Abstract
Bile acids play important roles in the regulation of lipid, glucose, and energy homeostasis. Recent studies suggest that glucose regulates gene transcription in the liver. The aim of this study was to investigate the potential role of glucose in regulation of bile acid synthesis in human hepatocytes. High glucose stimulated bile acid synthesis and induced mRNA expression of cholesterol 7α-hydroxylase (CYP7A1), the key regulatory gene in bile acid synthesis. Activation of an AMP-activated protein kinase (AMPK) decreased CYP7A1 mRNA, hepatocyte nuclear factor 4α (HNF4α) protein, and binding to CYP7A1 chromatin. Glucose increased ATP levels to inhibit AMPK and induce HNF4α to stimulate CYP7A1 gene transcription. Furthermore, glucose increased histone acetylation and decreased H3K9 di- and tri-methylation in the CYP7A1 chromatin. Knockdown of ATP-citrate lyase, which converts citrate to acetyl-CoA, decreased histone acetylation and attenuated glucose induction of CYP7A1 mRNA expression. These results suggest that glucose signaling also induces CYP7A1 gene transcription by epigenetic regulation of the histone acetylation status. This study uncovers a novel link between hepatic glucose metabolism and bile acid synthesis. Glucose induction of bile acid synthesis may have an important implication in metabolic control of glucose, lipid, and energy homeostasis under normal and diabetic conditions.
Keywords: CYP7A1, bile acid synthesis, AMPK, nuclear receptors, glucose, epigenetic, cholestasis
Bile acids are synthesized from cholesterol in the liver (1). Bile acids are physiological detergents that play important roles in facilitating the absorption of dietary lipids and fat-soluble vitamins and biliary excretion and disposal of endogenous metabolites and xenobiotics. The primary bile acids synthesized in human liver are cholic acid and chenodeoxycholic acid, which are the endogenous ligands of nuclear receptors and signaling molecules that regulate lipid and glucose metabolism and have been implicated in fatty liver disease, obesity, and diabetes (2). In the liver, bile acids inhibit both triglyceride synthesis and gluconeogenesis (3). These effects of bile acids are mainly mediated through the bile acid-activated nuclear receptor farnesoid X receptor (FXR). Activation of FXR by bile acids or synthetic ligands improved insulin sensitivity and hyperlipidemia in diabetic mice (4). In addition, bile acids activate a G protein coupled receptor TGR5, which stimulates energy expenditure in brown adipocytes (5). TGR5 agonists have been shown to improve obesity and insulin resistance (6).
Bile acid synthesis is tightly regulated to maintain bile acid homeostasis and prevent bile acid toxicity. This is mainly achieved through the transcriptional regulation of cholesterol 7α-hydroxylase (CYP7A1), which encodes the rate-limiting enzyme in the classic bile acid biosynthetic pathway (7). CYP7A1 is repressed by bile acids returning to the liver via portal circulation. In the liver, FXR induces a negative nuclear receptor small heterodimer partner (SHP) to inhibit CYP7A1 (8). In the intestine, FXR induces fibroblast growth factor (FGF)15, which acts as an endocrine hormone to activate hepatic FGF receptor 4 to repress CYP7A1 gene transcription (9). In humans, bile acid synthesis increases during the postprandial period and decreases during fasting, which is completely opposite to increased bile acid synthesis at night and fasting, and decreased synthesis during the day in mice (10, 11). During the postprandial period, both insulin and glucose are increased. We reported previously that insulin induced while glucagon inhibited CYP7A1 gene expression and bile acid synthesis (12, 13). These recent studies may have pointed out a previously unknown but potentially important link between glucose signaling and bile acid synthesis in response to nutrition and energy status.
AMP-activated protein kinase (AMPK) is a serine/threonine protein kinase that acts as a master metabolic switch for cellular adaptation to nutritional and energy status. AMPK is allosterically activated by AMP when the cellular AMP:ATP ratio increases as a result of cellular ATP depletion. Hepatic AMPK can be activated during prolonged starvation, exercise, ischemia-reperfusion, and chronic alcohol consumption (14–16), and also by a cell permeable AMP analog 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR). In the liver, activation of AMPK inhibits the energy-consuming biosynthetic pathways including fatty acid synthesis and cholesterol synthesis by inhibiting acetyl-CoA carboxylase (ACC), fatty acid synthase, and HMG-CoA reductase (17, 18). Hepatic AMPK activation upon overexpression of a constitutive active AMPKα2 or AICAR treatment in rodent lowers triglyceride levels, whereas liver-specific deletion of AMPKα resulted in increased lipogenesis (19–21). The hypoglycemic effect of AMPK relies on both stimulation of glucose utilization, mainly in muscle and adipose, and reduction in hepatic glucose production (18). Activation of AMPK has recently emerged as a new strategy to treat diabetes and metabolic diseases (18).
It has been recognized that glucose signaling activates the transcription of a group of genes in response to carbohydrate overload. In hepatocytes, glucose inhibits AMPK and activates fatty acid synthesis, presumably via decreasing the cellular AMP:ATP ratio (22). Glucose activates a carbohydrate response element binding protein (ChREBP) to stimulate glycolysis and lipogenesis (23). The mechanism of ChREBP activation is not fully understood. It is thought that AMPK phosphorylates and inhibits ChREBP nuclear translocation, a process that is reversed by high glucose (24). Once activated, ChREBP induces hepatic glycolysis and lipogenic genes including L-pyruvate kinase (L-PK), fatty acid synthase, and ACC that direct acetyl-CoA for triglyceride synthesis (25).
Recent studies have found that hyperglycemia may cause changes in the epigenetic modification of the histone tails (acetylation/methylation) that lead to persistent alterations in gene expressions, an effect called “hyperglycemic memory” (26–28). It is reported that the glucose-induced epigenetic marks on the histone tails may be responsible for the activation of nuclear factor kB and its target genes in chronic inflammation and endothelial dysfunctions during diabetes. A recent study found that ATP-citrate lyase (ACL) is located in both the cytosol and the nucleus, and nuclear ACL converts citrate to acetyl-CoA, which is a substrate for histone acetyltransferases that acetylate the lysine residues (K) in histone 3 (H3) and H4 tails, and activates the genes involved in glucose metabolism (29).
In this study, we used primary human hepatocytes and HepG2 cells as models to study glucose regulation of CYP7A1 expression and bile acid synthesis in human livers. We found that glucose positively regulated the CYP7A1 gene and bile acid synthesis in hepatocytes. This glucose-mediated induction of CYP7A1 gene transcription may be mediated by both an AMPK-dependent pathway and epigenetic regulation of CYP7A1 chromatin structure. This study uncovered a novel mechanism of glucose-mediated regulation of bile acid synthesis and provided a link between hepatic bile acid synthesis and glucose metabolism.
