Abstract
Superoxide dismutase (SOD) catalyzes the conversion of superoxide radical to hydrogen peroxide. Periplasmic localization of bacterial Cu,Zn-SOD has suggested a role of this enzyme in defense against extracellular phagocyte-derived reactive oxygen species. Sequence analysis of regions flanking the Salmonella typhimurium sodC gene encoding Cu,Zn-SOD demonstrates significant homology to λ phage proteins, reflecting possible bacteriophage-mediated horizontal gene transfer of this determinant among pathogenic bacteria. Salmonella deficient in Cu,Zn-SOD has reduced survival in macrophages and attenuated virulence in mice, which can be restored by abrogation of either the phagocyte respiratory burst or inducible nitric oxide synthase. Moreover, a sodC mutant is extremely susceptible to the combination of superoxide and nitric oxide. These observations suggest that SOD protects periplasmic or inner membrane targets by diverting superoxide and limiting peroxynitrite formation, and they demonstrate the ability of the respiratory burst and nitric oxide synthase to synergistically kill microbial pathogens in vivo.
Cu,Zn-superoxide dismutase (SOD) was first identified in bovine erythrocytes (1) and recognized as a prokaryotic enzyme in Photobacterium leiognathi in 1974 (2). Although initially regarded as a biological curiosity, prokaryotic Cu,Zn-SOD was eventually found in a number of other bacterial species including Salmonella (3, 4). In part, the initial failure to recognize the presence of Cu,Zn-SOD in bacteria may have resulted from its low concentrations relative to Mn- and Fe-SODs (5). Periplasmic localization in prokaryotes (6, 7) led to the proposal that Cu,Zn-SOD may defend bacteria from extracellular oxidative stress (6), but its precise functional significance has not been established.
In the present study, the facultative intracellular pathogen Salmonella typhimurium was used to examine the role of Cu,Zn-SOD in defense against oxygen-dependent antimicrobial systems of mammalian phagocytic cells. The complete S. typhimurium sodC gene (4) encoding Cu,Zn-SOD and its flanking regions were cloned and sequenced. Superoxide dismutase activity of the SodC protein was confirmed, and insertional inactivation of the gene was achieved by homologous recombination. The SodC-deficient mutant strain and its isogenic parent were subsequently evaluated in chemical susceptibility studies, macrophage survival assays, and mouse virulence experiments to assess the functional importance of bacterial Cu,Zn-SOD both in vitro and in vivo.
MATERIALS AND METHODS
Media.
Bacteria were routinely grown in Luria–Bertani medium (10 mg/ml tryptone, 5 mg/ml yeast extract, 10 mg/ml NaCl) at 37°C, with penicillin (250 μg/ml) added as required. Chemical killing assays were performed in M9 minimal medium (7 mg/ml Na2HPO4, 3 mg/ml KH2PO4, 0.5 mg/ml NaCl, 1 mg/ml NH4Cl, 5 μg/ml thiamine, 0.12 mg/ml MgSO4, 0.015 mg/ml CaCl2) or PBS as indicated. Xylose-lysine-deoxycholate medium (Difco) was used for selection of recombinant Salmonella mutants. Agar (1.5%) was added to solid medium. Macrophages were cultivated in RPMI 1640 medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 10 mM Hepes buffer, and 1 mM sodium pyruvate.
Bacterial Strains and Plasmids.
Chemical susceptibility assays, macrophage killing assays, and virulence studies used wild-type Salmonella typhimurium ATCC14028s or its sodC mutant derivative MF1005. S. typhimurium XF1001 (katE/katG) is described in ref. 8. Escherichia coli S17–1 (9) was used to mobilize suicide vector pRR10(ΔtrfA) (10). Plasmid pBluescript KS (Stratagene) was used for construction of a S. typhimurium genomic library, and plasmid pET23a (Novagen) in E. coli BL21 (DE3) (11) was used for sodC overexpression under the control of T7 RNA Polymerase.
sodC Mutant Construction.
