Abstract
Purpose
To investigate whether the GLC1F locus is associated with normal tension glaucoma (NTG) in Japanese patients.
Methods
We recruited 242 unrelated Japanese subjects, including, 141 NTG patients and 101 healthy controls. The patients exhibiting a comparatively early onset were selected as they suggest that genetic factors may show stronger involvement. Genotyping and assessment of allelic diversity was performed on 11 highly polymorphic microsatellite markers in and around the GLC1F locus.
Results
Individuals carrying the 163 allele of D7S1277i had a statistically significant increased risk of NTG (p=0.0013, pc=0.016, OR=2.47, 95%CI=1.42–4.30). None of the other markers identified significant loci (pc>0.05) after Bonferroni’s correction.
Conclusions
These findings suggested that the genes in the GLC1F locus may be associated with the pathogenesis of NTG.
Introduction
Glaucoma causes permanent damage of the retina and optic nerve, leading to vision loss and blindness [1]. Primary open-angle glaucoma (POAG) is the most common type of glaucoma and normal tension glaucoma (NTG) is an important subset of POAG; while many POAG patients have high intraocular pressure (IOP) [2], NTG patients have statistically normal IOP [3-5]. NTG is more prevalent in the Asian population, in particular in the Japanese population [6-8]. The diagnosis of glaucoma is based on a combination of factors including optic nerve damage and specific field defects, with IOP being the only treatable risk factor. However, NTG is underdiagnosed: it usually presents late in life after loss of the visual field because it may be asymptomatic with normal IOP.
Glaucoma is recognized as a multi-factorial disorder [9]. Many genes are associated with glaucoma, and several are specifically involved in open-angle glaucoma [10-14]. The GLC1F locus on chromosome 7q35-q36 is the sixth gene locus for POAG [15].
Although NTG may be associated with the GLC1F locus, such an association has not been investigated. Thus, we performed microsatellite (MS) mapping around the GLC1F locus in Japanese NTG patients.
Methods
Subjects
We recruited 242 Japanese subjects from Yokohama City University, Yamanashi University, Gifu University, Kobe University, Yamaguchi University, Kumamoto University, Hokkaido University, Tokyo University, Niigata University, Kanazawa University, Hiroshima University, Tajimi Municipal Hospital, and Tokai University, all in Japan. Of these subjects, 141 had NTG, and 101 were control subjects. The criteria used for the diagnosis of NTG are previously described [16]. The mean age of the patients was 47.30±1.29 years old, and the male: female ratio was 0.92. The mean refraction value was −3.75±0.35 diopters (D), and the mean deviation observed in the Humphrey® static visual field determination (Carl Zeiss Meditec, Oberkochen, Germany) was –10.19±0.94 dB. The age- and sex-matched controls were not affected by glaucoma or any local or systemic illnesses known to cause optic disc or visual field changes. The control cases had no myopia or had mild myopia with refractive errors of –3.00 D or less. Therefore, the groups were somewhat different mean refraction errors. All subjects had similar social background and resided in the same urban area, and provided informed consent. The study was conducted in accordance with the Declaration of Helsinki and subsequent revisions thereof.
Analysis of the 11 microsatelllite loci
Genomic DNA was extracted from blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) or with the standard guanidine method. Eleven MS markers within the GLC1F locus were selected based on the National Center for Biotechnology Information (Figure 1). Polymerase chain reaction (PCR) was performed in a 12.5 µl reaction mixture comprising PCR buffer, genomic DNA, 0.2 mM dinucleotide triphosphates (dNTPs), 0.5 µM primers, and 0.35 U Taq polymerase. The PCR conditions were as follows: 94 °C for 5 min, followed by 30 cycles of denaturation at 94 °C for 30 s, annealing at 56 °C for 30 s, extension at 72 °C for 1 min, and a final elongation step at 72 °C for 10 min. The reaction was performed in a PCR thermal cycler (GeneAmp System 9700; Applied Biosystems, Foster City, CA). The forward primers were labeled at the 5′ ends with 6-FAM, 3-PET, 2-VIC, or 2-NED fluorescent dye (Sigma-Aldrich, St. Louis, Mo; Table 1). To determine the number of MS repeats, the PCR products were denatured at 97 °C for 2 min, mixed with formamide, and electrophoresed using an ABI3130 Genetic Analyzer (Applied Biosystems). The number of MS repeats was estimated by the Southern method with a GS500 TAMRA size marker (Applied Biosystems) and automated with GeneScan 672 software (Applied Biosystems).