MATERIALS AND METHODS
Cell culture
The human hepatoblastoma cell line, HepG2, was purchased from American Type Culture Collection (Manassas, VA). The cells were maintained in DMEM and F-12 (Sigma, St. Louis, MO) supplemented with 100U/ml penicillin G/streptomycin sulfate (Mediatech, Herndon, VA) and 10% (v/v) heat inactivated FBS (Irvine Scientific, Santa Ana, CA). Primary human hepatocytes were isolated from human donors and were obtained through the Liver Tissue and Cells Distribution System of National Institute of Diabetes and Digestive and Kidney Diseases (S. Strom, University of Pittsburgh, Pittsburgh, PA). Cells were maintained in Hepatocyte Maintenance Medium (HMM) (Clonetics, Cambrex Bioscience, Walkersville, MD). The use of human hepatocytes has been approved by Institutional Review Board and Liver Tissue and Cell Distribution System. For glucose treatment, cells were first cultured in glucose/pyruvate-free DMEM media (Gibco) supplemented with 5.5 mM glucose for 24 h followed by glucose treatment for indicated time periods and concentrations. For activation of AMPK by AICAR or Ad-AMPKα2, cells were maintained in media containing 27.5 mM glucose.
Transient transfection assay
Wild-type human CYP7A1/Luc reporter construct was described previously (30, 31). The β-galactosidase expression plasmid (pCMV-β) was obtained from Clonetech (Palo Alto, CA). The 4×HNF4/TK/Luc contains four copies of hepatocyte nuclear factor 4α (HNF4α) binding site cloned upstream of a TK promoter and luciferase gene. For reporter assays, HepG2 cells were grown to approximately 80% confluence in 24-well tissue culture plates. Luciferase reporter and β-galactosidase expression plasmids were transfected using Lipofectamin 2000 reagent (Life Technologies, Inc., Gaithersburg, MD) following manufacturer's instructions. Luciferase and β-galactosidase activities were assayed and expressed as relative luciferase activity as described previously (32). Assays were performed in triplicates and expressed as mean ± SD.
RNA isolation and quantitative real-time PCR
RNA isolation, reverse transcription reactions, and real time PCR were performed as described previously (13). All primers/probe used were TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA). Amplification of Ubiquitin C (UBC) was used in the same reactions of all samples as an internal control. Relative mRNA expression was quantified using the comparative CT (Ct) method and expressed as 2 −ΔΔCt.
Measurement of cellular ATP levels
HepG2 cells were precultured in media containing 5.5 mM glucose for 24 h, followed by 27.5 mM glucose treatment for 12 and 24 h. Cellular ATP levels were determined with an ATP bioluminescence assay kit (Roche Applied Science, Mannheim, Germany) following manufacturer's instruction.
Quantification of bile acids
Total bile acids from cells and culture media were extracted with Sep-Pak C18 cartridge and quantified by a total bile acid colorimetric assay kit (Genzyme Diagnostics, Framingham, MA) following the manufacturer's instruction.
ChIP assay
HepG2 cells were cultured in T175 tissue culture flasks. Chromatin immunoprecipitation (ChIP) assays were performed as described previously (32). Antibodies against acetyl-H3 and acetyl-H4 (Upstate Biotechnology Inc., Charlottesville, VA), HNF4α, (Santa Cruz Biotechnology, Santa Cruz, CA) and di-methyl- H3K4, di-methyl-H3K9, and tri-methyl H3K9 (Cell Signaling Technology, Danvers, MA) were used to immuno-precipitate chromatin. Taqman PCR primers/MGB probe sets were designed to detect the proximal promoter region of CYP7A1 by real-time PCR as previously described (33). Non-immune IgG was used as a measure of nonspecific background in immunoprecipitation. Chromatin purified from 10% sonicated whole cell lysate was used in PCR as “input”, and the value was set as “1”. ChIP assays were performed in triplicates and expressed as mean ± SD.
Immunoblotting analysis
Total cell lysates were analyzed by SDS-PAGE. An antibody against HNF4α was purchased from Santa Cruz Biotechnology, CA. Antibodies against ACC, phospho-ACC, AMPKα, phospho-AMPKα (Thr172), ACL, and AceCS1 were purchased from Cell Signaling Technology.
Recombinant adenovirus
Adeno-EGFP was obtained from Dr. Li Wang (University of Utah, Salt Lake City, UT). Mouse HNF4α cDNA was cloned into pShuttle-IRES-hrGFP vector (Stratagene, CA) and the recombinant adenovirus expressing mouse HNF4α was generated as described previously (4). Adenovirus expressing wild-type AMPKα2 (Ad-AMPKα2) and a dominant negative form of AMPKα2 (Ad-DN- AMPKα2) were kindly provided by Dr. Morris Birnbaum (University of Pennsylvania, Philadelphia, PA). Recombinant adeno viruses were amplified in HEK293A cells and purified with Adeno-X Virus mini purification kit (BD Biosciences, San Jose, CA). Virus titer was determined by adeno-X rapid titer kit (BD Biosciences).
RNA interference
SMART pool siRNA probes targeting to human ACL, human AceCS1, and control siRNA were purchased from Dharmacon (Lafayette, CO). siRNA were transfected with Lipofectamine RNAiMAX (invitrogen) following the manufacturer's instruction. Briefly, HepG2 cells were cultured to 50% confluent and siRNA were transfected at a final concentration of 50 nM. Four h after transfection, cells were cultured in 5.5 mM glucose for 24 h followed by either 5.5 mM or 27.5 mM glucose treatment for an additional 24 h. Cells were subsequently used for real-time PCR, immunoblot, or ChIP assay analysis.
Statistical analysis
All results were expressed as mean ± SD. Real-time PCR results of primary human hepatocytes were analyzed with paired t-test. ChIP assay results were analyzed with paired t-test. Real-time PCR and reporter assays in HepG2 cells were performed in triplicates and analyzed with student's t-test. A p-value < 0.05 was considered as significant.