A 262-bp internal sodC fragment (nucleotides 228–489) was amplified by PCR using oligonucleotide primers 5′-TATGCCGGGAATGAAAGACGGTAA-3′ and 5′-CAGTGGAGCAGGTTTATCGGAGTA-3′ derived from the published partial S. typhimurium sodC sequence (4). The fragment was first cloned into the SmaI site of pBluescript (Stratagene), then subcloned into the EcoRI and BamHI sites of suicide vector pRR10(ΔtrfA) (10), and mobilized from E. coli S17–1 into S. typhimurium 14028s, with selection of transconjugants on xylose-lysine-deoxycholate agar containing penicillin 250 μg/ml. Homologous recombination of the plasmid into the Salmonella chromosome created an interruption of sodC, which resulted in deletion of 14 aa from the carboxyl terminus of the SodC protein; sodC interruption was confirmed by Southern hybridization (not shown) (12). The sodC mutant S. typhimurium strain was designated MF1005.
Cloning of the S. typhimurium sodC Gene.
The S. typhimurium sodC gene fragment described above (4) was used as a probe to identify a 3.3-kbp sodC-containing chromosomal fragment (GenBank accession no. AF007380) from an S. typhimurium 14028s genomic library. Sequencing was performed on an ABI373A automated fluorescent sequencer (Applied Biosystems).
Detection of Superoxide Dismutase Activity.
Sonicated cell extracts from E. coli overexpressing the S. typhimurium sodC gene from plasmid pET23a-sodC or carrying the pET vector alone were subjected to 15% PAGE under nondenaturing conditions at neutral pH. SOD activity was demonstrated as an achromatic zone after photoactivation of riboflavin and reaction with nitroblue tetrazolium (13). Sodium cyanide (1 mM) was added to confirm that S. typhimurium SodC is a copper, zinc-containing superoxide dismutase (14).
Chemical Susceptibility Assays.
Disk diffusion susceptibility assays (15) were performed with 106 stationary phase bacteria on M9–0.2% glucose agar using 15 μl of 3% hydrogen peroxide, 1.9% methyl viologen, 500 mM S-nitrosoglutathione (GSNO), or 500 mM SIN-1 (3-morpholinosydnonimine hydrochloride) (all reagents from Sigma); GSNO was synthesized as described (15). Other killing assays were performed with 0.1 unit/ml xanthine oxidase (Sigma) and 250 μM hypoxanthine added to overnight cultures diluted 1:100 in PBS. Serial dilutions were plated in triplicate at timed intervals for quantitation of colony-forming units. 2,2′-(Hydroxynitrosohydrazono)bisethanamine (SPER/NO, 1 mM, Alexis Biochemicals, San Diego) was added as indicated.
Macrophage Killing Assays.
Murine peritoneal exudate cells were harvested in RPMI 1640 medium with 10% fetal calf serum 4 days after intraperitoneal injection of 5 mM sodium periodate (Sigma), which stimulates both the phagocyte respiratory burst and inducible NO synthase (16). Cells were plated in 96-well flat-bottomed microtiter plates and allowed to adhere overnight in the presence of 20 units/ml interferon γ (IFN-γ). S. typhimurium opsonized with normal mouse serum were spun onto adherent cells at 10:1 multiplicity of infection (moi), internalized for 15 min, and washed with medium containing 6 μg/ml gentamicin to kill extracellular bacteria. NG-d-monomethyl arginine (250 μM) (control, Sigma), 250 μM NG-l-monomethyl arginine (NO synthase inhibitor, Sigma), or 250 μM acetovanillone (apocynin, 4-hydroxy-3-methoxyacetophenone; NADPH-oxidase inhibitor, Aldrich) were added to wells at 0 h as indicated. In separate experiments, 250 μM NG-l-monomethyl arginine was found to effectively block NO2− production by IFN-γ-stimulated macrophages as measured by the Griess reaction (17), 250 μM acetovanillone was found to abrogate stimulation of H2O2 production (18) by 20 ng/ml PMA (phorbol 12-myristate 13-acetate, Sigma) without affecting NO2− production, and 250 μM NG-d-monomethyl arginine was found to affect neither H2O2 nor NO2− production (data not shown). At 2-, 6-, and 24-h time points, cells were resuspended in PBS and lysed with 0.5% deoxycholate. Serial dilutions were plated for enumeration of colony-forming units.
