Abstract
Human fibroblasts from normal subjects and Niemann-Pick A (NPA) disease patients were fed with two labeled metabolic precursors of sphingomyelin (SM), [3H]choline and photoactivable sphingosine, that entered into the biosynthetic pathway allowing the synthesis of radioactive phosphatidylcholine and SM, and of radioactive and photoactivable SM ([3H]SM-N3). Detergent resistant membrane (DRM) fractions prepared from normal and NPA fibroblasts resulted as highly enriched in [3H]SM-N3. However, lipid and protein analysis showed strong differences between the two cell types. After cross-linking, different patterns of SM-protein complexes were found, mainly associated with the detergent soluble fraction of the gradient containing most cell proteins. After cell surface biotinylation, DRMs were immunoprecipitated using streptavidin. In conditions that maintain the integrity of domain, SM-protein complexes were detectable only in normal fibroblasts, whereas disrupting the membrane organization, these complexes were not recovered in the immunoprecipitate, suggesting that they involve proteins belonging to the inner membrane layer. These data suggest that differences in lipid and protein compositions of these cell lines determine specific lipid-protein interactions and different clustering within plasma membrane. In addition, our experiments show that photoactivable sphingolipids metabolically synthesized in cells can be used to study sphingolipid protein environments and sphingolipid-protein interactions.
Keywords: gangliosides, lipid rafts, membranes, Niemann-Pick disease, sphingolipids, sphingomyelin, photo labeling
Sphingolipids (SLs) are cell membrane components highly enriched in lipid domains, which are portions of the membrane also enriched in cholesterol, dipalmitoylphosphatidylcholine, and proteins necessary for the cell signaling processes (1). Much information is available on the composition of membrane lipid domains of cells in culture (2, 3) and tissues (4). On the other hand, there is somewhat scant information on how lipids are capable of modulating protein functions and on the distribution of components within the two membrane layers. A possible and widely accepted working hypothesis on this latter point is that the fine tuning of receptor functions is achieved by dynamic lateral interactions with SLs (5). These interactions would induce conformational changes in the receptors, directly (e.g., changing its intrinsic tyrosine kinase activity) (6, 7) or indirectly (e.g., changing its association with regulator or substrate proteins) (8), thus affecting their biological functions. As SLs, glycosphingolipids (GSLs) attracted the interest of scientists and many papers are available on membrane ganglioside-protein interactions and on the regulatory effect exerted by gangliosides on several membrane proteins (5). In addition, in recent years, sphingomyelin (SM) was suggested to have a specific role in membrane organization (9, 10). In addition, the availability of a membrane SM synthase besides membrane sphingomyelinases suggests the existence of a physiological process capable of modifying the SM:ceramide ratio at the membrane level and, as a consequence, some membrane properties like membrane curvature and membrane organization, according to the necessity of the cell (11).
Studies on ganglioside-protein environment and ganglioside-protein interactions were performed in the past by loading the cell plasma membranes with photoactivable and tritium labeled compounds that after illumination, form covalent linkages with closely locating proteins (12–14). As a result, radioactive ganglioside-protein complexes could be isolated and analyzed.
Loading the plasma membrane with photoactivable SM results in difficulties due to the amphiphilic properties of the molecule. In fact, SM is much more hydrophobic than gangliosides and in aqueous solution, forms large aggregates that are in equilibrium with a very scant number of monomers. Only monomers become components of the membranes, whereas the large aggregates are phagocyted and then catabolyzed in the lysosomes (15, 16). To overcome this and to study SM-protein interactions, cultures of human fibroblasts prepared from normal subjects and from Niemann-Pick type A (NPA) disease patients were fed with photoactivable sphingosine and tritiated choline, both precursors for the biosynthesis of [3H]SM-N3. NPA disease is a genetic SM lysosomal storage disease due to the lack of acid sphingomyelinase activity (17). These cells have a different SM content and make it possible to study the role of the excess of SM in membrane organization.
MATERIALS AND METHODS
Cell cultures
Normal human skin fibroblasts were obtained by the punch technique from normal children (age 6–12 months), cultured and propagated as described (18) in 60 mm dishes (0.42 ± 0.10 mg protein/dish), using Minimal Essential Medium with Earle's salt (EMEM) supplemented with 10% FBS. Pathological human fibroblasts, provided by Istituto Nazionale Neurologico “Carlo Besta”, were prepared as above from a 7-year-old NPA patient. Pathological cells had an acidic sphingomyelinase activity on the artificial substrate 2-N-hexadecanoylamino-4-nitrophenylphosphorylcholine lower than 4% and a 6-fold increase of SM with respect to control cells. For the experiments, cells were used at confluence.
Synthesis of Sph-N3
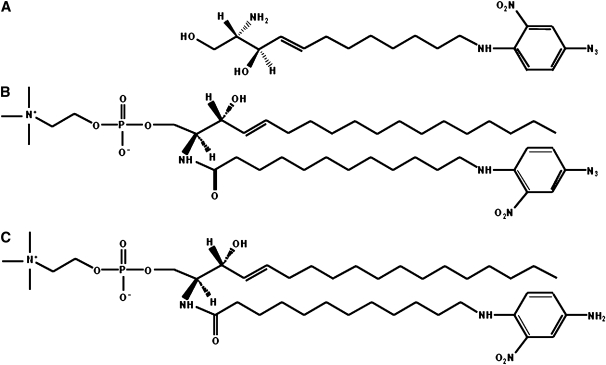
Sphingosine (Sph) derivative, containing the amine group at the terminal carbon 12 (Sph-NH2), was prepared as reported (19–21). The nitrophenylazide was then coupled to the N-terminal group and the protection group was removed in order to obtain photoactivable sphingosine (Sph-N3) (Fig. 1A). The reactions details are described in the supplementary data.