RESULTS
Glucose induces CYP7A1 mRNA expression and bile acid synthesis in human hepatocytes
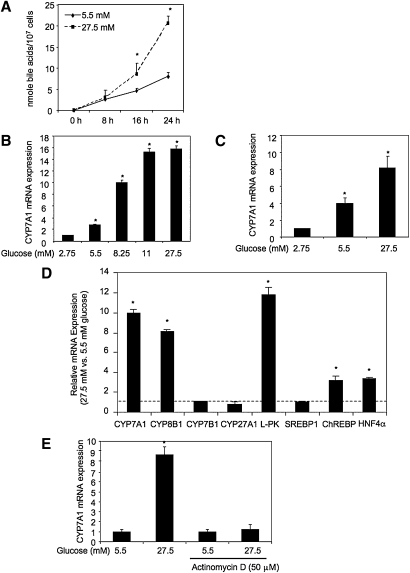
Current studies suggest important links between bile acid homeostasis and hepatic metabolism (3, 5, 10, 12, 13, 34). In this study, we investigated whether cellular glucose regulates bile acid synthesis in hepatocytes. HepG2 cells cultured in media containing high glucose (27.5 mM) showed about a 2-fold increase in bile acid synthesis rate compared with HepG2 cells cultured in the media containing normal glucose concentration (5.5 mM). The rates of bile acid synthesis in HepG2 cells (107) treated with 5.5 mM glucose and 27.5 mM glucose were determined to be about 0.32 nmole/hr and 0.73 nmole/hr, respectively (Fig. 1A). We then cultured primary human hepatocytes and HepG2 cells in media containing low (2.75 mM), normal (5.5 mM) or high (8.25–27.5 mM) glucose and measured the CYP7A1 mRNA expression by real-time PCR. Glucose dose-dependently induced CYP7A1 mRNA expression in HepG2 cells (Fig. 1B) and human hepatocytes (Fig. 1C). The high concentrations (11 and 27.5 mM) found in hyperglycemia markedly induced CYP7A1 mRNA expression in HepG2 and primary human hepatocytes by more than 10-fold. Glucose (27.5 mM) also stimulated mRNA expression of sterol 12α-hydroxylase (CYP8B1), which is involved in the synthesis of cholic acid. However, glucose had no effect on mitochondrial sterol 27-hydroxylase (CYP27A1), which oxidizes the steroid side-chain and initiates the alternative bile acid synthesis pathway, and oxysterol 7α-hydroxylase (CYP7B1) (Fig. 1D). CYP27A1 and CYP7B1 are widely expressed in most tissues and are involved in oxysterol synthesis. Glucose also induced nuclear receptor HNF4α, an important transcriptional activator of CYP7A1 and CYP8B1 in HepG2 cells (Fig. 1D). Glucose induced L-PK and ChREBP involved in glucose metabolism as expected, but had no effect on the lipogenic gene sterol response element binding protein-1 (SREBP-1). These results suggest that the glucose signaling regulates the expression of important regulatory genes involved in bile acid synthesis, in addition to glucose metabolism. The glucose-mediated induction of CYP7A1 was likely via transcriptional activation because a transcriptional inhibitor actinomycin D completely abolished glucose induction on CYP7A1 mRNA expression in HepG2 cells (Fig. 1E).
Fig. 1.
Glucose induces CYP7A1 mRNA expression and bile acid synthesis. A: Effect of glucose on bile acid synthesis in HepG2 cells. B: Effect of glucose on CYP7A1 mRNA expression in HepG2 cells. C: Effect of glucose on CYP7A1 mRNA expression in primary human hepatocytes cells. D: Effect of glucose on relative mRNA expression levels of the genes involved in bile acid synthetic and glucose metabolism in HepG2 cells. The ratios of relative mRNA expression levels in cells cultured in 27.5 mM versus 5.5 mM glucose are presented. E: Effect of actinomycin D on glucose induction of CYP7A1 mRNA expression in HepG2 cells. HepG2 cells (A, B, D, and E) or primary human hepatocytes (C) were cultured in glucose/pyruvate-free DMEM media supplemented with glucose at concentration and time indicated. Real-time PCR was used to assay relative mRNA expression. Bile acids in the culture media and cell lysate were quantified as described under Materials and Methods. * indicates statistical significance, n = 3, P < 0.05, 27.5 mM versus 5.5 mM glucose (A, D, and E); glucose concentrations versus 2.75 mM glucose (B and C).
AMPK represses CYP7A1 gene transcription and bile acid synthesis in human hepatocytes
To identify the molecular mechanism that mediates the glucose-dependent induction of the CYP7A1 gene transcription, we first tested the effect of glucose-activated transcriptional factor ChREBP on CYP7A1 gene expression. Analysis of human CYP7A1 gene sequence failed to identify potential ChREBP binding site E-box sequences. Cotransfection of a ChREBP expression plasmid stimulated the reporter activity of an L-PK promoter/reporter, but not a human CYP7A1 promoter/reporter in HepG2 cells cultured in 5.5 mM or 27.5 mM glucose (data not shown), suggesting that ChREBP does not mediate the glucose effect on CYP7A1 gene expression.
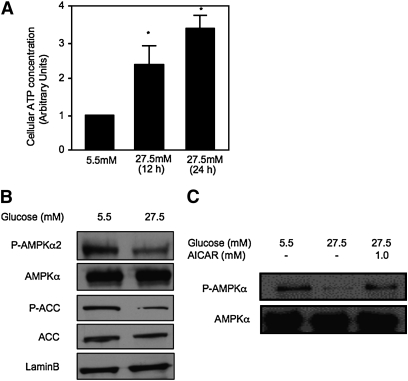
AMPK is activated by an increased cellular AMP to ATP ratio. It has been reported that high glucose represses AMPK activation in HepG2 cells (22, 35). Consistent with this report, we found that culturing HepG2 cells in 27.5 mM glucose significantly increased the cellular ATP levels (Fig. 2A) indicating a decreased AMP to ATP ratio. Glucose reduced phosphorylation of both AMPK and an AMPK target gene ACC (Fig. 2B). A similar reduction of phospho-AMPKα was also seen when primary human hepatocytes were treated with 27.5 mM glucose, and this effect was reversed by AICAR (1 mM), an AMP analog and AMPK activator (Fig. 2C).
Fig. 2.
Glucose inhibits AMPK activation. HepG2 cells (A and B) or primary human hepatocytes (C) were cultured in glucose/pyruvate-free DMEM media supplemented with 5.5 mM glucose for 24 h. Cells were then treated with glucose and/or AICAR for 24 h at concentrations indicated. A: Cellular ATP levels were determined as described under Materials and Methods and expressed as mean ± SD. * indicates statistical significance, n = 3, P < 0.05, 27.5 mM versus 5.5 mM glucose. Protein expressions were determined by immunoblot using specific antibodies.
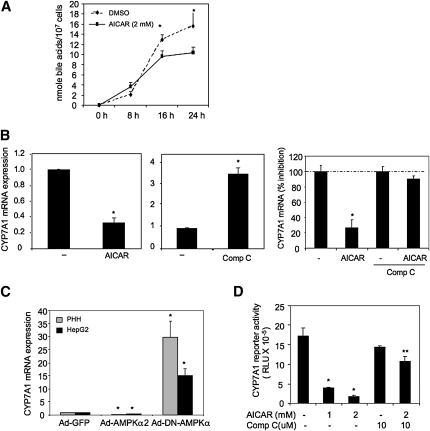
It is well known that activation of AMPK inhibits energy-consuming biosynthetic pathways, such as cholesterol and fatty acid synthesis. To investigate if AMPK also inhibits hepatic bile acid synthesis, we treated HepG2 cells with AICAR. AICAR treatment significantly repressed bile acid synthesis. The rates of bile acid synthesis in HepG2 cells (107) treated with DMSO or AICAR (2 mM) were determined to be about 0.67 nmole/hr and 0.48 nmole/hr, respectively (Fig. 3A). AICAR dose and time dependently repressed CYP7A1 mRNA expression in HepG2 cells (supplementary Fig. IA). In human primary hepatocytes, AICAR (2 mM) significantly repressed CYP7A1 mRNA by more than 60% (Fig. 3B, left panel), whereas an AMPK inhibitor Compound C (10 μM) induced CYP7A1 mRNA by ∼3-fold (middle panel). The repressive effect of AICAR on CYP7A1 mRNA was completely abolished by compound C (10 μM) in HepG2 cells (Fig. 3B, right panel). AICAR did not have any effect on CYP27A1 mRNA expression (supplementary Fig. IB), consistent with no effect of glucose on CYP7A1 mRNA expression (Fig. 1D). Furthermore, adenovirus-mediated expression of a constitutively active form of AMPK (Ad-AMPKα2) inhibited CYP7A1 mRNA in both primary human hepatocytes and HepG2 cells, while expressing a dominant negative form of AMPKα2 (Ad-DN-AMPK) induced CYP7A1 mRNA levels in these cells (Fig. 3C). Consistently, transient transfection assay showed that AICAR repressed a human CYP7A1 promoter/reporter activity, and this repressive effect was abrogated by compound C (Fig. 3D). These results suggest that glucose may stimulate bile acid synthesis and CYP7A1 gene transcription by inhibiting AMPK activity.