Mouse Virulence Assays.
C57BL/6 (Itys, genetically Salmonella-susceptible) mice, congenic phox-91 ko (knock-out) mice (19), or C3H/HeN (Ityr, genetically Salmonella-resistant) mice were intraperitoneally challenged with wild-type or mutant S. typhimurium diluted in 200 μl PBS. Inocula were determined by dilutional plating. Mutant bacteria were recovered from dead mice to confirm in vivo stability of insertional mutations. Each virulence assay was performed at least twice using five mice per experimental group, with identical results.
RESULTS
Sequence Analysis of S. typhimurium sodC and Flanking Regions.
Analysis of sodC and flanking regions revealed an adjacent countertranscribed ORF encoding a predicted protein with 20–64% homology to members of a family of outer membrane proteins that includes Lom of bacteriophage λ (20) and Ail of Yersinia enterocolitica (21), as well other ORFs encoding proteins highly homologous (53–86%) to bacteriophage λ tail proteins (Fig. 1A). The predicted full-length SodC protein is only 55% identical to the E. coli SodC protein (22) (Fig. 1B), and a typical hydrophobic NH2 terminus signal sequence was identified, consistent with secretion of SodC into the periplasmic space. Hybridization studies using the internal sodC fragment described in Materials and Methods as a probe demonstrated the presence of the sodC gene in Salmonella enteritidis, S. choleraesuis, S. newport, S. heidelburg, and S. typhisuis (not shown).
Figure 1.
(A) Map of S. typhimurium sodC region. An ORF encoding a predicted protein with homology to the Ail protein of Yersinia enterocolitica and ORFs highly homologous to genes that encode bacteriophage λ tail proteins are designated. (B) Sequence alignment of S. typhimurium and E. coli SodC proteins. The full-length SodC proteins from S. typhimurium and E. coli (22) are 54% identical at the amino acid level. The S. typhimurium sodC gene encodes a protein of 18.3 kDa predicted size. The protein possesses a typical hydrophobic NH2 terminus signal sequence (underlined); cleavage would produce a mature protein of 16.3 kDa predicted size.
Demonstration of SOD Activity.
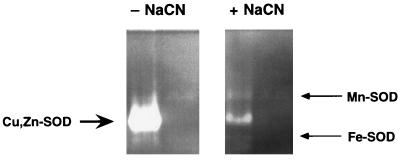
Overexpression of the cloned S. typhimurium sodC gene revealed SOD activity (13) (Fig. 2), inhibitable by sodium cyanide as is characteristic of copper, zinc-containing superoxide dismutases. E. coli Cu,Zn-SOD activity is not visible in extracts from cells carrying the plasmid vector alone, reflecting its low concentration relative to that of Fe-SOD and Mn-SOD (5).
Figure 2.
Confirmation of SodC enzymatic activity. Sonicated cell extracts from E. coli overexpressing the S. typhimurium sodC gene (first and third lanes) or carrying the plasmid vector alone (second and fourth lanes) were subjected to SOD activity gel analysis (13). The left arrow designates SodC, and right arrows designate Fe- and Mn-SOD proteins, respectively. Inhibition of SodC activity by 1 mM sodium cyanide is characteristic of Cu,Zn-SODs (14).
Susceptibility to Chemically Generated Reactive Oxygen and Nitrogen Species.
