Abstract
Bacterial cells are highly organized with many protein complexes and DNA loci dynamically positioned to distinct subcellular sites over the course of a cell cycle. Such dynamic protein localization is essential for polar organelle development, establishment of asymmetry, and chromosome replication during the Caulobacter crescentus cell cycle. We used a fluorescence microscopy screen optimized for high-throughput to find strains with anomalous temporal or spatial protein localization patterns in transposon-generated mutant libraries. Automated image acquisition and analysis allowed us to identify genes that affect the localization of two polar cell cycle histidine kinases, PleC and DivJ, and the pole-specific pili protein CpaE, each tagged with a different fluorescent marker in a single strain. Four metrics characterizing the observed localization patterns of each of the three labeled proteins were extracted for hundreds of cell images from each of 854 mapped mutant strains. Using cluster analysis of the resulting set of 12-element vectors for each of these strains, we identified 52 strains with mutations that affected the localization pattern of the three tagged proteins. This information, combined with quantitative localization data from epitasis experiments, also identified all previously known proteins affecting such localization. These studies provide insights into factors affecting the PleC/DivJ localization network and into regulatory links between the localization of the pili assembly protein CpaE and the kinase localization pathway. Our high-throughput screening methodology can be adapted readily to any sequenced bacterial species, opening the potential for databases of localization regulatory networks across species, and investigation of localization network phylogenies.
Keywords: automated fluorescence microscopy, high content screening, asymmetric cell division, systems biology, Caulobacter crescentus
In prokaryotes, many important functions, including the establishment and maintenance of cell polarity, chromosome replication and segregation, cell division, and asymmetric positioning of polar organelles require dynamic localization of protein complexes (1–8). Despite advances in automated fluorescence microscopy and quantitative image analysis techniques (9–11), phenomena involving protein localization are still most commonly assessed using manually operated microscopes and interpreted by visual examination of the images. This is inherently an arduous and low-throughput approach with relatively low accuracy and sensitivity. High-throughput screening of large numbers of images requires higher throughputs for image collection and image analysis and more quantitative descriptions of subcellular localization patterns and cell characteristics. We hypothesized that high-throughput systematic and quantitative phenotypic analysis of protein localization patterns would enable classification of mutant strains by the pathway affected.
Here we describe the design and application of automated quantitative fluorescence microscopy and an image analysis platform optimized for screening mutagenized bacterial cells to detect strains with protein localization defects. The goal was to rapidly detect genes that affect localization of multiple target proteins and classify them by pathway in a single experiment. We used Caulobacter crescentus (Caulobacter hereafter) as the target organism because localized proteins play a central role in its cell cycle regulation (12, 13). More than 10% of all Caulobacter proteins, including many signaling components, exhibit a nonuniform subcellular distribution (14, 15). Caulobacter divides asymmetrically into a motile swarmer cell and a replication competent stalked cell. After a brief period as a motile cell, the swarmer cell differentiates to a stalked cell by shedding its flagellum, growing a stalk, and initiating DNA replication (Fig. 1A).
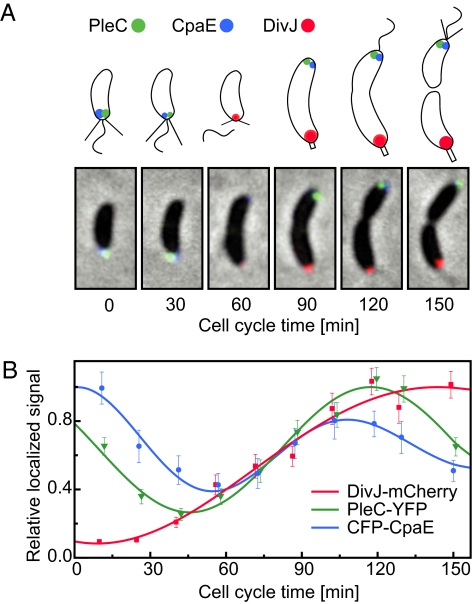
Fig. 1.
Cell cycle localization patterns of PleC, DivJ, and CpaE. (A) The composite microscope images and cell schematics show the position of DivJ-mCherry (red), PleC-YFP (green), and CFP-CpaE (blue) foci within Caulobacter cells at 30-min time intervals. (B) Normalized average localized fluorescence signal of DivJ-mCherry (red squares), PleC-YFP (green triangles), and CFP-CpaE (blue circles) are shown as a function of the cell cycle, fit to an exponentially damped sine function.