Fig. 1.
Chemical structures. A: Photoactivable sphingosine derivate, Sph-N3. B: Photoactivable sphingomyelin, SM-N3. C: Reduced form of SM-N3, SM-NH2.
Synthesis of photoactivable SM (SM-N3) and its reduced form SM-NH2
SM containing a photoactivable fatty acid chain (see Fig. 1B) was synthesized under red safelight. A total of 286 μmoles of 12-aminododecanoic acid (Merck GmbH) were dissolved in 5 ml of anhydrous dimethylformamide. An equimolar quantity of triethylamine and a 2-fold molar quantity of 4-F-3-NO2-phenylazide (Sigma), dissolved in 5.8 ml of anhydrous methanol were added. The reaction was stirred overnight at 80°C. The mixture was dried and the photoactivable fatty acid was purified by two flash chromatographic columns (silica gel 60, hexane/EtOAc, 1:2 by vol and silica gel 60, hexane/EtOAc, 2:1 by vol) (80% yield).
2-Chloro-1-methylpyridinium iodide (Sigma), pentafluorophenol (Merck GmbH), photoactivable 12-aminododecanoic acid, and dry tributylamine in the molar ratio of 1.2:1.1:1:2.4 were solubilized in this order in 230 μl of dry dichloromethane in a three neck flask provided with an N2 septum-inlet adaptor and a condenser. The reaction mixture was refluxed for 3 h under continuous stirring, then cooled and evaporated under vacuum. The product was purified by flash chromatography with silica gel 60 column equilibrated and sequentially eluted with hexane/EtOAc, 24:1 by vol and hexane/EtOAc 1:4 (90% yield).
To a solution of 210 μmoles of sphingosylphosphorylcholine (Sigma) in 2.5 ml of chloroform:methanol, 1:2 by vol, photoactivable 12-aminolauric acid pentafluorphenylester, 1-hydroxybenzotyriazole (Sigma), and tributylamine in the molar ratio of 1.1:1.8:1.5 with respect to fatty acid derivative, were added. After vigorous stirring for 12 h at room temperature, the reaction mixture was dried and SM-N3 purified by a silica gel 60 flash column (1 × 15 cm), equilibrated, and eluted with chloroform/methanol/water, 70:25:3 by vol.
To a solution of 100 μmoles of SM-N3 in 4 ml of tetrahydrofurane (THF) and 1ml of 100 mM Tris-HCl, pH 7.4, dithiothreitol (Sigma) was added to a 200 mM final concentration. The reaction mixture was stirred for 48 h at room temperature and the SM-NH2 synthesized was purified on C-18 reverse-phase silica gel flash column (1 × 15 cm), equilibrated and eluted with methanol/water/triethylamine, 15:2:1 by vol (22). Structural analysis of SM-NH2 was performed by mass spectrometry.
Cell proliferation and viability after Sph and Sph-N3 treatment
A total of 25 × 103 cells of normal or NPA fibroblasts were plated in 60 mm cell culture plates. After 6 h, the cells were treated with 38 nM of Sph or Sph-N3 for up to 72 h. After 24, 48, and 72 h, cells were detached with trypsin, incubated in 0.25% Trypan blue solution for 2 min, and counted using a Bürker chamber. The dead cells were determined to be Trypan blue-positive cells.
Feeding of cells with Sph-N3 followed by [3-3H(Sph)]GM1 or [3-3H(Sph)]SM administration
Confluent 60 mm cell plates of normal and NPA fibroblasts were shifted in a dark room under red safelight and incubated for 2 h with 38 nM Sph-N3 solubilized in EMEM containing 10% FBS.
[3-3H(Sph)]GM1 (1.2 Ci/mmol) or [3-3H(Sph)]SM (0.375 Ci/mmol) (23) were solubilized in an appropriate volume of prewarmed (37°C) EMEM to obtain a Sph concentration of 5 × 10−7 M and 15 × 10−7 M, respectively. The medium from normal and NPA fibroblasts, fed or not with Sph-N3, was removed and the cells rapidly washed with EMEM. A total of 2 ml of the medium containing the radioactive lipids was added to each plate and the cells were incubated for 4 h pulse at 37°C. After pulse, cells were washed twice with serum-free culture medium and then maintained in normal cell culture medium for 15 h chase. Cells were washed twice with PBS and harvested in water. After lyophilization, cells were submitted to lipid extraction. The incorporation of radioactivity was determined by liquid scintillation counting and the radioactive lipids were separated by high-performance thin-layer chromatography (HPTLC) with the solvent system CHCl3/CH3OH/(CH3)2CHOH/0.2% aqueous CaCl2, 20:60:20:4 by vol and analyzed by radioimaging.
Feeding of cells with Sph-N3 and [methyl-3H]choline
Confluent cell cultures were shifted in a dark room under red safelight and incubated for 2 h with 38 nM Sph-N3 solubilized in EMEM containing 10% FBS. Cells were then maintained in serum-free EMEM for 12 h and in serum-free EMEM containing 40 μM zinc chloride for other 12 h (24). Then cells were fed with 56 μM [methyl-3H]choline (Amersham) solubilized in serum-free EMEM for 12 h. Finally medium was substituted with EMEM containing 10% FBS for the last 12 h of incubation.