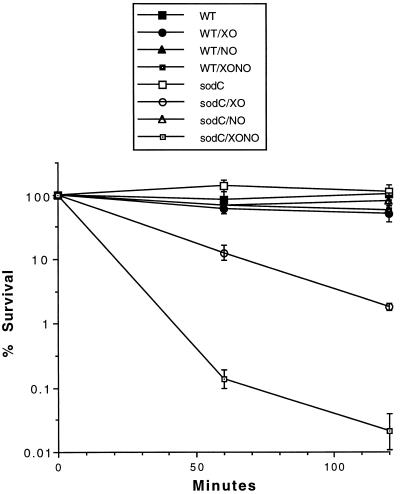
The chemical susceptibility of sodC mutant S. typhimurium was compared with that of isogenic wild-type bacteria by disk diffusion assay (15). Wild-type and sodC mutant S. typhimurium were found to have identical susceptibility to hydrogen peroxide, methyl viologen (paraquat), GSNO, and SIN-1 (data not shown). However, the sodC mutant has increased (29×) susceptibility to extracellular superoxide generated by xanthine oxidase (Fig. 3). Susceptibility to xanthine oxidase but not the intracellular superoxide-generator methyl viologen is consistent with earlier observations indicating that the inner membrane of Gram-negative bacteria is impermeable to superoxide (6, 23). The sodC mutation was observed to dramatically increase (2,700×) susceptibility to synergistic killing by xanthine oxidase (Fig. 3) and the NO donor SPER/NO, although SPER/NO by itself has no significant antimicrobial activity against either wild-type or sodC mutant Salmonella.
Figure 3.
In vitro susceptibility of sodC mutant S. typhimurium to extracellular superoxide and NO. Wild-type or sodC mutant (MF1005) bacteria in PBS with 250 μM hypoxanthine were treated with 0.1 unit/ml xanthine oxidase (XO), 1 mM SPER/NO (NO), or the combination of xanthine oxidase and SPER/NO (XONO) as described in Materials and Methods.
Susceptibility to Macrophage Killing.
The sodC S. typhimurium mutant has increased susceptibility to killing by activated murine macrophages (Fig. 4), but this hypersusceptibility requires the presence of both the respiratory burst and nitric oxide synthase. Enhanced killing can be blocked by either the NO synthase inhibitor NG-l-monomethyl arginine (24) or by the respiratory burst inhibitor acetovanillone (18). Macrophage killing assays were performed three times with samples in triplicate, with identical results.
Figure 4.
Susceptibility of sodC mutant S. typhimurium to killing by murine peritoneal macrophages. IFN-γ-stimulated periodate-elicited peritoneal exudate cells from C3H/HeN mice were used to kill wild-type (WT) or sodC mutant (MF1005) S. typhimurium. NG-d-monomethyl arginine (250 μM) was added to control wells, 250 μM NG-l-monomethyl arginine (MMA) was added to inhibit NO synthase, and 250 μM acetovanillone (AV) was added to inhibit the phagocyte NADPH-oxidase.
Murine Virulence.
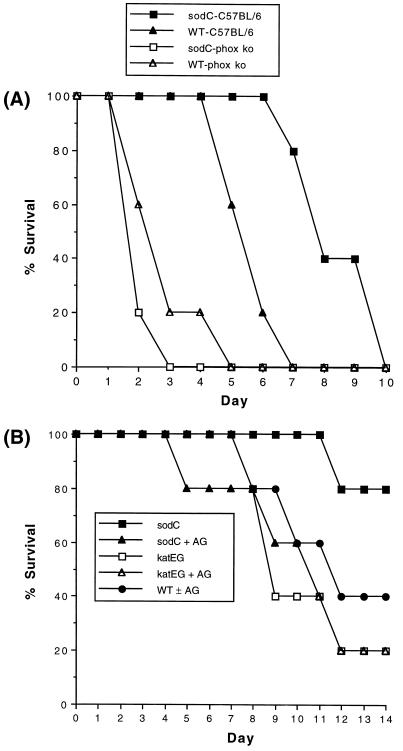
Further evidence of the biological relevance of bacterial Cu,Zn-SOD was obtained in murine virulence studies. Six-week-old C57BL/6 (Itys) and congenic phox-91 ko mice (19) were challenged with 50 intraperitoneal wild-type or sodC mutant S. typhimurium organisms. Attenuated virulence of the sodC mutant was completely restored by abrogation of the respiratory burst (Fig. 5A). In a parallel experiment, 6-week-old C3H/HeN (Ityr) mice were challenged with 2 × 103 intraperitoneal wild-type, sodC mutant, or catalase-deficient katE katG mutant (8) S. typhimurium. Separate groups of mice were administered 2.5% (wt/vol) aminoguanidine (Sigma), an inhibitor of inducible NO synthase (25). Attenuated virulence of the sodC mutant strain was restored by inhibition of NO synthase (Fig. 5B). In contrast, the catalase-deficient strain was fully virulent, and its virulence was not significantly affected by coadministration of aminoguanidine. Mouse virulence studies were performed a minimum of two times each; results of representative experiments are shown.