The regulatory pathways identified in a protein mislocalization screen will depend on the choice of fluorescently-tagged localized proteins. We used a strain with three different fluorescent tags fused to the cell cycle-regulated polar-localized proteins, DivJ, PleC, and CpaE. This triple-labeling strategy allowed us to perform three screens simultaneously. DivJ and PleC are histidine kinases that are dynamically localized to opposite poles of the predivisional cell (16) (Fig. 1A). After cell division, the kinase activities of PleC and DivJ are restricted to different cell compartments where they affect different developmental programs (17). During the swarmer to stalked cell transition, PleC is released from the flagellated pole of the swarmer cell and reassembled at the opposite pole. After elimination of the flagellum and pili, DivJ localizes to the stalked pole vacated by PleC (Fig. 1 A and B). The polar CpaE protein is required for polar pili biogenesis and is localized to the incipient flagellated pole of the predivisional cell (18, 19) (Fig. 1 A and B).
To increase throughput and information content, we optimized all aspects of the screening process (Fig. S1–S5). Using an engineered Tn5 element we carried out conditional transposon mutagenesis. We used a colony picking robot to array 20,000 independent mutants into 96-well growth plates. Then we used a custom 384-pin tool to inoculate mutants in a precise pattern onto two soft agar plates containing different substrates. This facilitated a sensitive automated phenotypic prescreen for lead candidates that were then arrayed onto 48 sample microscope slides. Custom software was used for full microscope automation and fast image acquisition. Automated image analysis was used to analyze several hundred individual cells per image field, yielding population statistics for subcellular fluorescence distributions. By identifying mutants with fluorescence distributions distinctly different from the unmutagenized control, we identified regulatory links between the localization of the CpaE pili assembly protein and the kinase localization pathway and another role for the DivJ kinase in the polar localization of DivJ, PleC, and CpaE. We also found all of the previously known upstream genes in this single experiment. This combined approach to identify pathways controlling protein localization is applicable to any bacterial species.
Results
Conditional Transposon Mutagenesis of a Triply Labeled Strain.
The three labeled proteins (PleC, DivJ, and CpaE) in Fig. 1A were chosen because of their central importance in regulation of the cell cycle and establishment of asymmetry and because some factors involved in their localization were already known. Using a two-step homologous recombination protocol, the yellow fluorescent protein (yfp) (20) or red fluorescent protein (mcherry) was fused to the chromosomal copy of pleC and divJ, respectively. We introduced an additional swarmer pole specific marker, the pilus assembly gene cpaE fused to the cyan fluorescent protein (cfp). Because the fluorescence spectra of CFP, YFP, and mCherry do not overlap, we could image the three corresponding PleC, DivJ, and CpaE reporter fusions in the same experiment.
The fluorescent-tagged reporter cells were mutagenized using two engineered Tn5 derivatives Tn5Pvan or Tn5Pxyl (SI Text). The Tn5 derivatives contain outward-facing Pxyl or Pvan promoters that allow conditional expression of the adjacent gene(s) (Fig. S2). Twenty thousand independent transposon-generated mutants were arrayed by a colony-picking robot into 96-well plates and grown in the presence of the inducers vanillate or xylose. We assumed that transposon insertions that disrupt cell polarity would partially mimic the nonmotile or filamentous phenotypes of pleC and divJ null mutations, and we designed a growth and motility assay to screen for such mutant candidates. Nonmotile or slow growth mutants form relatively small colonies on soft agar plates. If the phenotype was due to a conditional transposon insertion, it would be dependent on the presence of vanillate or xylose in the media. Using a custom designed tool with 384 floating pins, the transposon mutant library was replica printed onto large soft agar plates that either contained or lacked inducer (Fig. S5). Colony sizes on plates with and without inducer were compared to identify mutant candidates showing either conditional or nonconditional changes in colony size (Fig. S5). This yielded 1,374 candidate mutants that were screened for mislocalization of any one of the tagged polarity markers. As a control, we included 195 samples of the unmutagenized reporter strain.
Automated Quantification of Protein Localization Within Single Bacterial Cells.