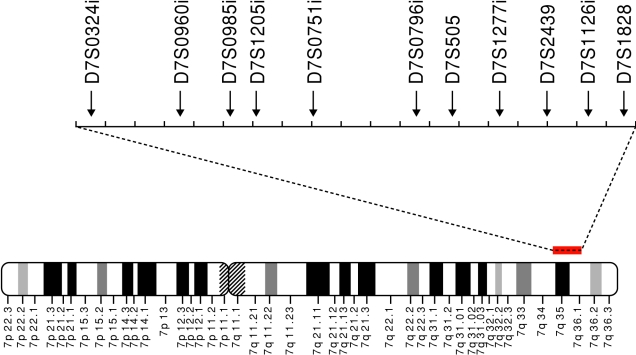
Figure 1.
Location of the 11 microsatellite markers used in this study. All the markers were located on chromosome 7q35–36.
Table 1. Primer sequences of 11 microsatellite markers used in this study.
| Locus | Dye | Orientation | Sequence (5′ to 3′) |
|---|---|---|---|
| D7S0324i | FAM | F | TTTAACAGTTCACACCATGTAAATC |
| R | TTCGAAGCACTACCAAGTCTAC | ||
| D7S0960i | PET | F | GCAATTGTATTCGTTTCATTAG |
| R | CATTACAGGCGTGAGCTAC | ||
| D7S0985i | VIC | F | CTGGCTAACATGGTGAAAC |
| R | GAGAACCTCAGTTGAGGTAGAG | ||
| D7S1205i | NED | F | TCTGCCTAGCACTAGAAAGAAG |
| R | GGCTCTGTTTACTATACTGGAGG | ||
| D7S0751i | FAM | F | TGGCATAAGCTATTTGTATGTTTA |
| R | TACAGTGAGCTATGATGGCAC | ||
| D7S0796i | NED | F | CGTATGGATACCTATGTAACAAAC |
| R | TAATTAGACCACATTTAACCAGAC | ||
| D7S505 | VIC | F | ACTGGCCTGGCAGAGTCT |
| R | CAGCCATTCGAGAGGTGT | ||
| D7S1277i | PET | F | TGTCTTCTGAGACTGTAAGATGTTC |
| R | TGGGAGAACAGTAGGATGG | ||
| D7S2439 | FAM | F | CAGCAAAAGGTACAGCAATTTC |
| R | AAAGTCTACGCCGCATTC | ||
| D7S1126i | FAM | F | CATGCTGAGCCTCAACTAC |
| R | CTGTTGGACTCGTACTAAGATTAC | ||
| D7S1828 | PET | F | TCTTTCCTTTCCTGCATCAC |
| R | AGAATCTTGACATTATCTGACTTCA |
Statistical analysis
Allele and phenotype frequencies were estimated by direct counting. The allelic frequencies between the patients and controls were evaluated using Fisher’s exact test. The probability of association was corrected using Bonferroni’s inequality method. A corrected p (pc) value of <0.05 was considered statistically significant. Statistical analyses were performed with SPSS software (version 10.1; SPSS Science, Chicago, IL).
Results
The proportion of allele frequencies and phenotype frequencies for the markers were consistent with Hardy–Weinberg equilibrium. Table 2 shows the allele frequencies of MS markers, with each allele designated by its amplification size. The frequency of the 163 allele of D7S1277i was higher in the patients than in the controls (p=0.0049, OR=1.97, 95%CI=1.22–3.17); however, after Bonferroni’s correction, the difference between the groups was insignificant (pc=0.063).