Figure 5.
(A) Virulence of sodC mutant S. typhimurium in wild-type and respiratory burst-deficient mice. C57BL/6 (Itys) and congenic phox-91 ko mice (19) were intraperitoneally challenged with wild-type or sodC mutant (MF1005) S. typhimurium. The time to death for wild-type and sodC mutant organisms was significantly different in C57BL/6 mice (P < 0.01) but not in phox ko (knock-out) mice (P > 0.05). (B) Virulence of sodC and katEG mutant S. typhimurium and the effect of NO synthase inhibition. C3H/HeN (Ityr) mice were intraperitoneally challenged with wild-type, sodC mutant (MF1005), or katEG mutant (XF1001) S. typhimurium. Separate groups of mice received drinking water treated with 2.5% (wt/vol) aminoguanidine (25) (AG), an inhibitor of inducible NO synthase. The survival of untreated mice challenged with sodC mutant organisms was significantly different from the other experimental groups (P < 0.01) after adjustment for multiple comparisons.
DISCUSSION
Both clinical observations in patients with chronic granulomatous disease and experimental studies with murine phagocytes indicate that the phagocyte respiratory burst with production of superoxide is a critical component of host defense against salmonellosis (26, 27), as it is in many other infections (28). Studies in experimental animals also suggest a beneficial role of inducible NO synthase during Salmonella infection (25), and elevated production of NO has been detected in patients with bacterial gastroenteritis (29). The present observations indicate that synergistic antimicrobial activity can result from the combination of the phagocyte respiratory burst and NO synthase in vivo as well as in vitro. This study also demonstrates a critical role of periplasmic Cu,Zn-SOD encoded by the sodC gene in defending Salmonella from products of both the respiratory burst NADPH-oxidase and inducible NO synthase. This contrasts with the function of the more abundant cytosolic Mn-SOD enzyme, which is not required for Salmonella virulence despite its importance in detoxification of intracellular superoxide (30) or limiting production of intracellular peroxynitrite generated from SIN-1 (15).
We have detected sodC in other virulent Salmonella serovars in addition to S. typhimurium, and this gene has also been detected in bacterial pathogens such as E. coli, Brucella, Actinobacillus, Pseudomonas, Haemophilus, Pasteurella, Neisseria, and Legionella (2, 4, 5, 31). Flanking sequences with homology to bacteriophage genes indicate a possible mechanism for horizontal gene transfer of the Salmonella Cu,Zn-SOD among pathogenic bacteria, as has been described for other virulence determinants (32). An ORF immediately downstream of sodC encodes a predicted protein with homology to Ail, a Yersinia enterocolitica outer membrane protein (21) implicated in eukaryotic cell invasion, serum resistance, and virulence. This suggests that sodC may reside in a cluster of virulence-associated genes. Lateral acquisition may account for the surprisingly distant relatedness of the S. typhimurium sodC gene and its counterpart in E. coli (22). The Salmonella SodC protein described in this study is actually as closely related to SodC from P. leiognathi (55% identity) (33), a non-enteric member of the Vibrionaceae.