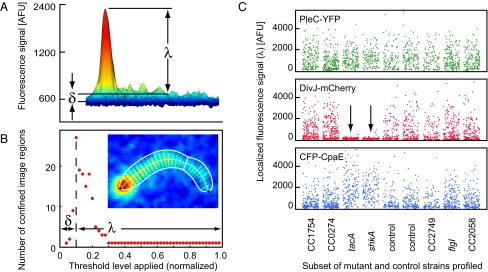
We used custom software [Kontrol of Automated MicroScope (KAMS)] to determine the position of the tagged proteins fluorescent loci within cell images (Figs. S3 and S4). The KAMS acquisition module acquired fluorescence and phase contrast fields populated by 200–300 individual cells from 48-wells on the sample slide. The localized fluorescence signal (λ) and dispersed fluorescence signal (δ) in each fluorescent channel are extracted from individual cells using a dynamic thresholding routine (Fig. 2 A and B and SI Text). The KAMS analysis module determined the cell outline, centerline, and division plane from the phase contrast cell image (Fig. 2B Inset). For each fluorescent channel, individual cells were classified as having localized fluorescence signals at one pole, both poles, or neither pole.
Fig. 2.
Single cell quantification of subcellular protein distribution. (A) The fluorescence intensity profile of a single cell expressing a chromosomal DivJ-mCherry fusion is shown, with the peak intensity mapped to red and lowest intensity to blue. The localized fluorescence signal (λ) is the difference between the peak level and dispersed fluorescence signal (δ) after background correction. (B) Cell shape parameters estimated from the phase contrast image are shown overlaid on the fluorescence intensity profile (Inset). Changing numbers of discrete image regions found by dynamic thresholding are plotted as a function of the relative threshold level applied. The maximum number of confined image regions found is a measure of the amount of dispersed fluorescence signal (δ) present in the cell (SI Text). (C) Each dot indicates the localized fluorescence signal (λ) from a single cell for the PleC-YFP (green), DivJ-mCherry (red), and CFP-CpaE (blue) reporters. The transposon insertions in tacA and shkA (arrows) led to a significant decrease in localized DivJ-mCherry and increased localized CFP-CpaE fluorescence signal as compared to the unmutagenized strains (control).
To validate KAMS, the unmutagenized triply-labeled reporter strain was synchronized and culture samples taken periodically during the cell cycle were analyzed. Fig. 1B shows the temporal pattern of normalized intensities of the three foci averaged over many cells. The fluorescence foci from PleC and CpaE localized at the flagellated pole disappear during the swarmer to stalked cell transition and then reappear at the incipient swarmer pole of the predivisional cell (Fig. 1A). In contrast, there is no DivJ focus in the swarmer cell and a DivJ focus begins to accumulate at the stalked pole after the swarmer to stalked cell transition (Fig. 1A).
Detection of Mislocalization Mutants.
For each candidate mutant strain selected in the prescreen, 100–300 cells were analyzed. For each cell image the localized and dispersed fluorescence signals (λ, δ) were measured as in Figs. 2 A and B. Because DivJ, PleC, and CpaE are dynamically localized to the cell poles over the cell cycle (Fig. 1A), individual cells at different cell cycle stages exhibit different spatial patterns of fluorescence, and the population sample of many cells found in a single field has a signal distribution characteristic of the strain in that field. Mutant strains with transposon insertions in genes that regulate the subcellular recruitment of DivJ, PleC, or CpaE should show altered localized fluorescence intensities and thus an altered characteristic signal distribution for the cell population. For example, populations of cells containing either of two Tn5 insertions mapping to the tacA and shkA genes exhibited a decreased localized fluorescence signal for DivJ-mCherry, and an increased localized fluorescence signal in CFP-CpaE, as compared to the unmutagenized control strain (Fig. 2C). To characterize mislocalization phenotypes and detect mutations that affect the dispersed fluorescence signal as well, we calculated the average localized (L-metric) and dispersed (D-metric) fluorescence signals for the cell populations in the image fields collected for each candidate mutant strain (SI Text). Another class of mislocalization mutants might impair the polar release of both PleC and CpaE during the swarmer to stalked cell transition and thus exhibit a bipolar PleC and CpaE distribution. To detect this phenotype, we calculated for each mutant strain the fraction of cells exhibiting bipolar (B-metric) or monopolar (M-metric) localization patterns (SI Text). This produced four metrics for each strain imaged for each of the three labeled proteins.