Fig. 3.
Activation of AMPK inhibits bile acid synthesis and CYP7A1 mRNA expression in hepatocytes. A: HepG2 cells were treated with vehicle (DMSO) or AICAR (2 mM) for the time and doses indicated. Total bile acids in culture media and cell lysates were quantified. B: Primary human hepatocytes (n = 3) were treated with vehicle (DMSO), 2 mM AICAR (left panel) or (10 μM) Compound C (middle panel), and HepG2 cells were pretreated with 10 μM compound C for 1 h followed by DMSO or AICAR (2 mM) (right panel) for 24 h. CYP7A1 mRNA expression levels were determined by real-time PCR. * indicates statistical significance, n = 3, P < 0.05, AICAR or Compound C-treated versus vehicle control (DMSO). C: Primary human hepatocytes (n = 3) or HepG2 cells were infected with Ad-GFP, Ad-AMPKα2, or Ad-DN-AMPK at MOI of 10 for 24 h. * indicates statistical significance, n = 3, P < 0.05, Ad-DNAMPKα2, or Ad-AMPKα2 versus Ad-GFP (control). D: Effect of AICAR and/or Compound C on human CYP7A1 reporter activity. HepG2 cells were transfected with a human CYP7A1 reporter (ph-1887-luc CYP7A1), and treated with AICAR and/or Compound C in the concentrations indicated. Reporter activities were assayed as described under Material and Methods. CYP7A1 mRNA expression was determined by real-time PCR. * indicates statistical significance, n = 3, P < 0.05, AICAR versus vehicle (DMSO) control; ** indicates statistical significance, n = 3, P < 0.05, Compound C + AICAR versus AICAR.
AMPK inhibits CYP7A1 by reducing HNF4α protein abundance
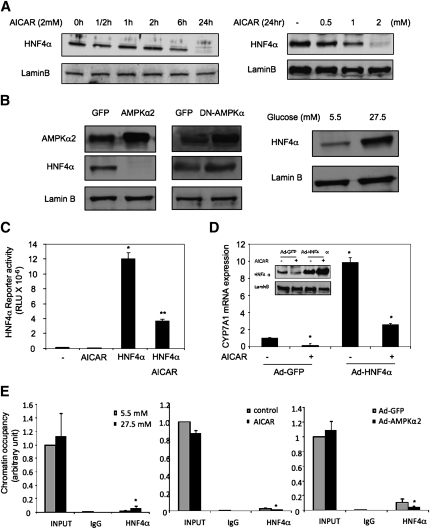
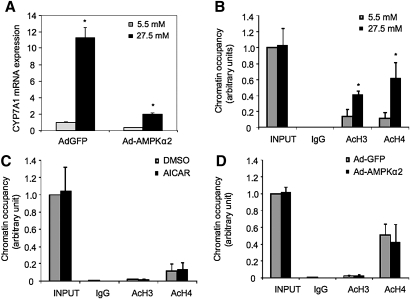
Our previous studies suggest that HNF4α is a critical activator of the human CYP7A1 gene and plays an important role in mediating CYP7A1 gene regulation in response to various cellular signals (33, 36). Because AMPK is known to phosphorylate and inhibit HNF4α trans-activation and DNA binding activity (37, 38), we tested if AMPK targets HNF4α to inhibit CYP7A1 gene transcription. AICAR treatment or adenovirus-mediated transduction of either an active or a dominant negative AMPKα2 did not affect HNF4α mRNA levels in HepG2 cells (supplementary Fig. IC). However, AICAR dramatically reduced HNF4α protein levels in a time and dose-dependent manner (Fig. 4A). Consistently, Ad-AMPKα2 decreased (Fig. 4B, left panel), whereas Ad-DN-AMPK increased HNF4α protein levels (Fig. 4B, middle panel) in HepG2 cells. These treatment conditions did not cause cytotoxicity. These results are consistent with several previous studies on AICAR inhibition of HNF4α protein levels in HepG2 cells (38–40). Furthermore, glucose markedly increased HNF4α protein levels (Fig. 4B, right panel), consistent with glucose inhibition of AMPK. To test if AMPK affects HNF4α transactivating activity, we performed transient transfection assay using a luciferase reporter construct containing four copies of the HNF4α binding site. Cotransfection of an HNF4α expression plasmid stimulated the reporter activity, which was significantly repressed by AICAR treatment (Fig. 4C). This is consistent with a previous report that AMPK reduced HNF4α trans-activating and DNA binding activity by phosphorylation (38). To further test the role of HNF4α in mediating AMPK inhibition of CYP7A1, we over-expressed HNF4α in HepG2 cells by adenovirus-mediated gene transduction. Figure 4D shows that increased HNF4α expression (inset figure) induced CYP7A1 mRNA expression, which was markedly reduced by AICAR. Quantitative ChIP assays showed that high glucose increased (Fig. 4E, left panel), whereas AICAR (Fig. 4E, middle panel) or Ad-AMPKα2 (Fig. 4E, right panel) decreased HNF4α occupancy in the CYP7A1 chromatin. HepG2 cells have been shown to express very low levels of PGC-1α and SRC-1(41), which are important coactivators of HNF4α. Thus, the inhibition of CYP7A1 by AMPK may be mainly through strong inhibition of HNF4α protein, and was unlikely via indirect modulation of coactivators. In summary, these data suggest that AMPK inhibits HNF4α trans-activation of the CYP7A1 gene.
Fig. 4.
AMPK inhibits CYP7A1 gene expression via inhibition of HNF4α protein and transactivation activity. A: HepG2 cells were treated with AICAR (2 mM) for the time and dose indicated and HNF4α protein levels were determined by immunoblot. B: HepG2 cells were infected by either Ad-GFP, Ad-AMPKα2, or Ad-DN-AMPK at an MOI of ∼10 (left panel), or treated with glucose (right panel) for 24 h. Immunoblot analyses were performed using antibodies indicated. C: AICAR inhibits HNF4α reporter activity in transient transfection assays. HepG2 cells were transfected with a heterologous HNF4α reporter (4XHNF4α-tk-Luc) and a HNF4α expression plasmid. Cells were treated with AICAR (2 mM) as indicated. D: HepG2 cells were infected with Ad-GFP or Ad-HNF4α at an MOI of ∼10 for 24 h, followed by an additional 24 h treatment of vehicle (DMSO) or AICAR (2 mM). HNF4α protein was measured by immunoblot (inset). CYP7A1 mRNA expression was determined by real-time PCR. * indicates statistical significance, n = 3, P < 0.05, AICAR-treated versus control (DMSO treated or Ad-GFP infected). E: HepG2 cells were treated with glucose (5.5 mM or 27.5 mM), AICAR (2 mM) or infected with Ad-GFP or Ad-AMPKα2 (MOI = 10) for 24 h as indicated. The occupancy of HNF4α in CYP7A1 chromatin was determined by ChIP assays as described under Materials and Methods.