Table 2. Allele and phenotype frequencies of 11 microsatellite markers in NTG cases and controls.
| Frequency, n (%) | |||||||
|---|---|---|---|---|---|---|---|
| Marker | Number of detected allele | Allele* | Cases (n=141) | Controls (n=101) | P | Pc | Odds ratio (95%CI) |
| Allele | |||||||
| D7S0324i | 10 | 171 | 50 (17.7) | 27 (13.4) | 0.20 | ||
| D7S0960i | 3 | 429 | 51 (18.1) | 48 (23.8) | 0.13 | ||
| D7S0985i | 6 | 462 | 4 (1.4) | 8 (4.0) | 0.076 | ||
| D7S1205i | 6 | 158 | 81 (28.7) | 61 (30.2) | 0.73 | ||
| D7S0751i | 8 | 359 | 25 (8.9) | 23 (11.4) | 0.36 | ||
| D7S0796i | 18 | 418 | 44 (15.6) | 20 (9.9) | 0.068 | ||
| D7S505 | 9 | 265 | 8 (2.8) | 3 (1.5) | 0.33 | ||
| D7S1277i | 13 | 163 | 70 (24.8) | 29 (14.4) | 0.0049 | 0.063 | 1.97 (1.22–3.17) |
| D7S2439 | 13 | 210 | 28 (9.9) | 11 (5.5) | 0.074 | ||
| D7S1126i | 9 | 276 | 123 (43.6) | 99 (49.0) | 0.24 | ||
| D7S1828 | 6 | 365 | 100 (35.5) | 89 (44.1) | 0.056 | 0.34 | 0.70 (0.48–1.00) |
| Phenotype** | |||||||
| D7S1277i | 13 | 163 | 65 (46.1) | 26 (25.7) | 0.0013 | 0.016 | 2.47 (1.42–4.30) |
| D7S1828 | 6 | 369 | 97 (68.8) | 57 (56.4) | 0.049 | 0.29 | 1.70 (1.00–2.89) |
The asterisk indicates that alleles with >1% of frequency and found to have the most difference in allele frequency between cases and controls in each marker are shown. Each allele was named by the size of its amplification. The double asterisk indicates that only alleles with a p<0.05 are listed.
Table 2 also shows the phenotype frequencies of D7S1277i and D7S1828 with p<0.05. The 163 allele of D7S1277i and the 369 allele of D7S1828 were associated with the risk of NTG (p=0.0013, OR=2.47, 95%CI=1.42–4.30, p=0.049, OR=1.70, 95%CI=1.00–2.89, respectively). After Bonferroni’s correction, the frequency of D7S1277i 163 allele remained significantly different between the groups (pc=0.016).
Discussion
The allele frequencies of GLC1F significantly differed between POAG patients and healthy controls in the American population [15]. The prevalence of POAG was unrelated to the GLC1F locus in a Finnish family, but the relationship between GLC1F and NTG was not studied [17]. We selected NTG patients with precise criteria because the existence of various clinical entities can lead to a lack of statistical validity in case-control analyses. With used 11 MS markers located in the GLC1F locus to detect an association between the GLC1F locus and NTG, and identified a markedly increased frequency of the D7S1277i 163 allele in patients, compared to controls (46.1% versus 25.7%).
Our data suggested that the gene(s) in the D7S1277i locus on 7q36.1 may be significantly associated with the risk of NTG. D7S1277i is located in the intron of the AGAP3 (ArfGAP with GTPase domain, ankyrin repeat and PH domain 3; also known as CRAG) gene, a GTPase activating protein for ADP ribosylation factors. AGAP3 is a novel GTPase that accelerates the degradation of abnormally elongated polyglutamine proteins through intranuclear inclusion body formation. AGAP3 is induced by reactive oxygen species and inhibits the progression of polyglutamine disease [18]. Although the association between AGAP3 and glaucoma is unclear, AGAP3 polymorphisms may act as risk factors in the development of NTG.