Hierarchical Clustering Identifies Functional Protein Localization Modules.
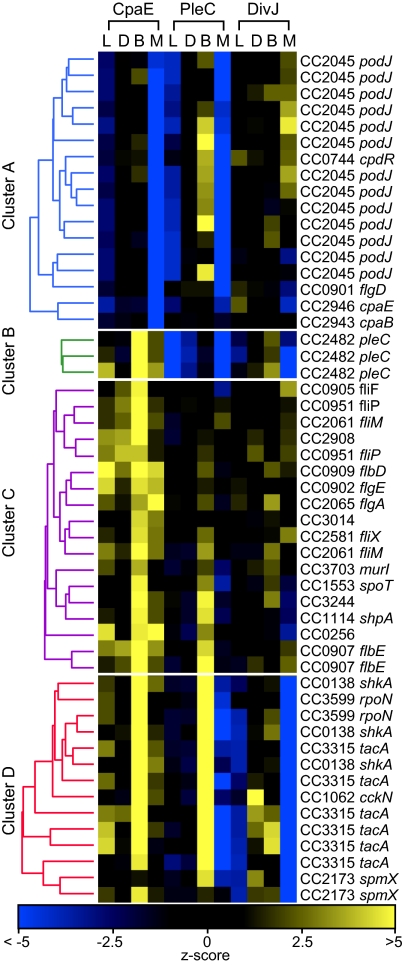
The Tn5 insertion sites for each of the 1,374 candidate mutants selected in the prescreen were mapped by sequencing. Of the 1,374 candidates, we kept those that unambiguously mapped to the genome with base pair accuracy. This resulted in 854 transposon hits that affected 565 genes. To identify gene disruptions showing similar phenotypic profiles and discover potential functional modules, data from the 854 insertion mutants were processed using hierarchical clustering. Data sets were converted to standard scores (z-scores) using the unmutagenized control strain as a standard (SI Text). The localization phenotype of each mutant was represented by a 12-dimensional vector composed of the four z-scores for each of the three fluorophores characterized by the L-, D-, B-, and M-metrics (SI Text). Hierarchical clustering was performed to identify gene clusters with similar localization patterns for the three labeled proteins. The four statistically significant clusters shown in Fig. 3 comprise 52 mutant strains among the 854 mapped mutants with a distinctive mislocation phenotype for one or more of the labeled reporters (Fig. S6). The remaining group of 802 strains was enriched for mutations in flagellar or chemotaxis genes affecting motility and thus colony size.
Fig. 3.
Identification of gene modules coordinating temporal and spatial protein localization by hierarchical clustering analysis. Heat map representation of the z-scores (SI Text) for localized (L), dispersed (D), bipolar (B), and monopolar (M) metrics for CFP-CpaE, PleC-YFP, and DivJ-mCherry are shown for four statistically significant clusters of 52 localization mutants (SI Text). Dendrograms representing the linkage of mutant alleles within cluster A (blue), cluster B (green), cluster C (purple) and the cluster D (red) are shown.
Cluster A includes 13 independent transposon-generated mutants within podJ and single insertions in cpaE, cpaB, cpdR, and flgD that affected CpaE and PleC localization. PodJ is known to be required for correct PleC localization, as well as for positioning of polar pili components (19, 21). Twelve of the 13 transposon mutations that mapped to the cytoplasmic portion of podJ showed a 60% reduction in polar localized PleC and CpaE signals (Fig. 3 and Fig. S6). One podJ mutant exhibited a marginal decrease in PleC localization. The transposon insertion in that strain caused a frame shift at A683 leaving the cytoplasmic portion, as well as the predicted transmembrane domain of PodJ intact (Fig. S6). A previous report suggested that the cytoplasmic domain together with a significant portion of the periplasmic domain of PodJ is required for polar PleC localization (22). However, our results show that only the membrane-anchored cytoplasmic domain of PodJ is required for PleC localization.