Cell surface biotinylation and cell illumination
Cells fed with Sph-N3 and [methyl-3H]choline were washed five times with culture medium containing 10% FBS and twice with PBS. Cells were then maintained for 1 h in serum starvation, washed twice with PBS, and incubated with 0.25 mg/ml of sulfo-NHS-biotin (Pierce) in PBS, pH 7.4 (5 ml/dish) for 30 min at 4°C (25). Under these experimental conditions, the internalization of the biotin derivative does not occur and biotinylation is restricted to the cell surface (26). After biotin labeling, cells were rinsed twice with ice-cold PBS, 4 ml of ice-cold PBS were added, and cells were illuminated for 45 min under ultraviolet light (λ=360 nm) (27–30). Cell viability was assessed by the trypan blue exclusion method (31).
Preparation of DRM fractions by sucrose gradient centrifugation
After photolabeling and biotinylation, cells were subjected to homogenization and ultracentrifugation on discontinuous sucrose gradient, as previously described (2). Briefly, cells were harvested, lysed in 1% Triton X-100 in TNEV (10 mM Tris-HCl buffer, pH 7.5, 150 mM NaCl, 5 mM EDTA) in the presence of 1 mM Na3VO4, 1 mM PMSF, and 75 mU/ml aprotinin, and Dounce homogenized (10 strokes, tight). Cell lysate (1 mg of cell protein/ml) was centrifugated 5 min at 1,300 g to remove nuclei and cellular debris. The postnuclear fraction was mixed with an equal volume of 85% sucrose (w/v) in TNEV, placed at the bottom of a discontinuous sucrose gradient (30–5%), and centrifugated for 17 h at 200,000 g at 4°C. After ultracentrifugation, 11 fractions were collected starting from the top of the tube. Equal amounts of the low density fractions 4, 5, and 6 were put together to obtain the detergent-resistant membrane (DRM) fraction, whereas equal amounts of the high density (HD) fractions 10 and 11 were put together to obtain the HD fraction. The entire procedure was performed at 0–4°C in ice immersion.
Immunoprecipitation experiments
A total of 300 μl of DRM fraction containing 5–10 μg of proteins was immunoprecipitated with 50 μl of streptavidin-coupled magnetic beads (Dynal) previously washed twice with PBS. The mixtures were stirred overnight at 4°C, then the immunoprecipitate (IP) was recovered by centrifugation (32). Under these conditions (domain-preserving conditions), we preserved the organization of lipid domains (32, 33). In some experiments, IP samples were treated with 1% SDS in lysis buffer at 100°C for 5 min, then diluted 10-fold with lysis buffer (0.1% SDS final concentration) and reimmunoprecipitated as described previously, obtaining a new IP. These conditions (domain-disrupting conditions) are known to break up the membrane organization and to allow the disaggregation of the DRM domains (34).
Analysis of protein patterns
Total cell homogenate, sucrose gradient fractions, IP, and supernatant remaining after immunoprecipitation were analyzed by SDS-PAGE. Total protein patterns were determined by silver-staining directly to the gel, according to manufacturer's instructions. Radioactive proteins due to the cross-linkage with [3H]SM-N3 were analyzed by digital autoradiography after SDS-PAGE and blotting on a polyvinyldifluoride (PVDF) membrane. Biotinylated proteins were recognized with horseradish peroxidase-conjugated streptavidin (Vector) and enhanced chemiluminescence detection (Pierce Supersignal).
Lipid analysis
Aliquots of cell homogenate, low density (4, 5, 6,) and high density fractions (10, 11,) were dialyzed and lyophilized. These samples were subjected to lipid extraction with CHCl3/CH3OH/H2O, 2:1:0.1 by vol (35). The total lipid extract was analyzed by thin layer chromatography (HPTLC Kieselgel 60, 10 × 10 cm) with the solvent system CHCl3/CH3OH/0.2% aqueous CaCl2, 50:40:8 by vol. The HPTLC visualization was obtained by digital autoradiography and the identification of lipids was accomplished by comigration with standard lipids, the lipid mixture characterization having been previously established (2).
To verify the biosynthesis of [3H]SM-N3, cells fed with [methyl-3H]-choline and Sph-N3 were subjected to detergent homogenization, sucrose gradient fractionation, and lipid analysis under red safelight. Then, again under red safelight, the total lipid extracts obtained from cell homogenate, DRM, and HD fractions were dried and dissolved in 40 μl of THF. 10 μl of 100 mM Tris-HCl containing 200 mM dithiothreitol were added and the reaction mixture was stirred for 72 h at room temperature. Radioactive SM-NH2 was analyzed by HPTLC performed with two sequential runs in CHCl3/CH3OH/H2O, 50:40:8 by vol, followed by digital autoradiography.
Aliquots of the total lipid extracts from cell homogenates, DRM, and HD fractions were subjected to a two-phase partitioning (35).
The organic phases were dried under nitrogen flow and resuspended with 100 μl of CHCl3 and 100 μl of 0.6 M NaOH in CH3OH and allowed to stand at 37°C for 3 h and overnight at room temperature. This alkaline treatment allows for removal of glycerophospholipids from the organic phases breaking their ester bonds, and maintaining unaltered the amide linkage of sphingolipids. The reaction was blocked by adding 120 μl 0.5 M HCl in CH3OH.
Finally, after phase separation (by adding 1 ml of CHCl3/CH3OH/H2O, 70:18:12 by vol), the new organic phases were loaded on HPTLC plates and run in CHCl3/CH3OH/CH3COOH/H2O, 30:20:2:1 by vol. SM was detected by spraying the TLC with a molybdate reagent (36).