D7S1277i may be associated with other gene(s) but not AGAP3 since MS markers are highly polymorphic and generally show wide linkage disequilibriums within 100–200 kb [19-24]. Twenty genes are located within the 200 kb region of D7S1277i. NOS3 (nitric oxide synthase 3) is located ~100 kb centromeric of D7S1277i and has been reported as a POAG candidate gene [15]. NOS catalyzes the production of nitric oxide (NO). NO is involved in vasodilation and the regulation of ocular flow [25]. Ocular NO production may be important in the neuroprotection of retinal ganglion cells [26]. A NOS3 polymorphism was significantly associated with glaucoma with migraine, but not with NTG or POAG in a case-control study [27]. Other NOS3 polymorphisms also showed significant association with female POAG with high IOP in a case-control study [28]. Therefore, NOS3 could affect the phenotype of glaucoma patients, and a disease phenotype-stratified analysis of NOS3 in our NTG patients is required.
In conclusion, we performed an association analysis of the GLC1F locus using MS markers in NTG patients and detected an NTG-associated region in the GLC1F locus. Further studies in the region might help identify the pathogenic gene(s) of NTG.
Acknowledgments
This study was supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan; a grant-in-aid from the Ministry of Health, Labour, and Welfare, Japan; and a grant from the Johnson & Johnson KK Vision Care Company.
References
- 1.Foster PJ, Buhrmann R, Quigley HA, Johnson GJ. The definition and classification of glaucoma in prevalence surveys. Br J Ophthalmol. 2002;86:238–42. doi: 10.1136/bjo.86.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quigley HA. Open-angle glaucoma. N Engl J Med. 1993;328:1097–106. doi: 10.1056/NEJM199304153281507. [DOI] [PubMed] [Google Scholar]
- 3.Hitchings RA, Anderson SA. A comparative study of visual field defects seen in patients with low-tension glaucoma and chronic simple glaucoma. Br J Ophthalmol. 1983;67:818–21. doi: 10.1136/bjo.67.12.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hitchings RA. Low-tension glaucoma – its place in modern glaucoma practice. Br J Ophthalmol. 1992;76:494–6. doi: 10.1136/bjo.76.8.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Werner EB. Normal-tension glaucoma. In: Ritch R, Shields MB, Krupin T, editors. The Glaucomas. 2nd ed. St. Louis: Mosby; 1996. p. 769–97. [Google Scholar]
- 6.Iwase A, Suzuki Y, Araie M, Yamamoto T, Abe H, Shirato S, Kuwayama Y, Mishima HK, Shimizu H, Tomita G, Inoue Y. Kitazawa Y for the Tajimi Study Group, Japan Glaucoma Society. The prevalence of primary open-angle glaucoma in Japanese: the Tajimi Study. Ophthalmology. 2004;111:1641–8. doi: 10.1016/j.ophtha.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 7.Oku Y, Oku H, Park M, Hayashi K, Takahashi H, Shouji T, Chihara E. Long axial length as risk factor for normal tension glaucoma. Graefes Arch Clin Exp Ophthalmol. 2009;247:781–7. doi: 10.1007/s00417-009-1045-2. [DOI] [PubMed] [Google Scholar]
- 8.Shiose Y, Kitazawa Y, Tsukahara S, Akamatsu T, Mizokami K, Futa R, Katsushima H, Kosaki H. Epidemiology of glaucoma in Japan–a nationwide glaucoma survey. Jpn J Ophthalmol. 1991;35:133–55. [PubMed] [Google Scholar]