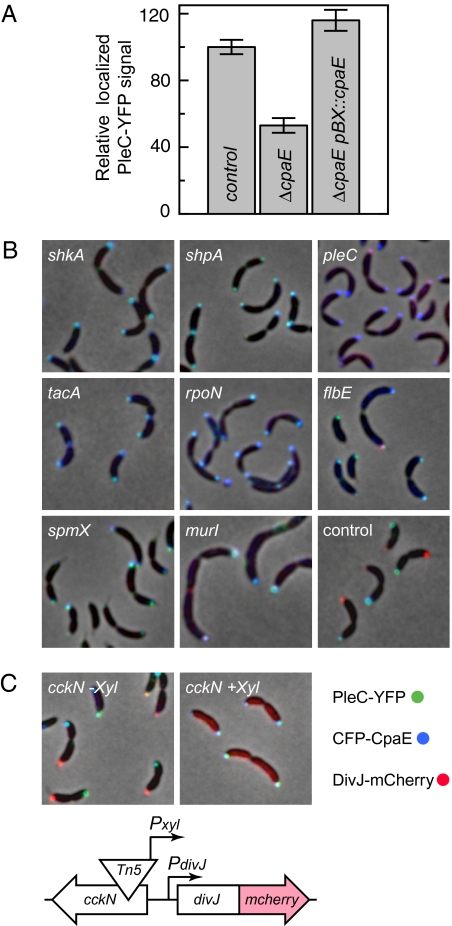
In the mutant with a transposon insertion in cpaE, CFP-CpaE was dispersed throughout the cell with no accumulation at the cell pole (Figs. 3 and 4A). In this case, the Tn5 insertion led to truncation of the last 102 C-terminal amino acids and left 80% of CpaE, as well as the N-terminal CFP reporter, intact. The mislocation of CpaE in this strain suggests that the localization signal of CpaE may be within its C-terminal region, which likely mediates peripheral membrane anchoring by means of an amphipathic membrane targeting sequence. Similar to podJ mutants, the cpaE mutation also showed reduced localized PleC levels (Figs. 3 and 4A and Fig. S6). Together with other pili components, CpaE is encoded within an operon. To determine if CpaE is the only pilus protein required for efficient PleC localization and to exclude potential polar effects on genes in the operon, a pleC-yfp reporter was introduced into cells bearing individual in-frame deletions of each pilus gene in the operon (18). Measurement of the localized PleC signal suggested that an in-frame deletion of cpaE caused a 47% (±4%) decrease in PleC recruitment, similar to the isolated cpaE::Tn5Pvan insertion, although mutations in other pilus genes affected PleC localization only marginally (Fig. 4A and Table S1). In addition, providing cpaE on a plasmid restored wild-type PleC localization levels to the cpaE deletion mutant (Fig. 4A and Table S1). Decreased PleC localization in the cpaE deletion strain was not a consequence of reduced pleC expression because the total cellular PleC-YFP concentration was not altered (Table S1). These results show that both cpaE and podJ contribute to PleC localization to the cell pole.
Fig. 4.
Protein localization patterns of a set of mutants identified by the automated fluorescence microscopy screen. (A) An in-frame deletion of cpaE showed a 47% reduction in polar localized PleC-YFP levels as compared to the control. Providing cpaE (pBX::cpaE) on a plasmid restored PleC-YFP localization. (B) Subcellular fluorescence intensities of DivJ-mCherry (red), PleC-YFP (green), and CFP-CpaE (blue) are merged onto the phase contrast images (gray). Transposon insertions in shkA, shpA, pleC, tacA, rpoN, flbE, spmX, and murI alter the localization pattern of PleC, CpaE, and DivJ as compared to the control. (C) A transposon insertion within cckN caused conditional loss of DivJ-mCherry localization and bipolar localization of PleC-YFP and CFP-CpaE upon addition of xylose. The orientation of the Tn5Pxyl insertion in cckN positioned the Pxyl promoter toward the neighboring divJ gene yielding constitutive DivJ expression.
Cluster B contains 3 mutants with transposon insertions at different locations in pleC. Consistent with previous reports (16, 23), all Tn5 insertions in pleC exhibited bipolar CpaE localization (Figs. 3 and 4B).
Cluster C contains mutants with multiple insertions within class II flagellar genes. These mutants exhibited increased amounts of localized (L-metric) and bipolar (B-Metric) CpaE (Fig. 3), indicating a connection between the flagellar assembly hierarchy and polar pili biogenesis. A subgroup of the mutants in cluster C, composed of two strains with independent flbE insertions and with insertions in shpA and CC3244, showed localization phenotypes similar to those of mutants shown in cluster D (Fig. 3 and 4B and Fig. S6). This suggests that genes from both sets are functionally interlinked.