Glucose induces histone acetylation of CYP7A1 promoter chromatin
Our results suggest that glucose may induce CYP7A1 via inactivation of AMPK. However, over-expression of a constitutively active AMPKα2 by adenovirus did not completely abrogate glucose induction of CYP7A1 mRNA expression (Fig. 5A), suggesting that AMPK-independent mechanisms may be involved. Several recent studies have suggested that glucose induction of gene transcription is associated with epigenetic modifications of histone (26–28). Interestingly, ChIP assays showed that high glucose (27.5 mM) increased acetylation of H3 and H4 in the CYP7A1 promoter chromatin (Fig. 5B), suggesting that increased histone acetylation may lead to activation of the CYP7A1 gene. AICAR or Ad-AMPKα2 did not affect acetylation of H3 and H4 in CYP7A chromatin (Fig. 5C, D). These results suggest that glucose-induced epigenetic activation of CYP7A1 is a different mechanism from glucose inhibition of AMPK activity.
Fig. 5.
Glucose induces histone acetylation in CYP7A1 chromatin. A: HepG2 cells were infected by Ad-GFP or Ad-AMPKα2 at an MOI of ∼10 for 24 h followed by incubation in media supplemented with 5.5 mM or 27.5 mM glucose for additional 24 h. CYP7A1 mRNA levels were determined by real time PCR. * indicates statistical significance, n = 3, P < 0.05, 27.5 mM versus 5.5 mM glucose. B: HepG2 cells were treated with glucose (5.5 mM or 27.5 mM) for 24 h as indicated. The acetyl-H3 (Ac-H3) and acetyl-H4 (Ac-H4) in CYP7A1 gene chromatin were determined by ChIP assays as described under Materials and Methods. C: HepG2 cells were treated with vehicle control or AICAR (2 mM) for 24 h, and Ac-H3 and Ac-H4 occupancy in CYP7A1 chromatin was determined by ChIP assays. D: HepG2 cells were infected with Ad-GPF or Ad-AMPKα2 for 24 h. Ac-H3 and Ac-H4 in CYP7A1 chromatin were determined by ChIP assay.
ACL is required for histone acetylation of CYP7A1 chromatin in response to glucose
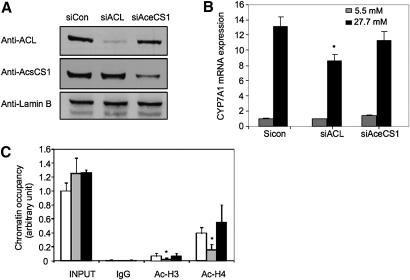
It is reported recently that glucose stimulates glycolysis and lipogenesis in mammalian cells, and nuclear ACL, which converts citrate to acetyl-CoA, is required for histone acetylation and induction of glucose-regulated genes (29). In contrast, acetyl-CoA synthase (AceCS1), which converts fatty acid-derived acetate to acetyl-CoA in the mitochondria, did not affect the acetylation status of histones. To test the role of ACL and AceCS1 in the glucose-mediated epigenetic regulation of the CYP7A1 gene, we used siRNA probes to knock down ACL and AceCS1 protein expression in HepG2 cells (Fig. 6A). Knockdown of ACL significantly reduced glucose induction of CYP7A1 (Fig. 6B) and ChREBP (positive control) mRNA expression (supplementary Fig. II), whereas knockdown of AceCS1 did not show any effect on glucose induction of CYP7A1 (Fig. 6B), and ChREBP (supplementary Fig. II). Consistent with these results, ChIP assays showed that knockdown of ACL, but not AceCS1, decreased glucose-mediated acetylation of H3 and H4 in the CYP7A1 chromatin (Fig. 6C). Thus, ACL is required for glucose-induced histone acetylation of CYP7A1 chromatin.
Fig. 6.
ACL is required for glucose induction of histone acetylation in CYP7A1. A: Immunoblot of ACL and AceCS1 in HepG2 cells transfected with siRNA control (sicon), siRNA to ACL (siACL) or siRNA to AceCS1 (siAceCS1). B: HepG2 cells were transfected with sicon, siACL, or siAceCS1 probe and treated with 5.5 mM or 27.5 mM glucose for 24 h as described under Materials and Methods. The mRNA expression levels were determined by real-time PCR. * indicates statistical significance, n = 3, P < 0.05, siACL versus sicon. C: HepG2 cells were transfected with sicon (open bar), siACL (gray bar), or siAceCS1 (closed bar), and cultured in 27.5 mM glucose for 24 h. ChIP assays were performed to detect Ac-H3 and Ac-H4 in CYP7A1 chromatin.
Glucose decreased H3K9 methylation in CYP7A1 chromatin
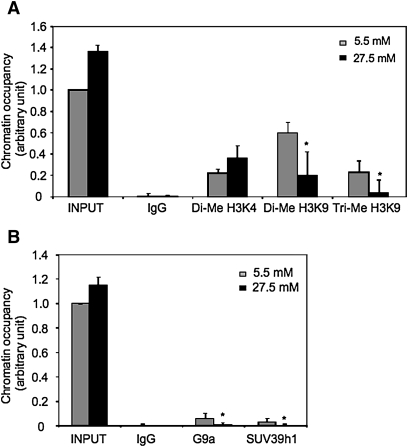
Recent studies showed that histone methylation at H3K4 and H3K9 are important epigenetic marks, whereas H3K9 methylation is associated with gene silencing and methylation of H3K4 usually causes gene activation (42, 43). Interestingly, it has been shown that bile acids inhibited the CYP7A1 gene by increasing the recruitment of histone methyltransferase G9a, which leads to increased methylation at H3K9 in CYP7A1 chromatin (44). Furthermore, these epigenetic marks could be altered by glucose in diabetes (26, 45). Our ChIP assays detected significant dimethylation of H3K4 and H3K9 and trimethylation of H3K9 within CYP7A1 promoter chromatin (Fig. 7A). Glucose at 27.5 mM had no effect on dimethylation of H3K4, but significantly decreased dimethylation and trimethylation of H3K9 on CYP7A1 chromatin. These epigenetic modifications of H3K9 by glucose in CYP7A1 gene chromatin are consistent with glucose activation of the CYP7A1 gene. Because H3K9 methylation is mainly mediated by histone methyltransferase G9a and SUV39 h1 (46, 47), we performed ChIP assays to test the binding of G9a and SUV39 h1 on the CYP7A1 chromatin. As shown in Fig. 7B, we detected G9a and SUV39 h1 binding to the CYP7A1 chromatin when HepG2 cells were cultured in 5.5 mM glucose, whereas G9a and SUV39 h1 binding was dramatically decreased to background levels by high glucose treatment. These results suggest that glucose signaling activates CYP7A1 gene transcription by inhibiting methyltransferase-mediated H3K9 di- and tri-methylation of CYP7A1 chromatin.