- 9.Suzuki Y, Iwase A, Araie M, Yamamoto T, Abe H, Shirato S, Kuwayama Y, Mishima HK, Shimizu H, Tomita G, Inoue Y. Kitazawa; Tajimi Study Group. Risk factor for open-angle glaucoma in a Japanese population: the Tajimi Study. Ophthalmology. 2006;113:1613–7. doi: 10.1016/j.ophtha.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 10.Tang S, Toda Y, Kashiwagi K, Mabuchi F, Iijima H, Tsukahara S, Yamagata Z. The association between Japanese primary open-angle glaucoma and normal tension glaucoma patients and the optineurin gene. Hum Genet. 2003;113:276–9. doi: 10.1007/s00439-003-0964-y. [DOI] [PubMed] [Google Scholar]
- 11.Monemi S, Spaeth G, DaSilva A, Popinchalk S, Ilitchev E, Liebmann J, Ritch R, Héon E, Crick RP, Child A, Sarfarazi M. Identification of a novel adult-onset primary open-angle glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 2005;14:725–33. doi: 10.1093/hmg/ddi068. [DOI] [PubMed] [Google Scholar]
- 12.Stone EM, Fingert JH, Alward WL, Nguyen TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A, Nichols BE, Mackey DA, Ritch R, Kalenak JW, Craven ER, Sheffield VC. Identification of a gene that causes primary open angle glaucoma. Science. 1997;275:668–70. doi: 10.1126/science.275.5300.668. [DOI] [PubMed] [Google Scholar]
- 13.Wang CY, Shen YC, Lo FY, Su CH, Lee SH, Lin KH, Tsai HY, Kuo NW, Fan SS. Polymorphism in the IL-1alpha (−889) locus associated with elevated risk of primary open angle glaucoma. Mol Vis. 2006;12:1380–5. [PubMed] [Google Scholar]
- 14.Park S, Jamshidi Y, Vaideanu D, Bitner-Glindzicz M, Fraser S, Sowden JC. Genetic risk for primary open-angle glaucoma determined by LMX1B haplotypes. Invest Ophthalmol Vis Sci. 2009;50:1522–30. doi: 10.1167/iovs.08-2483. [DOI] [PubMed] [Google Scholar]
- 15.Wirtz MK, Samples JR, Rust K, Lie J, Nordling L, Schilling K, Acott TS, Kramer PL. GLC1F, a new primary open-angle glaucoma locus, maps to 7q35-q36. Arch Ophthalmol. 1999;117:237–41. doi: 10.1001/archopht.117.2.237. [DOI] [PubMed] [Google Scholar]
- 16.Shibuya E, Meguro A, Ota M, Kashiwagi K, Mabuchi F, Iijima H, Kawase K, Yamamoto T, Nakamura M, Negi A, Sagara T, Nishida T, Inatani M, Tanihara H, Aihara M, Araie M, Fukuchi T, Abe H, Higashide T, Sugiyama K, Kanamoto T, Kiuchi Y, Iwase A, Ohno S, Inoko H, Mizuki N. Association of Toll-like receptor 4 gene polymorphisms with normal tension glaucoma. Invest Ophthalmol Vis Sci. 2008;49:4453–7. doi: 10.1167/iovs.07-1575. [DOI] [PubMed] [Google Scholar]
- 17.Lemmelä S, Ylisaukko-oja T, Forsman E, Järvelä I. Exclusion of 14 candidate loci for primary open angle glaucoma in finnish families. Mol Vis. 2004;10:260–4. [PubMed] [Google Scholar]
- 18.Qin Q, Inatome R, Hotta A, Kojima M, Yamamura H, Hirai H, Yoshizawa T, Tanaka H, Fukami K, Yanagi S. A novel GTPase, CRAG, mediates promyelocytic leukemia protein-associated nuclear body formation and degradation of expanded polyglutamine protein. J Cell Biol. 2006;172:497–504. doi: 10.1083/jcb.200505079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ota M, Mizuki N, Katsuyama Y, Tamiya G, Shiina T, Oka A, Ando H, Kimura M, Goto K, Ohno S, Inoko H. The critical region for Behcet’s disease in the human major histocompatibility complex is reduced to a 46-kb segment centromeric of HLA-B, by association analysis using refined microsatellite mapping. Am J Hum Genet. 