Cluster D contains 14 transposon-generated mutants that exhibit bipolar distribution of PleC and CpaE as well as an increase in the dispersed DivJ signal (Figs. 3 and 4B and Fig. S6). Eleven of the transposon insertions were mapped to components of a sigma factor activator cascade composed of the hybrid kinase shkA, the sigma factor activator tacA, and rpoN. This sigma factor-activated cascade has been reported, together with shpA, to promote stalk biogenesis (24). Furthermore, two Tn5 insertions caused disruption of spmX, which encodes a lytic transglycosylase, whose expression is TacA-dependent and has been shown to mediate DivJ localization (25).
A Tn5Pxyl insertion in the cckN gene upstream of divJ showed significantly elevated dispersed DivJ levels, as well as a bipolar PleC and CpaE distribution in the presence of the xylose inducer (Figs. 3 and 4C). Sequence analysis showed that the Tn5Pxyl element within cckN had its Pxyl promoter oriented toward the neighboring divJ gene (schematic Fig. 4C), indicating that the divJ gene was likely constitutively expressed upon addition of xylose. In agreement with this hypothesis, we found that the Tn5Pxyl insertion within cckN exhibited a conditional mislocalization phenotype (Fig. 4C). In wild-type cells, divJ is expressed only transiently during the swarmer to stalked cell transition and is not expressed during the predivisional cell stage or in swarmer cells (26, 27). Based on these findings, we propose that temporally restricting divJ expression to the swarmer to stalked-cell transition contributes to proper localization of DivJ, PleC, and CpaE and thus to the establishment of asymmetric cell division.
Epistasis Analysis of the DivJ, PleC, and CpaE Localization Dependency Network.
Mutants mapping to shkA, shpA, tacA, rpoN, spmX and the conditional Tn5Pxyl insertion upstream of divJ all exhibited impaired DivJ localization and bipolar localization of PleC and CpaE (Fig. 3 and Fig. S6). We performed multiple epistasis experiments to identify the network of protein dependencies leading to DivJ, PleC, and CpaE localization. Transcription of a plasmid borne copy of tacA under the control of a vanillate-inducible promoter restored localization of DivJ and monopolar localization of PleC and CpaE to mutant strains with insertions in shkA, shpA, tacA, and also to the conditional Tn5Pxyl insertion upstream of divJ (Tables S2 and S3). In contrast, the DivJ mislocalization phenotype of rpoN or spmX mutants could only be complemented with a plasmid-borne copy of spmX (Table S2). Disruption of the rpoN gene prevents spmX transcription (25), and we showed that in the rpoN insertion mutant, PleC and CpaE are bipolar and DivJ is mislocalized (Fig. 4B). We found that transcription of plasmid-borne spmX only partially complemented the PleC and CpaE mislocalization phenotype of the rpoN disruption (Table S3). In addition, in a strain bearing a pleC mutation, although plasmid-borne transcription of tacA restored DivJ localization, it did not fully restore monopolar CpaE localization (Table S3), suggesting that additional factors are involved in the release of polar-localized proteins in the swarmer cell. Altogether, these results suggest that the ShkA-TacA-RpoN-SpmX localization dependency network plays a pivotal role in coordinating the localization of PleC and DivJ. The affected set of genes control the polar remodeling required for proper release of swarmer pole specific components as well as the recruitment of DivJ (Fig. 5).
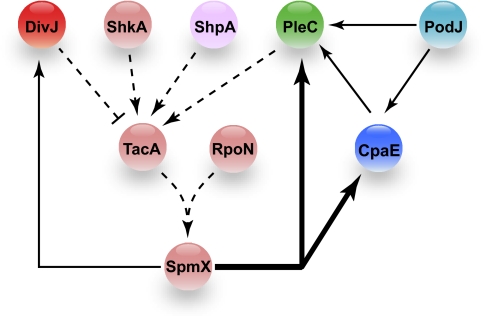
Fig. 5.