Damage to periplasmic or inner membrane proteins and lipids potentially could be mediated by superoxide, hydrogen peroxide, hydroxyl radical formed by the metal-catalyzed Haber–Weiss reaction, or a product of NO and superoxide, such as peroxynitrite (15, 34–36) (Fig. 6). Hydrogen peroxide and peroxynitrite can penetrate cell membranes and may also act on intracellular targets. However, the requirement of periplasmic SOD but not catalase for virulence suggests against a critical role of either hydrogen peroxide or hydroxyl. The restoration of virulence to a sodC mutant by abrogation of either NO synthase or the respiratory burst, as well as the profoundly increased susceptibility of sodC mutant Salmonella to the combination of xanthine oxidase and NO, implicate a product of NO and superoxide and, to a lesser extent, superoxide alone in bacterial killing. Cu,Zn-SOD appears most likely to protect Salmonella by detoxifying superoxide and reducing periplasmic formation of peroxynitrite or other reactive species (35) produced from superoxide and NO, although alternative mechanisms of synergistic interaction, as proposed for hydrogen peroxide and NO (37), cannot be excluded.
Figure 6.
Proposed mechanism of SodC-mediated protection against products of NO synthase and the respiratory burst NADPH-oxidase. NO·, nitric oxide; O2, superoxide; H2O2, hydrogen peroxide; ·OH, hydroxyl radical; SodC, periplasmic superoxide dismutase; KatG, periplasmic catalase.
Acknowledgments
We thank Irwin Fridovich and Jonathan Stamler for helpful discussions. This work was supported in part by the National Institutes of Health [AI01363 (M.A.D.), AI34397 (S.J.L.), AI39557 (F.C.F.), HL51967 (C.N.), GM07739 (M.S.)] and the U.S. Department of Agriculture [9401954 (F.C.F.)].
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: SOD, superoxide dismutase; ATCC, American Type Culture Collection; SIN-1, 3-morpholinosydnonimine hydrochloride; SPER/NO, 2,2′-(hydroxynitrosohydrazono)bisethanamine; IFN-γ, interferon γ; PMA, phorbol 12-myristate 13-acetate; MMA, NG-l-monomethyl arginine; AV, acetovanillone; ko, knock-out; Ity, genetic locus conferring immunity to Salmonella typhimurium (NRAMP1).
Data deposition: The sequence reported in this paper (S. typhimurium sodC) and flanking regions have been deposited in the GenBank database (accession no. AF007380).
References
- 1.McCord J M, Fridovich I. J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 2.Puget K, Michelson A M. Biochem Biophys Res Commun. 1974;58:830–838. doi: 10.1016/s0006-291x(74)80492-4. [DOI] [PubMed] [Google Scholar]
- 3.Kroll J S, Langford P R, Wilks K E, Keil A D. Microbiology. 1995;141:2271–2279. doi: 10.1099/13500872-141-9-2271. [DOI] [PubMed] [Google Scholar]
- 4.Canvin J, Langford P R, Wilks K E, Kroll J S. FEMS Microbiol Lett. 1996;136:215–220. doi: 10.1111/j.1574-6968.1996.tb08052.x. [DOI] [PubMed] [Google Scholar]
- 5.Benov L T, Fridovich I. J Biol Chem. 1994;269:25310–25314. [PubMed] [Google Scholar]
- 6.Steinman H M, Ely B. J Bacteriol. 1995;172:2901–2910. doi: 10.1128/jb.172.6.2901-2910.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benov L, Chang L Y, Day B, Fridovich I. Arch Biochem Biophys. 1995;319:508–511. doi: 10.1006/abbi.1995.1324. [DOI] [PubMed] [Google Scholar]
- 8.Buchmeier N A, Libby S J, Xu Y, Loewen P C, Switala J, Guiney D G, Fang F C. J Clin Invest. 1995;95:1047–1053. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon R, O’Connell M, Labes M, Pühler A. Methods Enzymol. 1986;118:640–659. doi: 10.1016/0076-6879(86)18106-7. [DOI] [PubMed] [Google Scholar]