Fig. 7.
Glucose decreased H3K9 methylation and decreased G9a and SUV39 h1 recruitment under CYP7A1 chromatin. HepG2 cells were treated with 5.5 mM or 27.5 mM glucose as described under Materials and Methods. ChIP assays were performed to detect di-methyl H3K4, di- methyl H3K9, tri-methyl H3K9, G9a and SUV39 h1 in CYP7A1 chromatin.
DISCUSSION
This study uncovered a cross-regulation between cellular glucose metabolism and hepatic bile acid synthesis. Bile acids are known to regulate glucose homeostasis, and glucose in turn regulates bile acid synthesis by inducing CYP7A1 gene transcription. In the current study, we demonstrated that glucose increased bile acid synthesis and induced CYP7A1 gene transcription through inactivation of AMPK. At the molecular level, activation of AMPK decreases HNF4α protein stability and transactivating activity, which leads to the downregulation of CYP7A1 gene transcription. This study also revealed that high glucose increases histone acetylation and decreases histone methylation in CYP7A1 promoter chromatin. Acetyl-CoA originated from glucose via nuclear ACL may mediate these epigenetic modifications of the CYP7A1 gene.
Recent studies provided evidence that bile acid synthesis could be affected by the metabolic and nutritional states. In humans, the postprandial bile acid synthesis rate is much higher than that at night and during fasting (10). From a metabolic point of view, it makes physiological sense that increased glucose levels induce bile acid synthesis to facilitate intestinal absorption of nutrients during the postprandial period. This is also consistent with our previous report that insulin induced CYP7A1 gene expression in human hepatocytes (12). Increased glucose levels may also indirectly induce CYP7A1 by promoting insulin stimulation of CYP7A1 gene expression in human livers. Furthermore, synchronizing bile acid synthesis with hepatic glucose metabolism could have a significant implication on overall lipid, glucose, and energy homeostasis. It has been reported that bile acids may mimic the insulin action by activating AKT signaling to stimulate glycogen synthesis (48). During the postprandial period and under a carbohydrate load, excess glucose in hepatocytes stimulates lipid synthesis including cholesterol, fatty acids, and bile acids, and also glycogen synthesis to store metabolic energy in liver, muscle, and adipocytes. Conversely, under the conditions of prolonged fasting and glucose deprivation, hepatic AMPK is activated to inhibit energy consuming pathways including fatty acid synthesis, cholesterol synthesis, and bile acid synthesis. It should be noted that HepG2 cells and human hepatocytes are different with regard to bile acid synthesis. We consistently observed lower levels of CYP7A1 mRNA expression in HepG2 cells than in primary human hepatocytes, and HepG2 cells mainly synthesize CDCA but primary human hepatocytes synthesize both CA and CDCA. Thus, primary human hepatocyte is a better model than HepG2 cells for studying bile acid synthesis in human liver. Due to the difficulty in obtaining a sufficient amount of human hepatocytes for analysis, our study has been limited mainly to using HepG2 cells. In this study, we did not analyze the effect of glucose on bile acid composition, which will require a large amount of cells for analysis. Our preliminary study showed that high glucose treatment also strongly induced CYP8B1 mRNA expression in HepG2 cells (Fig. 1D). It is possible that glucose may increase CA production and thus increase the ratio of CA to CDCA, and the hydrophilicity of bile acids in the bile.
A recently discovered role of nuclear ACL in histone acetylation provides a potential link between glucose signaling and epigenetic regulation of gene transcription (29). It appears that acetyl-CoA derived from glycolysis and a product of nuclear ACL could significantly affect the acetylation status of histones not only in glycolytic genes but also in bile acid biosynthetic genes in hepatocytes. Our ChIP assays also revealed that H3K9 in CYP7A1 chromatin was extensively methylated, consistent with low basal expression of CYP7A1 in human hepatocytes. Glucose may induce CYP7A1 expression by both decreasing H3K9 methylation and increasing histone acetylation status in CYP7A1 chromatin. Thus, hepatic glucose metabolism could play a critical role in the epigenetic regulation of CYP7A1 expression to maintain cholesterol and bile acid homeostasis.
The understanding of bile acid synthesis in human diabetes is still lacking. It is largely limited by the complicated metabolic changes in diabetic conditions as well as the clear species differences in regulation of bile acid synthesis in human and rodent models. However, an early study reported that bile acid pool was significantly higher in human diabetic patients with uncontrolled hyperglycemia than in diabetic patients with controlled euglycemia (49). It was suggested that increased bile acid pool in hyperglycemic patients was likely due to an increased de novo bile acid synthesis. Our current study provides a molecular mechanism for glucose induction of bile acid synthesis in human hepatocytes. Based on this study, the increased bile acid synthesis in hyperglycemia could be due to persistent epigenetic changes induced by chronic exposure of hepatocytes to high glucose. Because of the distinct species differences in the regulation of bile acid synthesis between humans and rodents and a lack of a suitable in vivo model for studying glucose regulation of bile acid synthesis, future clinical study of hyperglycemic patients would provide further insight into the mechanism of cross- regulation of glucose and bile acid metabolism in humans.
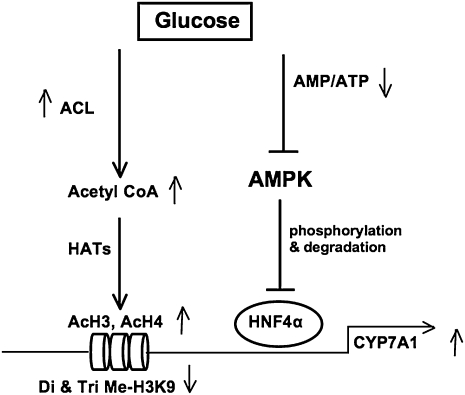
In summary, our current study provides a novel link between bile acid homeostasis and hepatic glucose and energy metabolisms. Glucose induces hepatic bile acid synthesis by two mechanisms (Fig. 8). Glucose increases ATP and decreases the AMP/ATP ratio to inhibit AMPK activity. This results in increasing HNF4α protein, which induces CYP7A1 gene expression. Glucose signal also induces CYP7A1 gene transcription by epigenetic modifications of the acetylation status of CYP7A1 chromatin structure. This is accomplished by increasing nuclear ACL, which stimulates histone acetyltransferease activity to acetylate H3 and H4 tails, and also reduces di- and tri-methylation of H3K9 by a methytransferase G9a, which is known to inhibit CYP7A1 gene transcription. Cross-regulation of glucose and bile acid metabolism may have important implications in metabolic control of glucose, lipid, and energy homeostasis under normal physiology and diabetic conditions.
Fig. 8.