Protein localization dependency network. The localization dependency network for PleC, DivJ, and CpaE inferred from the automated quantitative fluorescence microscopy and epistasis analyses is shown. The fluorescently tagged reporter proteins used are DivJ-mCherry (dark red), PleC-YFP (green), and CFP-CpaE (dark blue). Proteins belonging to cluster A (light blue), cluster C (purple), and the cluster D (light red) are shown. The thick solid line indicates that SpmX mediates the removal of PleC and CpaE from the incipient stalked pole. The thin solid lines indicate that SpmX mediates the localization of DivJ to the stalked pole, as previously reported (25). PodJ has been reported to be involved in the localization of PleC and CpaE (19, 21, 22), and here we show that PodJ partially mediates the polar localization of both PleC (60%) and CpaE (60%) (Fig. 3 and Fig. S6). We also found that CpaE partially facilitates the polar localization of PleC (47%) (Figs. 3 and 4 and Table S1). The dashed lines show a localization pathway derived from the results of the epistasis experiments (Tables S2 and S3) and DivJ, PleC and CpaE mislocalization phenotypes observed in Tn5 insertion mutants in shkA, shpA, tacA, rpoN, and spmX (Figs. 3 and 4).
Discussion
Functional domains and the peptide signals required for proper subcellular targeting of proteins can usually be identified using standard molecular biological methods. However, identifying interaction partners and regulatory pathways that orchestrate dynamic protein localization is more challenging. We addressed this challenge using conditional transposon mutagenesis in combination with high-throughput and automated quantitative live cell fluorescence microscopy techniques. The results allowed us to identify, in a single experiment, the genetic components of a localization network that coordinates the dynamic subcellular positioning of the cell fate determinants, DivJ and PleC, and the pili protein CpaE.
This screen for mislocalization mutants confirmed previous observations (19, 21) that PodJ is an important localization factor for PleC as well as for the pili assembly factor CpaE. Using quantitative image analysis, we measured the amount and distribution of the fluorescent signal in each of hundreds of cells (Figs. 2 A and B). This enabled detection of subtle changes in protein levels and positioning. For example, a 50% reduction in polar PleC recruitment in CpaE mutants occurred without a change in the total amount of PleC in the cell. The altered polar PleC accumulation was specific for CpaE, other structural pili proteins did not contribute significantly to polar PleC localization as shown by individual in-frame deletions of each pilus gene in the operon. Our results also uncovered the network of protein localization dependencies among CpaE, PodJ, and PleC, which coordinates the polar positioning of PleC (Fig. 5).
In summary, we used genome scale profiling of localization interdependencies to identify regulatory cascades that effect protein positioning in the cell. Multidimensional hierarchical clustering of localization phenotypes (Fig. 3) allowed us to identify multiple independent transposon-generated mutations that disrupted the phospho-signaling components ShkA, ShpA, PleC, the sigma factor enhancer TacA, RNA polymerase sigma 54 factor RpoN, and the lytic transglycosylase SpmX (Fig. 5). We also identified a conditional transposon insertion upstream of divJ, which simultaneously impaired polar release of PleC, as well as DivJ recruitment during the swarmer to stalked cell transition (Fig. 4C).
These results show that microscopy-based genetic screens can be radically accelerated through automation. Furthermore, postprocessing with automated image analysis to extract and quantify fluorescent cell images, and statistical processing of the resulting datasets, can quickly and sensitively find mutants with aberrant protein localization patterns. Epistasis experiments with this set of mutant strains allowed ordering of the protein localization network. This work shows that we can illuminate entire protein localization networks in microbial cells. The high-throughput screening methods we have applied are readily adaptable to any sequenced bacterial species so that databases of localization regulatory networks across species can be created to enable investigation of localization network phylogenies.
Materials and Methods
SI Materials and Methods contains descriptions of (i) strain, plasmid, and transposon element construction; (ii) image acquisition routines, analysis algorithms, and scoring metrics; and (iii) statistical analysis.
Supplementary Material
Acknowledgments
We thank Rafael Gómez-Sjöberg, Stephanie Weber, and Choon Sim for software and Antonio Iniesta for strains and plasmids. This work was funded by Department of Energy Grant DE-FG02-05ER64136 (to H.H.M.), and National Institutes of Health Grants R01 GM51426 R24, R01 GM32506, and GM073011-04 (to L.S.), and K25 GM070972-01A2 (to M.J.F.). B.C. was supported by the L&Th. La Roche Foundation and by the Swiss National Science Foundation Fellowship PA00P3-126243. N.J.H. was supported by Damon Runyon Cancer Research Foundation Fellowship DRG-1880-05, G.B. by National Institutes of Health Grant F32GM080008, and S.H. by a Samsung Scholarship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000846107/DCSupplemental.