- 10.Fang F C, Libby S J, Buchmeier N A, Loewen P, C, Switala J, Harwood J, Guiney D G. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Beauchamp C, Fridovich I. Anal Biochem. 1971;44:276–277. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 14.Borders C L J, Fridovich I. Arch Biochem Biophys. 1985;241:472–476. doi: 10.1016/0003-9861(85)90572-7. [DOI] [PubMed] [Google Scholar]
- 15.De Groote M A, Granger D, Xu Y, Campbell G, Prince R, Fang F C. Proc Natl Acad Sci USA. 1995;92:6399–6403. doi: 10.1073/pnas.92.14.6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding A H, Nathan C F, Stuehr D J. J Immunol. 1988;141:2407–2412. [PubMed] [Google Scholar]
- 17.Stuehr D J, Marletta M A. Proc Natl Acad Sci USA. 1985;82:7738–7742. doi: 10.1073/pnas.82.22.7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hart B A, Simons J M. Biotechnol Ther. 1992;3:119–135. [PubMed] [Google Scholar]
- 19.Pollock J D, Williams D A, Gifford M A C, Li L L, Du X, Fisherman J, Orkin S H, Doerschuk C M, Dinauer M C. Nat Genet. 1995;9:202–209. doi: 10.1038/ng0295-202. [DOI] [PubMed] [Google Scholar]
- 20.Barondess J J, Beckwith J. Nature (London) 1990;346:871–874. doi: 10.1038/346871a0. [DOI] [PubMed] [Google Scholar]
- 21.Miller V L, Bliska J B, Falkow S. J Bacteriol. 1990;172:1062–1069. doi: 10.1128/jb.172.2.1062-1069.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imlay K R, Imlay J A. J Bacteriol. 1996;178:2564–2571. doi: 10.1128/jb.178.9.2564-2571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nunoshiba T, deRojas-Walker T, Wishnok J S, Tannenbaum S R, Demple B. Proc Natl Acad Sci USA. 1993;90:9993–9997. doi: 10.1073/pnas.90.21.9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibbs J B, Jr, Vavrin Z, Taintor R R. J Immunol. 1987;138:550–565. [PubMed] [Google Scholar]
- 25.DeGroote M A, Testerman T, Xu Y, Stauffer G, Fang F C. Science. 1996;272:414–417. doi: 10.1126/science.272.5260.414. [DOI] [PubMed] [Google Scholar]
- 26.Mouy R, Fischer A, Vilmer E, Seger R, Griscelli C. J Pediatr. 1989;114:555–560. doi: 10.1016/s0022-3476(89)80693-6. [DOI] [PubMed] [Google Scholar]
- 27.Buchmeier N, Lipps C J, So M H Y, Heffron F. Mol Microbiol. 1993;7:933–936. doi: 10.1111/j.1365-2958.1993.tb01184.x. [DOI] [PubMed] [Google Scholar]
- 28.Miller R A, Britigan B E. Clin Microbiol Rev. 1997;10:1–18. doi: 10.1128/cmr.10.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dykhuizen R S, Masson J, McKnight G, Mowat A N G, Smith C C, Smith L, Benjamin N. Gut. 1996;39:393–395. doi: 10.1136/gut.39.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsolis R M, Bäumler A J, Heffron F. Infect Immun. 1995;63:1739–1744. doi: 10.1128/iai.63.5.1739-1744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.St. John G, Steinman H M. J Bacteriol. 1996;178:1578–1584. doi: 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheetham B F, Katz M E. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 33.Steinman H M. J Biol Chem. 1987;262:1882–1887. [PubMed] [Google Scholar]
- 34.Brunelli L, Crow J P, Beckman J S. Arch Biochem Biophys. 1995;316:327–334. doi: 10.1006/abbi.1995.1044. [DOI] [PubMed] [Google Scholar]
- 35.Wink D A, Cook J A, Kim S Y, Vodovotz Y, Pacelli R, Krishna M C, Russo A, Mitchell J B, Jourd’heuil D, Miles A M, Grisham M B. J Biol Chem. 1997;272:11147–11151. doi: 10.1074/jbc.272.17.11147. [DOI] [PubMed] [Google Scholar]
- 36.Fang F C. J Clin Invest. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pacelli R, Wink D A, Cook J A, Krishna M C, DeGraff W, Friedman N, Tsokos M, Samuni A, Mitchell J B. J Exp Med. 1995;182:1469–1479. doi: 10.1084/jem.182.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]