Other analytical methods
Radioactivity associated with cells, medium, gradient fractions, and lipid extracts was determined by liquid scintillation counting. The protein content was determined according to Lowry et al. (37) with BSA as reference standard. Mass spectrometry was carried out on a ThermoQuest Finnigan LCQdeca ESI-MS and a Xcalibur™ data system. Samples were dissolved in CH3OH at a concentration of 20–200 ng/μl and introduced through HPLC instrument. Ionization was performed under the following conditions: spray voltage, 4Kv; sheath gas flow rate, 50 arbitrary units; capillary temperature, 260°C; capillary voltage, -42 V. The scanning range was m/z 200–1600, and fragmentation voltage for collision-induced dissociation was 25–90%.
Statistical analysis
Experiments were run in triplicate, unless otherwise stated. Data are expressed as mean value ± SD.
RESULTS
Chemical synthesis
We synthesized Sph-N3, and SM-N3 reported in Fig. 1. In order to obtain Sph-N3 (Fig. 1A), the nitrophenylazide group was coupled to the N-terminal group of Sph-NH2 prepared as reported (19–21) with some modifications in order to increase the final yield. The reactions details are described in supplementary data. The synthesis of SM-N3 has been developed for the first time. The two products were 98% homogeneous and remained stable when stored in methanol at –20°C in a container protected from the light.
Effect of the Sph-N3 administration on normal and NPA fibroblasts
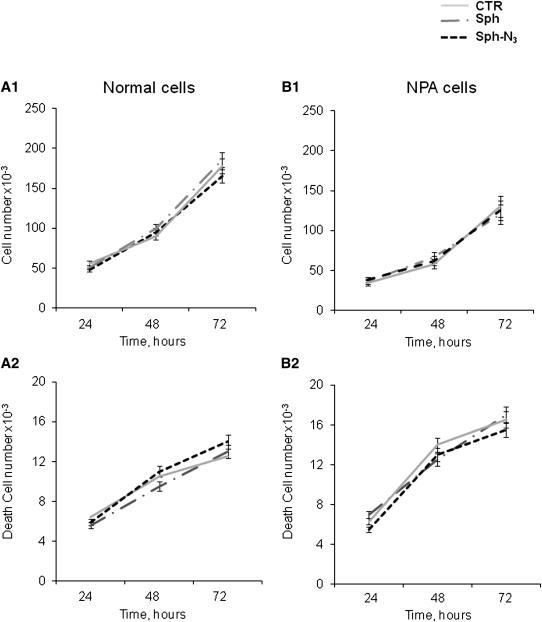
Normal and NPA fibroblasts were fed with Sph-N3 (Fig. 2) in order to verify cell toxicity. As shown in Figure 3, neither normal nor NPA fibroblasts present alterations in cell proliferation and death in the presence of both Sph and Sph-N3.
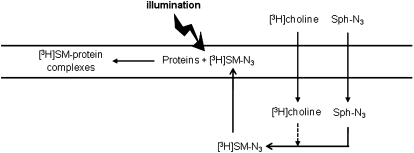
Fig. 2.
Experimental model scheme. Sph-N3 and [methyl-3H]choline were administered to human fibroblasts in culture with the aim of providing cells with photoactivable and radioactive precursors able to enter the sphingolipid metabolic pathways, allowing the production of radioactive and photoactivable sphingomyelin able to interact with neighboring proteins and to form, after cell illumination, stable radioactive SM-protein complexes.
Fig. 3.
Effect of normal and photoactivable sphingosine on proliferation and viability of cultured fibroblasts. Six hours after seeding, NPA and control fibroblasts were treated with normal (gray dotted line) or photoactivable (black dotted line) sphingosine solubilized in cell culture medium, both at the final concentration of 38 nM for up to 72 h. The proliferation and viability were compared with untreated cells (gray line). After 24, 48, and 72 h, the number of total cells (A1, B1) and of dead cells (A2 and B2, left) was evaluated by Trypan blue exclusion assay as described in Materials and Methods. Data are the means ± SD of three different experiments.
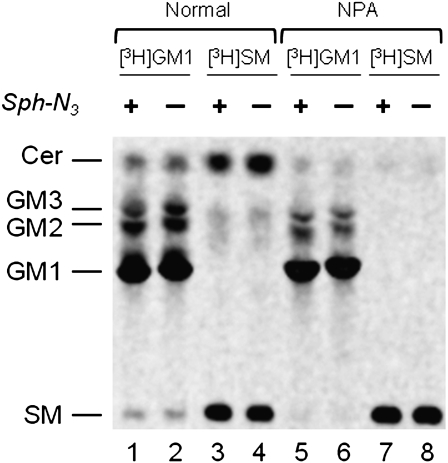
Feeding with Sph-N3 does not alter the capability of cells to take up GM1 or SM and insert them into the SL metabolic pathway. We had overlapping qualitative and quantitative results of radioactivity incorporation, lysosomal catabolism, and trafficking related to the recycling of sphingosine in normal and NPA fibroblasts treated or not with Sph-N3. The SL pattern obtained from cells treated with Sph-N3 was comparable and almost identical to that obtained from the corresponding control cells (Fig. 4). As shown in Fig. 4, the radioactive GM1 incorporated in normal fibroblasts is catabolyzed to GM2, GM3, and Cer. The catabolism also produced Sph, a part of which was recycled for the biosynthesis of SM (lane 1–2). In NPA fibroblasts, the metabolism of GM1 was lower than in normal cells, due to the lysosome impairment, but resulted in the same lipid pattern (lane 5–6). As expected, the radioactive SM is largely converted into Cer in normal fibroblasts (lane 3–4) (with some recycling of Sph for the biosynthesis of complex SLs), whereas this does not occur or is a very minor process in NPA fibroblasts (lane 7–8) due to the lack of the activity of lysosomal sphingomyelinase.