A model of glucose activation of CYP7A1 gene transcription in hpatocytes. Glucose induces CYP7A1 gene transcription by two mechanisms. First, high glucose decrease the cellular AMP to ATP ratio, thus results in inhibiting AMPK activity. AMPK is known to phosphorylate and inhibit HNF4α binding to DNA and dimerization. Inactivation of AMPK increases HNF4α recruitment to CYP7A1 chromatin to induce CYP7A1 gene transcription. Second, glucose stimulates CYP7A1 gene transcription by the epigenetic mechanisms. Glucose increases nuclear ACL activity to increase acetyl-CoA, which is a substrate of histone acetyltransferases (HATs). HAT acetylates H3 and H4 on CYP7A1 chromatin, and stimulates gene transcription. Furthermore, glucose reduces chromatin occupancy of a methyltransferase G9a, which is known to inhibit CYP7A1 gene transcription. This results in de-repression of CYP7A1 gene transcription by decreasing di-and tri-methylation of H3K9 in CYP7A1 chromatin.
Supplementary Material
Footnotes
Abbreviations:
- ACC
- acyl-CoA carboxylase
- ACL
- ATP-citrate lyase
- Ad
- adenovirus
- AICAR
- 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside
- AMPK
- AMP-activated protein kinase
- CA
- cholic acid
- CDCA
- chenodeoxycholic acid
- ChREBP
- carbohydrate response element
- CYP7A1
- cholesterol 7α-hydroxylase
- CYP8B1
- sterol 12α-hydroxylase
- FXR
- farnesoid X receptor
- FGF
- fibroblast growth factor
- HNF4α
- hepatocyte nuclear factor 4α
- L-PK
- liver pyruvate kinase
This study is supported by grants from National Institute of Diabetes and Digestive and Kidney Diseases DK 58379 and DK44442 (J.Y.L.C), American Heart Association Great River Affiliate Postdoctoral Fellowship (T.L.), and American Heart Association Scientist Development Grant (Y.Z.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Russell D. W., Setchell K. D. 1992. Bile acid biosynthesis. Biochemistry. 31: 4737–4749 [DOI] [PubMed] [Google Scholar]
- 2.Lefebvre P., Cariou B., Lien F., Kuipers F., Staels B. 2009. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89: 147–191 [DOI] [PubMed] [Google Scholar]
- 3.Watanabe M., Houten S. M., Wang L., Moschetta A., Mangelsdorf D. J., Heyman R. A., Moore D. D., Auwerx J. 2004. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Invest. 113: 1408–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Lee F. Y., Barrera G., Lee H., Vales C., Gonzalez F. J., Willson T. M., Edwards P. A. 2006. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc. Natl. Acad. Sci. USA. 103: 1006–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watanabe M., Houten S. M., Mataki C., Christoffolete M. A., Kim B. W., Sato H., Messaddeq N., Harney J. W., Ezaki O., Kodama T., et al. 2006. Bile acids induce energy expenditure by promoting intra cellular thyroid hormone activation. Nature. 439: 484–489 [DOI] [PubMed] [Google Scholar]
- 6.Houten S. M., Watanabe M., Auwerx J. 2006. Endocrine functions of bile acids. EMBO J. 25: 1419–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang J. Y.1998. Regulation of bile acid synthesis. Front. Biosci. 3: D176–D193 [DOI] [PubMed] [Google Scholar]
- 8.Goodwin B., Jones S. A., Price R. R., Watson M. A., McKee D. D., Moore L. B., Galardi C., Wilson J. G., Lewis M. C., Roth M. E., et al. 2000. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell. 6: 517–526 [DOI] [PubMed] [Google Scholar]
- 9.Inagaki T., Choi M., Moschetta A., Peng L., Cummins C. L., McDonald J. G., Luo G., Jones S. A., Goodwin B., Richardson J. A., et al. 2005. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2: 217–225 [DOI] [PubMed] [Google Scholar]
- 10.Galman C., Angelin B., Rudling M. 2005. Bile acid synthesis in humans has a rapid diurnal variation that is asynchronous with cholesterol synthesis. Gastroenterology. 129: 1445–1453 [DOI] [PubMed] [Google Scholar]
- 11.Kovar J., Lenicek V., Zimolova M., Vitek L., Jirsa M., Pitha P. Regulation of diurnal variation of cholesterol 7alpha-hydroxylase (CYP7A1) activity in healthy subjects. Physiol. Res. Epub ahead of print. June 19, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Li T., Kong X., Owsley E., Ellis E., Strom S., Chiang J. Y. 2006. Insulin regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes: roles of forkhead box O1 and sterol regulatory element-binding protein 1c. J. Biol. Chem. 281: 28745–28754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song K. H., Chiang J. Y. 2006. Glucagon and cAMP inhibit cholesterol 7alpha-hydroxylase (CYP7A1) gene expression in human hepatocytes: discordant regulation of bile acid synthesis and gluconeogenesis. Hepatology. 43: 117–125 [DOI] [PubMed] [Google Scholar]
- 14.Assifi M. M., Suchankova G., Constant S., Prentki M., Saha A. K., Ruderman N. B. 2005. AMP-activated protein kinase and coordination of hepatic fatty acid metabolism of starved/carbohydrate-refed rats. Am. J. Physiol. Endocrinol. Metab. 289: E794–E800 [DOI] [PubMed] [Google Scholar]
- 15.Peralta C., Bartrons R., Serafin A., Blazquez C., Guzman M., Prats N., Xaus C., Cutillas B., Gelpi E., Rosello-Catafau J. 2001. Adenosine monophosphate-activated protein kinase mediates the protective effects of ischemic preconditioning on hepatic ischemia-reperfusion injury in the rat. Hepatology. 34: 1164–1173 [DOI] [PubMed] [Google Scholar]
- 16.You M., Matsumoto M., Pacold C. M., Cho W. K., Crabb D. W. 2004. The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology. 127: 1798–1808 [DOI] [PubMed] [Google Scholar]
- 17.Clarke P. R., Hardie D. G. 1990. Regulation of HMG-CoA reductase: identification of the site phosphorylated by the AMP-activated protein kinase in vitro and in intact rat liver. EMBO J. 9: 2439–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viollet B., Lantier L., Devin-Leclerc J., Hebrard S., Amouyal C., Mounier R., Foretz M., Andreelli F. 2009. Targeting the AMPK pathway for the treatment of Type 2 diabetes. Front. Biosci. 14: 3380–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergeron R., Previs S. F., Cline G. W., Perret P., Russell R. R., 3rd, Young L. H., Shulman G. I. 2001. Effect of 5-aminoimidazole-4-carboxamide-1-beta-D-ribofuranoside infusion on in vivo glucose and lipid metabolism in lean and obese Zucker rats. Diabetes. 50: 1076–1082 [DOI] [PubMed] [Google Scholar]
- 20.Foretz M., Ancellin N., Andreelli F., Saintillan Y., Grondin P., Kahn A., Thorens B., Vaulont S., Viollet B. 2005. Short-term overexpression of a constitutively active form of AMP-activated protein kinase in the liver leads to mild hypoglycemia and fatty liver. Diabetes. 54: 1331–1339 [DOI] [PubMed] [Google Scholar]