References
- 1.Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- 2.Gordon GS, et al. Chromosome and low copy plasmid segregation in E. coli: Visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 3.Viollier PH, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arigoni F, Pogliano K, Webb CD, Stragier P, Losick R. Localization of protein implicated in establishment of cell type to sites of asymmetric division. Science. 1995;270:637–640. doi: 10.1126/science.270.5236.637. [DOI] [PubMed] [Google Scholar]
- 5.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 6.Raskin DM, de Boer PA. Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA. 1999;96:4971–4976. doi: 10.1073/pnas.96.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jensen RB, Shapiro L. Proteins on the move: Dynamic protein localization in prokaryotes. Trends Cell Biol. 2000;10:483–488. doi: 10.1016/s0962-8924(00)01840-7. [DOI] [PubMed] [Google Scholar]
- 8.Mignot T, Merlie JP, Jr., Zusman DR. Regulated pole-to-pole oscillations of a bacterial gliding motility protein. Science. 2005;310:855–857. doi: 10.1126/science.1119052. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, et al. Group II intron protein localization and insertion sites are affected by polyphosphate. PLoS Biol. 2008;6:e150. doi: 10.1371/journal.pbio.0060150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepperkok R, Ellenberg J. High-throughput fluorescence microscopy for systems biology. Nat Rev Mol Cell Biol. 2006;7:690–696. doi: 10.1038/nrm1979. [DOI] [PubMed] [Google Scholar]
- 11.Guberman JM, Fay A, Dworkin J, Wingreen NS, Gitai Z. PSICIC: Noise and asymmetry in bacterial division revealed by computational image analysis at sub-pixel resolution. PLOS Comput Biol. 2008;4:e1000233. doi: 10.1371/journal.pcbi.1000233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McAdams HH, Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301:1874–1877. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- 13.Collier J, Shapiro L. Spatial complexity and control of a bacterial cell cycle. Curr Opin Biotechnol. 2007;18:333–340. doi: 10.1016/j.copbio.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell JH, Keiler KC. Screen for localized proteins in Caulobacter crescentus. PLoS One. 2008;3:e1756. doi: 10.1371/journal.pone.0001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Werner JN, et al. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci USA. 2009;106:7858–7863. doi: 10.1073/pnas.0901781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wheeler RT, Shapiro L. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol Cell. 1999;4:683–694. doi: 10.1016/s1097-2765(00)80379-2. [DOI] [PubMed] [Google Scholar]
- 17.Matroule JY, Lam H, Burnette DT, Jacobs-Wagner C. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell. 2004;118:579–590. doi: 10.1016/j.cell.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Skerker JM, Shapiro L. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 2000;19:3223–3234. doi: 10.1093/emboj/19.13.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viollier PH, Sternheim N, Shapiro L. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc Natl Acad Sci USA. 2002;99:13831–13836. doi: 10.1073/pnas.182411999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinz AJ, Larson DE, Smith CS, Brun YV. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol Microbiol. 2003;47:929–941. doi: 10.1046/j.1365-2958.2003.03349.x. [DOI] [PubMed] [Google Scholar]
- 22.Lawler ML, Larson DE, Hinz AJ, Klein D, Brun YV. Dissection of functional domains of the polar localization factor PodJ in Caulobacter crescentus. Mol Microbiol. 2006;59:301–316. doi: 10.1111/j.1365-2958.2005.04935.x. [DOI] [PubMed] [Google Scholar]
- 23.Viollier PH, Sternheim N, Shapiro L. A dynamically localized histidine kinase controls the asymmetric distribution of polar pili proteins. EMBO J. 2002;21:4420–4428. doi: 10.1093/emboj/cdf454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biondi EG, et al. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2006;59:386–401. doi: 10.1111/j.1365-2958.2005.04970.x. [DOI] [PubMed] [Google Scholar]
- 25.Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGrath PT, et al. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- 27.Ohta N, Lane T, Ninfa EG, Sommer JM, Newton A. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc Natl Acad Sci USA. 1992;89:10297–10301. doi: 10.1073/pnas.89.21.10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.