby repulsing α1 and disorganizing the nucleotide-binding pocket (24) (Fig. S8C). However, β1 is buried in the Ypt1-GDI complex, and GDI binding hampers formation of the β-sheet zipper (Fig. S8A). For the Rab5-specific VPS9 domain GEFs, two helices from the VPS9 domain are wedged into the space between switch I and switch II of Rab21, which disrupts magnesium and phosphate binding through insertion of an aspartic acid finger (25) (Fig. S8D). In the Rab-GDI complex, the nucleotide-binding pocket is locked, making insertion of the two helices from VPS9 spatially impossible (Fig. S8A). Similar to SidM, Sec2p recognizes switch I and induces its conformational change and displacement from the main body of Sec4p (26, 27) (Fig. S8E); however, the stiff long coiled-coil structure of Sec2p does not permit further switch II binding and reorientation when Sec4p is bound to GDI. The multimeric membrane-tethering TRAPP complex catalyzes the nucleotide exchange of Ypt1 (28). The TRAPP-binding regions on Ypt1 are buried in the Ypt1-GDI complex (Fig. S8 A and F). The locked nucleotide-binding pocket of Ypt1 on GDI binding is not available for insertion of the flexible C terminus of the Bet3p subunit of the TRAPP complex. These comparisons highlight the unique mode of Rab binding and conformational reorganization of the switch regions observed with SidM, and also provide structural explanations as to why other eukaryotic GEFs cannot function similarly as SidM to catalyze GDI displacement in addition to nucleotide exchange.
Finally, while this manuscript was being considered for publication, Suh et al. (29) reported the crystal structure of the Rab1-activation domain of SidM in complex with a Rab1 N121I mutant. Although the conformational changes of Rab1 observed by Suh et al. are largely consistent with our data, whether these conformational changes are caused by the intrinsic nucleotide-binding deficiency of the Rab1 N121I mutant is a serious question. In addition, our structures contain the N-terminal domain and the C-terminal P4M domain, and our Rab1-free structure identifies the conformationally changed loop LA that is critical for binding to and disorganizing the P-loop. Our structural and functional analyses also complement those of Suh et al. on mechanistic insights into the dual GDF and GEF activities of SidM, particularly the coupling of the two reactions.
Materials and Methods
SidM DNA was amplified from genomic DNA of L. pneumophila strain Lp02. cDNAs for human Rab1a and RabGDI2 were amplified from a HeLa cDNA library. Recombinant proteins for crystallization were expressed and purified from Escherichia coli. Crystal structures of SidM317–647 alone and the SidM193–550–Rab1 complex were determined by Se-SAD. Detailed information on reagents, expression and purification of recombinant proteins, crystallization, data collection, structural determination, and biochemical assays is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Yusuke Yamada and his staff at the KEK Synchrotron Facility (Tsukuba, Japan) for assistance with data collection. We also thank members of the Shao laboratory for helpful discussions and technical assistance. This work was supported by Chinese Ministry of Science and Technology Grant 2008AA022309 and National Basic Research Plan of China Grant 2006CB806502 (to F.S.).
Footnotes
The authors declare no conflict of interest.
Data deposition: Structural coordinates used in this paper have been deposited in the Protein Data Bank, www.pdb.org (accession code 3L0M for SidM317–647 and 3L0I for the SidM193–550–Rab1 complex).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0914231107/DCSupplemental.