Fig. 4.
Radioactive lipids analysis of normal (lanes 1, 2, 3, 4) or NPA (lanes 5, 6, 7, 8) cells treated (lanes 1, 3, 5, 7) or not (lanes 2, 4, 6, 8) with Sph-N3 and fed with [3-3H(Sph)]GM1(lanes 1, 2, 5, 6) or [3-3H(Sph)]SM (lanes 3, 4, 7, 8). One thousand dpm of the total lipids extracted from normal and NPA fibroblasts fed with Sph-N3 followed by radioactive GM1 or SM administration were separated by HPTLC using CHCl3/CH3OH/(CH3)2CHOH/0.2% aqueous CaCl2, 20:60:20:4 by vol. as solvent system. Radioactive lipids were detected by digital autoradiography performed with a Biospace β-imager instrument for 48 h.
[3H]SM-N3 biosynthesis
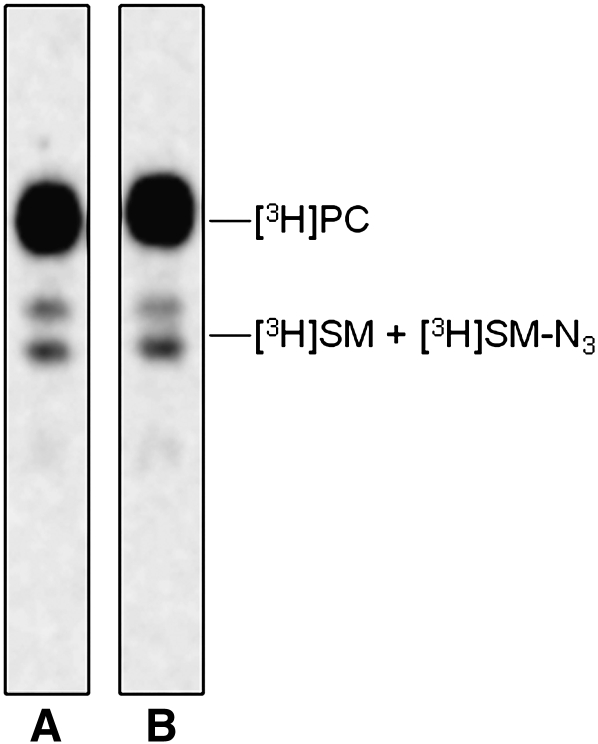
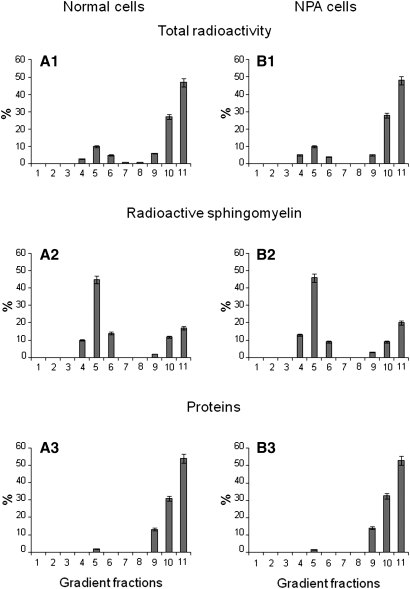
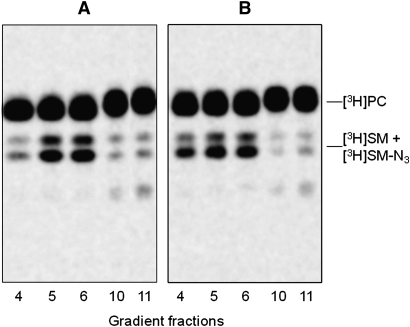
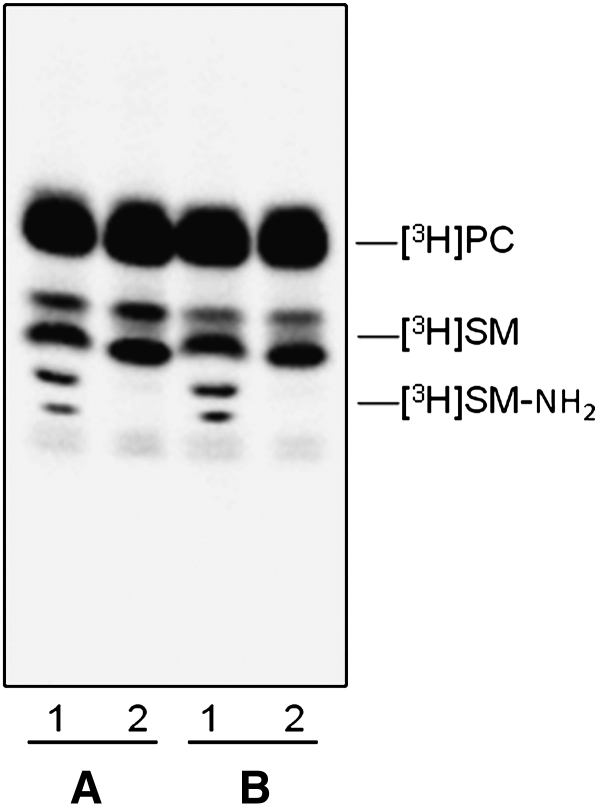
We verified that the endogenous SM content in NPA fibroblasts was strongly increased with respect normal cells: 109 versus 16 nmoles/mg cell proteins. Sph-N3 and [methyl-3H]choline were administered to NPA and normal fibroblasts in culture with the aim to give cell precursors for the biosynthesis of tritiated and [3H]SM-N3, as explained in Fig. 2. To follow the [3H]SM-N3 distribution within gradient fractions, cell homogenates were prepared from fibroblasts fed under red safelight with the labeled precursors and were then loaded on a sucrose gradient. The 11 fractions obtained by ultracentrifugation were maintained in a dark room and subjected to lipid analysis. Both cell lines incorporated an equivalent amount of radioactivity that was mainly associated with phosphatidylcholine (PC) and SM, which are the two membrane lipids having choline as head group. As shown in Figure 5, the 90% of the total radioactivity was associated with PC and the remaining with SM. As shown in Figure 6, cell radioactivity was largely associated with fractions 9–11 (panel A1 and B1), corresponding to the HD fractions that contain membranes solubilized by the detergent and the higher quantity of cell proteins (panel A3 and B3). Moreover, some radioactivity was also associated with fractions 4–6 corresponding to the low density fractions that are resistant to the detergent solubilization. TLC analysis of the lipids extracts showed different contents of PC and SM in the 11 fractions (Fig. 7). PC was distributed within all fractions, being much more abundant in the high density fractions 9–11, whereas SM was highly concentrated in the low density detergent-resistant fractions 4–6. In particular, as shown in Fig. 6 (Panel A2 and B2), around 70% of SM was associated with the fractions 4–6 