- 21.Andreelli F., Foretz M., Knauf C., Cani P. D., Perrin C., Iglesias M. A., Pillot B., Bado A., Tronche F., Mithieux G., et al. 2006. Liver adenosine monophosphate-activated kinase-alpha2 catalytic subunit is a key target for the control of hepatic glucose production by adiponectin and leptin but not insulin. Endocrinology. 147: 2432–2441 [DOI] [PubMed] [Google Scholar]
- 22.Zang M., Zuccollo A., Hou X., Nagata D., Walsh K., Herscovitz H., Brecher P., Ruderman N. B., Cohen R. A. 2004. AMP-activated protein kinase is required for the lipid-lowering effect of metformin in insulin-resistant human HepG2 cells. J. Biol. Chem. 279: 47898–47905 [DOI] [PubMed] [Google Scholar]
- 23.Yamashita H., Takenoshita M., Sakurai M., Bruick R. K., Henzel W. J., Shillinglaw W., Arnot D., Uyeda K. 2001. A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci. USA. 98: 9116–9121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaguchi T., Takenoshita M., Kabashima T., Uyeda K. 2001. Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc. Natl. Acad. Sci. USA. 98: 13710–13715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Towle H. C.2005. Glucose as a regulator of eukaryotic gene transcription. Trends Endocrinol. Metab. 16: 489–494 [DOI] [PubMed] [Google Scholar]
- 26.Villeneuve L. M., Reddy M. A., Lanting L. L., Wang M., Meng L., Natarajan R. 2008. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc. Natl. Acad. Sci. USA. 105: 9047–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Osta A., Brasacchio D., Yao D., Pocai A., Jones P. L., Roeder R. G., Cooper M. E., Brownlee M. 2008. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J. Exp. Med. 205: 2409–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brasacchio D., Okabe J., Tikellis C., Balcerczyk A., George P., Baker E. K., Calkin A. C., Brownlee M., Cooper M. E., El-Osta A. 2009. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 58: 1229–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., Thompson C. B. 2009. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 324: 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crestani M., Stroup D., Chiang J. Y. 1995. Hormonal regulation of the cholesterol 7 alpha-hydroxylase gene (CYP7). J. Lipid Res. 36: 2419–2432 [PubMed] [Google Scholar]
- 31.Wang D. P., Stroup D., Marrapodi M., Crestani M., Galli G., Chiang J. Y. L. 1996. Transcriptional regulation of the human cholesterol 7α-hydroxylase gene (CYP7A) in HepG2 cells. J. Lipid Res. 37: 1831–1841 [PubMed] [Google Scholar]
- 32.Li T., Chiang J. Y. 2005. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7 alpha-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 288: G74–G84 [DOI] [PubMed] [Google Scholar]
- 33.Li T., Chiang J. Y. 2007. A novel role of transforming growth factor beta1 in transcriptional repression of human cholesterol 7alpha-hydroxylase gene. Gastroenterology. 133: 1660–1669 [DOI] [PubMed] [Google Scholar]
- 34.Lee F. Y., Lee H., Hubbert M. L., Edwards P. A., Zhang Y. 2006. FXR, a multipurpose nuclear receptor. Trends Biochem. Sci. 31: 572–580 [DOI] [PubMed] [Google Scholar]
- 35.Zang M., Xu S., Maitland-Toolan K. A., Zuccollo A., Hou X., Jiang B., Wierzbicki M., Verbeuren T. J., Cohen R. A. 2006. Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes. 55: 2180–2191 [DOI] [PubMed] [Google Scholar]
- 36.Li T., Jahan A., Chiang J. Y. 2006. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the JNK/c-jun pathway in human liver cells. Hepatology. 43: 1202–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leclerc I., Lenzner C., Gourdon L., Vaulont S., Kahn A., Viollet B. 2001. Hepatocyte nuclear factor-4alpha involved in type 1 maturity-onset diabetes of the young is a novel target of AMP-activated protein kinase. Diabetes. 50: 1515–1521 [DOI] [PubMed] [Google Scholar]
- 38.Hong Y. H., Varanasi U. S., Yang W., Leff T. (2003) AMP-activated protein kinase regulates HNF4alpha transcriptional activity by inhibiting dimer formation and decreasing protein stability. J. Biol. Chem. 278: 27495–27501 [DOI] [PubMed] [Google Scholar]
- 39.Prieur X., Schaap F. G., Coste H., Rodriguez J. C. 2005. Hepatocyte nuclear factor-4alpha regulates the human apolipoprotein AV gene: identification of a novel response element and involvement in the control by peroxisome proliferator-activated receptor-gamma coactivator-1alpha, AMP-activated protein kinase, and mitogen-activated protein kinase pathway. Mol. Endocrinol. 19: 3107–3125 [DOI] [PubMed] [Google Scholar]
- 40.Horike N., Sakoda H., Kushiyama A., Ono H., Fujishiro M., Kamata H., Nishiyama K., Uchijima Y., Kurihara Y., Kurihara H., et al. 2008. AMP-activated protein kinase activation increases phosphorylation of glycogen synthase kinase 3beta and thereby reduces cAMP-responsive element transcriptional activity and phosphoenolpyruvate carboxykinase C gene expression in the liver. J. Biol. Chem. 283: 33902–33910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Jimenez C. P., Gomez-Lechon M. J., Castell J. V., Jover R. 2006. Underexpressed coactivators PGC1alpha and SRC1 impair hepatocyte nuclear factor 4 alpha function and promote dedifferentiation in human hepatoma cells. J. Biol. Chem. 281: 29840–29849 [DOI] [PubMed] [Google Scholar]
- 42.Jenuwein T., Allis C. D. 2001. Translating the histone code. Science. 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- 43.Martin C., Zhang Y. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6: 838–849 [DOI] [PubMed] [Google Scholar]
- 44.Fang S., Miao J., Xiang L., Ponugoti B., Treuter E., Kemper J. K. 2007. Coordinated recruitment of histone methyltransferase G9a and other chromatin-modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol. Cell. Biol. 27: 1407–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reddy M. A., Villeneuve L. M., Wang M., Lanting L., Natarajan R. 2008. Role of the lysine-specific demethylase 1 in the proinflammatory phenotype of vascular smooth muscle cells of diabetic mice. Circ. Res. 103: 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Rea S., Eisenhaber F., O'Carroll D., Strahl B. D., Sun Z. W., Schmid M., Opravil S., Mechtler K., Ponting C. P., Allis C. D., et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 406: 593–599 [DOI] [PubMed] [Google Scholar]
- 47.Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., et al. 2002. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 16: 1779–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han S. I., Studer E., Gupta S., Fang Y., Qiao L., Li W., Grant S., Hylemon P. B., Dent P. 2004. Bile acids enhance the activity of the insulin receptor and glycogen synthase in primary rodent hepatocytes. Hepatology. 39: 456–463 [DOI] [PubMed] [Google Scholar]
- 49.Bennion L. J., Grundy S. M. 1977. Effects of diabetes mellitus on cholesterol metabolism in man. N. Engl. J. Med. 296: 1365–1371 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.