References
- 1.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 2.Pfeffer SR. Rab GTPases: Specifying and deciphering organelle identity and function. Trends Cell Biol. 2001;11:487–491. doi: 10.1016/s0962-8924(01)02147-x. [DOI] [PubMed] [Google Scholar]
- 3.Dirac-Svejstrup AB, Sumizawa T, Pfeffer SR. Identification of a GDI displacement factor that releases endosomal Rab GTPases from Rab-GDI. EMBO J. 1997;16:465–472. doi: 10.1093/emboj/16.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivars U, Aivazian D, Pfeffer SR. Yip3 catalyses the dissociation of endosomal Rab-GDI complexes. Nature. 2003;425:856–859. doi: 10.1038/nature02057. [DOI] [PubMed] [Google Scholar]
- 5.Brumell JH, Scidmore MA. Manipulation of rab GTPase function by intracellular bacterial pathogens. Microbiol Mol Biol Rev. 2007;71:636–652. doi: 10.1128/MMBR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salcedo SP, Holden DW. Bacterial interactions with the eukaryotic secretory pathway. Curr Opin Microbiol. 2005;8:92–98. doi: 10.1016/j.mib.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 7.de Felipe KS, et al. Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog. 2008;4:e1000117. doi: 10.1371/journal.ppat.1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: Making a cozy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila . Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 10.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 11.Murata T, et al. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol. 2006;8:971–977. doi: 10.1038/ncb1463. [DOI] [PubMed] [Google Scholar]
- 12.Brombacher E, et al. Rab1 guanine nucleotide exchange factor SidM is a major phosphatidylinositol 4-phosphate–binding effector protein of Legionella pneumophila . J Biol Chem. 2009;284:4846–4856. doi: 10.1074/jbc.M807505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin J, Liang Z, Zhang Z, Li G. Membrane topography and topogenesis of prenylated Rab acceptor (PRA1) J Biol Chem. 2001;276:41733–41741. doi: 10.1074/jbc.M103475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutt DM, Da-Silva LF, Chang LH, Prosser DC, Ngsee JK. PRA1 inhibits the extraction of membrane-bound rab GTPase by GDI1. J Biol Chem. 2000;275:18511–18519. doi: 10.1074/jbc.M909309199. [DOI] [PubMed] [Google Scholar]
- 15.Calero M, Collins RN. Saccharomyces cerevisiae Pra1p/Yip3p interacts with Yip1p and Rab proteins. Biochem Biophys Res Commun. 2002;290:676–681. doi: 10.1006/bbrc.2001.6242. [DOI] [PubMed] [Google Scholar]
- 16.Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 17.Nagai H, et al. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci USA. 2005;102:826–831. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burstein D, et al. Genome-scale identification of Legionella pneumophila effectors using a machine-learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cambronne ED, Roy CR. The Legionella pneumophila IcmSW complex interacts with multiple Dot/Icm effectors to facilitate type IV translocation. PLoS Pathog. 2007;3:1837–1848. doi: 10.1371/journal.ppat.0030188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amor JC, et al. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem. 2005;280:1392–1400. doi: 10.1074/jbc.M410820200. [DOI] [PubMed] [Google Scholar]
- 21.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: Definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 22.Rak A, et al. Structure of Rab GDP-dissociation inhibitor in complex with prenylated YPT1 GTPase. Science. 2003;302:646–650. doi: 10.1126/science.1087761. [DOI] [PubMed] [Google Scholar]
- 23.Valencia A, Chardin P, Wittinghofer A, Sander C. The ras protein family: Evolutionary tree and role of conserved amino acids. Biochemistry. 1991;30:4637–4648. doi: 10.1021/bi00233a001. [DOI] [PubMed] [Google Scholar]
- 24.Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding: Structure of Rab8 in complex with MSS4. EMBO J. 2006;25:1445–1455. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delprato A, Lambright DG. Structural basis for Rab GTPase activation by VPS9 domain exchange factors. Nat Struct Mol Biol. 2007;14:406–412. doi: 10.1038/nsmb1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato Y, Fukai S, Ishitani R, Nureki O. Crystal structure of the Sec4p.Sec2p complex in the nucleotide exchanging intermediate state. Proc Natl Acad Sci USA. 2007;104:8305–8310. doi: 10.1073/pnas.0701550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong G, Medkova M, Novick P, Reinisch KM. A catalytic coiled coil: Structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol Cell. 2007;25:455–462. doi: 10.1016/j.molcel.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai Y, et al. The structural basis for activation of the Rab Ypt1p by the TRAPP membrane–tethering complexes. Cell. 2008;133:1202–1213. doi: 10.1016/j.cell.2008.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suh HY, et al. Structural insights into the dual nucleotide exchange and GDI displacement activity of SidM/DrrA. EMBO J. 2010;29:496–504. doi: 10.1038/emboj.2009.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.