in both cell lines. The [3H]SM recognized on the TLC plates as a double spot due to heterogeneity in the fatty acids content, is the sum of [3H]SM and [3H]SM-N3, the two compounds displaying a very similar chromatographic behavior. To analyze the ratio between [3H]SM and [3H]SM-N3, fractions were subjected to chemical reduction as described above, in order to convert SM-N3 into SM-NH2, which has a different chromatographic behavior and can be separated from SM-N3. Figure 8 shows the TLC separation of the radioactive lipids from low density-det ergent resistant fractions treated by chemical reduction. After quantification of the digital autoradiography, we calculated that the ratio between [3H]SM and [3H]SM-N3 in DRM of both normal and NPA cells was about 5:1.
Fig. 5.
Radioactive lipids analysis. TLC separation of the total lipids extracted from normal (lane A) and NPA (lane B) fibroblasts fed with Sph-N3 and [methyl-3H]choline. One thousand dpm were applied on a 4 mm line for each sample. TLC was run in CHCl3/CH3OH/0.2% aqueous CaCl2, 50:40:8 by vol. Digital autoradiography was performed with a Biospace β-imager instrument for 48 h.
Fig. 6.
Gradient fractions analysis. Gradient fractions distribution (abscissa axis) of total radioactivity (A1, B1), radioactive sphingomyelin (A2, B2), and cell proteins (A3, B3) in normal and NPA cells. Data are the means ± SD of three different experiments.
Fig. 7.
Radioactive lipids distributions within gradient fractions. TLC separation of radioactive total lipid extracts obtained from sucrose gradient fractions 4, 5, 6, 10, and 11 prepared from normal (A) and NPA (B) fibroblasts. One thousand dpm were applied on a 4 mm line for each sample. TLC was run in CHCl3/CH3OH/0.2% aqueous CaCl2, 50:40:8 by vol. Digital autoradiography was performed with a Biospace β-imager instrument for 48 h.
Fig. 8.
[3H]SM-N3 analysis. TLC separation of radioactive total lipid extracts obtained from DRM fractions prepared from normal (A) and NPA (B) fibroblasts. A total of 1000 dpm of control lipid extracts (lane 2) and 1000 dpm of lipid extracts previously subjected to chemical reduction with dithiothreitol (lane 1) were applied on a 4 mm line. TLC was performed with two sequential runs in CHCl3/CH3OH/H2O, 50:40:8 by vol. The radioactivity was detected by digital autoradiography using a Biospace β-imager instrument for 48 h.
This result confirms that cells fed with photoactivable precursors are able to biosynthesize photoactivable SLs and in particular, that the use of tritiated choline in combination with Sph-N3 yields [3H]SM-N3.