1999;64:1406–10. doi: 10.1086/302364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuki N, Ota M, Yabuki K, Katsuyama Y, Ando H, Palimeris GD, Kaklamani E, Accorinti M, Pivetti-Pezzi P, Ohno S, Inoko H. Localization of the pathogenic gene of Behcet's disease by microsatellite analysis of three different populations. Invest Ophthalmol Vis Sci. 2000;41:3702–8. [PubMed] [Google Scholar]
- 21.Oka A, Tamiya G, Tomizawa M, Ota M, Katsuyama Y, Makino S, Shiina T, Yoshitome M, Iizuka M, Sasao Y, Iwashita K, Kawakubo Y, Sugai J, Ozawa A, Ohkido M, Kimura M, Bahram S, Inoko H. Association analysis using refined microsatellite markers localizes a susceptible locus for psoriasis vulgaris within a 111 kb segment telomeric of the HLA-C gene. Hum Mol Genet. 1999;8:2165–70. doi: 10.1093/hmg/8.12.2165. [DOI] [PubMed] [Google Scholar]
- 22.Keicho N, Ohashi J, Tamiya G, Nakata K, Taguchi Y, Azuma A, Ohishi N, Emi M, Park MH, Inoko H, Tokunaga K, Kudoh S. Fine localization of a major disease-susceptibility locus for diffuse panbronchiolitis. Am J Hum Genet. 2000;66:501–7. doi: 10.1086/302786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ota M, Katsuyama Y, Kimura A, Tsuchiya K, Kondo M, Nause T, Mizuki N, Itoh K, Sasazuki T, Inoko H. A second susceptibility gene for developing rheumatoid arthritis in the human MHC is localized within a 70 kb interval telomeric of the TNF genes in the HLA class Ш region. Genomics. 2001;71:263–70. doi: 10.1006/geno.2000.6371. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Leaves NI, Anderson GG, Ponting CP, Broxholme J, Holt R, Edser P, Bhattacharyya S, Dunham A, Adcock IM, Pulleyn L, Barnes PJ, Harper JI, Abecasis G, Cardon L, White M, Burton J, Matthews L, Mott R, Ross M, Cox R, Moffatt MF, Cookson WO. Positional cloning of a quantitative trait locus on chromosome 13q14 that influences immunoglobulin E levels and asthma. Nat Genet. 2003;34:181–6. doi: 10.1038/ng1166. [DOI] [PubMed] [Google Scholar]
- 25.Koss MC. Functional role of nitric oxide in regulation of ocular blood flow. Eur J Pharmacol. 1999;374:161–74. doi: 10.1016/s0014-2999(99)00242-3. [DOI] [PubMed] [Google Scholar]
- 26.Polak K, Luksch A, Berisha F, Fuchsjaeger-Mayrl G, Dallinger S, Schmetterer L. Altered nitric oxide system in patients with open-angle glaucoma. Arch Ophthalmol. 2007;125:494–8. doi: 10.1001/archopht.125.4.494. [DOI] [PubMed] [Google Scholar]
- 27.Logan JF, Chakravarthy U, Hughes AE, Patterson CC, Jackson JA, Rankin SJ. Evidence for association of endothelial nitric oxide synthase gene in subjects with glaucoma and a history of migraine. Invest Ophthalmol Vis Sci. 2005;46:3221–6. doi: 10.1167/iovs.05-0368. [DOI] [PubMed] [Google Scholar]
- 28.Kang JH, Wiggs JL, Rosner BA, Hankinson SE, Abdrabou W, Fan BJ, Haines J, Pasquale LR. Endothelial nitric oxide synthase gene variants and primary open-angle glaucoma: interactions with sex and postmenopausal hormone use. Invest Ophthalmol Vis Sci. 2010;51:971–9. doi: 10.1167/iovs.09-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]