Photolabeling and immunoprecipitation experiments
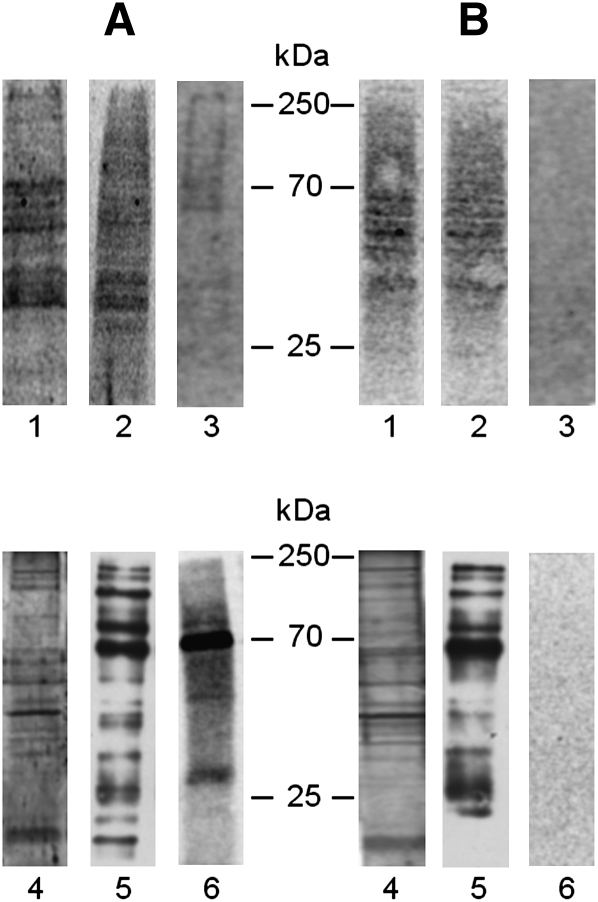
Normal and NPA fibroblasts fed with Sph-N3 and [methyl-3H]choline were biotinylated, illuminated under ultraviolet light, and then 11 fractions were separated by ultracentrifugation of the cell homogenates loaded on a sucrose gradient as described in Materials and Methods. Proteins from total homogenates, low- and high-density sucrose gradient fractions were separated by SDS-PAGE, blotted on PVDF membranes, and submitted to radioimaging for recognition of the tritiated SM-protein complexes. Figure 9 (A, B, lane 1) shows that many complexes are formed in the cells after illumination, suggesting that many proteins are in proximity of SM. Analysis of the sucrose gradient fractions shows that these complexes belong to the HD detergent-soluble fractions (Fig. 9, A, B, lane 2) that comprise those membranes containing a minor portion of SM-N3 (about 30%) and 98% of cell proteins. Instead, radioactive proteins were hardly detectable in the low density fractions (DRM) enriched in SM and containing only 1.5–2% of the total cell proteins. Table 1 reports data on the radioactivity associated with cell lipids and proteins. About 0.01% of total cell radioactivity was associated with the proteins belonging to the low-density detergent-resistant fractions prepared from normal cells fed with radioactive choline and Sph-N3.
Fig. 9.
Proteins cross-linked with [3H]SM-N3 from cell homogenates (lane 1), HD fraction (lane 2) and DRM fraction (lane 3) prepared from normal (A) and NPA (B) fibroblasts were separated by 10% SDS-PAGE, blotted on a polyvinyldifluoride (PVDF) membrane, and visualized by digital autoradiography for 120 h. Proteins recovered in the immunoprecipitation experiments performed in domain preserving conditions starting from aliquots of DRM fractions prepared from normal (A) and NPA (B) fibroblasts were separated by 10% SDS-PAGE. The proteins were directly detected by silver staining (lane 4) or blotted on a PVDF membrane and then visualized by Western blot using HRP-streptavidin (lane 5) or by digital autoradiography for 120 h (lane 6). Data are the means of three different experiments.
TABLE 1.
Radioactivity associated with normal and NPA fibroblasts.
| Lipid Radioactivity |
Protein Radioactivity |
||||
|---|---|---|---|---|---|
| Cell Radioactivity |
DRM |
HD |
DRM |
HD |
|
| (dpm × 10) | |||||
| NPA cells | 15,158 ± 242 | 2,268 ± 35 | 12,857 ± 220 | 0.0022 ± 0.0005 | 148.940 ± 5.06 |
| Normal cells | 18,040 ± 330 | 2,868 ± 41.8 | 15,061 ± 264 | 1.984 ± 0.011 | 179.080 ± 7.48 |
DRM, detergent resistant membrane corresponding to the membrane low density fraction; HD, high density detergent-soluble membrane fraction. Data are the means ± SD of three different experiments.
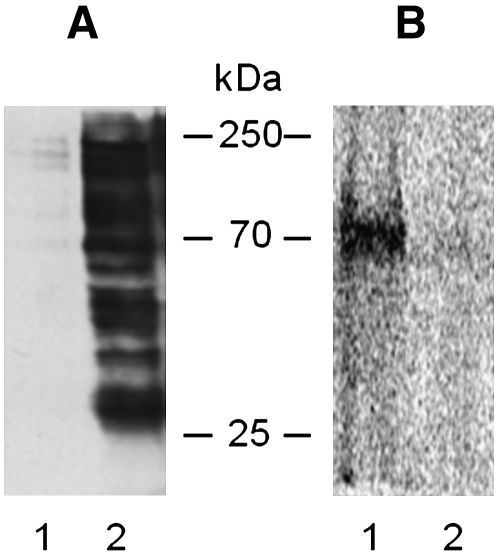
In a lipid membrane impoverished of proteins, [3H]SM-N3 preferentially binds lipids, not proteins. In order to have enough radioactivity associated with proteins, the DRM fractions (prepared from cells previously submitted to surface biotinylation) were subjected to immunoprecipitation with streptavidin-conjugated beads under conditions that preserve the membrane domain integrity. For both cell lines, we recovered more than 90% of biotinylated proteins in the IP samples (data not shown). The IPs were submitted to SDS-PAGE and analyzed for their content in total protein, biotinylated protein, and, by digital autoradiography, for [3H]SM-N3-protein complexes (Fig. 9). Figure 9, lanes 4 and 5, shows that many proteins detectable by silver or by streptavidin staining were present in the IP in both normal and NPA cells (panels A, B, respectively). Nevertheless, it is possible to observe qualitative and quantitative differences between total and biotinylated protein patterns. Radioactivity was associated with a few proteins in normal cells, the main with an apparent molecular mass of about 70 kDa (panel A, lane 6). Instead, no radioactive proteins could be detected in the immunoprecipitate from NPA cells (panel B, lane 6). To determine the membrane topology of the cross-linked proteins, the IP from normal cells was solubilized in boiling SDS solution and reimmunoprecipitated with streptavidin. Under these conditions, the membranes were solubilized by the detergent and then only biotinylated proteins were immunoprecipitated, whereas the proteins associated with the internal side of the plasma membrane were recovered in the surnatant (Fig. 10A). Figure 10B shows that under domain disrupting conditions, radioactive proteins remained in the supernatant and were not immunoprecipitated, suggesting that the SM-protein complexes are formed with proteins belonging to the inner layer of plasma membrane.
Fig. 10.
Topology of SM-protein complexes belong to detergent resistant membrane (DRM) of normal cells. The IP sample obtained in domain preserving conditions from DRM fraction of L40 fibroblasts was reimmunoprecipitated in domain disrupting conditions. Proteins recovered in the IP100 (lane 2) and in supernatant remaining after immunoprecipitation 100 (lane 1) were separated by 10% SDS-PAGE and blotted on a PVDF membrane, which was stained with HRP-streptavidin (A) and then visualized by digital autoradiography to detect the radioactive SM-protein complexes (B). Data are the means of three different experiments.
DISCUSSION
Several lipid derivatives containing fluorescent, paramagnetic, or photoactivable probes have been used in the past to study lipid metabolism, traffic, subcellular localization, and interactions with other molecules. The use of these derivatives implies the need to demonstrate that their behavior mimics as close as possible that of natural endogenous compounds. In the past, we synthesized radioactive and photoactivable gangliosides to study ganglioside-protein interactions. These gangliosides were administered to cells and taken up to become membrane components (12, 13). In previous papers (8, 12, 27, 38), we used photoactivable gangliosides and produced evidence that these derivatives, once taken up by cells, undergo metabolic processing and intracellular distribution similarly to the natural compounds. Indeed, the most critical step in the use of these derivatives is the initial association with cell membrane (38). Based on this experience, we designed a novel sphingosine derivative (Fig. 1A) that easily enters the cells and is subsequently used for the biosynthesis of complex SLs, including SM, carrying the photoactivable group.
Using this tool, we studied the SM-protein interactions in normal cells and in pathological cells, NPA disease cells, where SM accumulation could determine or influence the SM protein environment.
We performed experiments to verify cell toxicity for the Sph-N3. The cell proliferation and viability didn't change upon 72 h of Sph-N3 administration in either normal or NPA fibroblasts nor following the membrane insertion and endocytosis of radioactive GM1 and radioactive SM. Therefore, we verified that the plasma membrane functionality was not affected by Sph-N3 feeding.
Normal and NPA fibroblasts were fed with Sph-N3 and radioactive choline that entered into the biosynthetic pathway of SM allowing the metabolic synthesis of photoactivable and radioactive SM ([3H]SM-N3), useful, after cell illumination, to form SM-protein complexes (see the scheme in Fig. 2). The nitrene group, formed by illumination of the azide linked to the end of ceramide moiety of SM, preferentially binds lipids due to lipid-lipid interactions occurring in the lipid core of membrane, whereas binds proteins in minor quantity (39). This occurs particularly in lipid domains where lipids are strongly enriched with respect to proteins. Nevertheless, we were mainly interested to see the SM protein environment in these domains, membrane portions highly enriched in SLs and cholesterol and often called DRM or “lipid rafts,” where SM represents 60–70% of the total cell SM content. [methyl-3H] choline was taken up by the cells and as expected entered into the metabolic pathway of the choline-containing compounds. In fact, both PC and SM were found to be radioactive compounds, the first in much larger quantity (Fig. 5). Sph administered to fibroblasts in culture is rapidly taken up where its recycling is near 80% (39). Now, we show that fibroblasts in culture are capable of taking up Sph-N3 and inserting it into the SL biosynthetic pathway. In both normal and NPA cells, the administration of Sph-N3 in combination with [methyl-3H]choline allows the metabolic synthesis of a photoactivable and tritiated SM. [3H]SM-N3 and [3H]SM displayed a very similar TLC behavior and were recognized on the TLC plate as a double spot due to heterogeneity of the fatty acid moiety. To distinguish the [3H]SM-N3 from the [3H]SM, we synthesized a standard of SM-N3 (Fig. 1B) and, on this standard, we developed a reliable reduction procedure capable of converting the azide group into an aminic group in quantitative yield. The SM-NH2 produced (Fig. 1C), was characterized by mass spectrometry and used as chromatographic standard. By reducing the azide to amine, SM and SM-NH2 could be separated by TLC. Applying the reduction reaction to our samples, [3H]SM and [3H]SM-NH2 could be separated and resulted in a ratio of about 5:1. In addition to this, we verified that the topology of [3H]SM-N3 corresponds to that of [3H]SM; in fact, it was highly enriched into the low-density detergent-resistant fractions. This suggests that cells dilute Sph-N3 into the endogenous Sph and use it as the natural product. Therefore, the biosynthesized [3H]SM-N3 should mimic endogenous SM and can be considered a useful tool to recognize the protein SM environment through recognition of SM-protein complexes formed after cell illumination. After illumination, the radioactivity present in the DRM fraction was largely associated with lipids whereas the radioactivity associated with proteins was hardly detectable by digital autoradiography after SDS-PAGE separations and blotting on a PVDF membrane. The problem was resolved by concentrating the plasma membrane lipid domains by cell surface protein biotinylation followed by immunoprecipitation of the DRM fractions using streptavidin-coupled magnetic beads performed under domain preserving conditions. Surprisingly, we found that SM-protein complexes were formed in normal cells while no radioactive bands could be identified in the IP from NPA cells (Fig. 9, lanes 6). In addition to this, several differences were recognized in the protein patterns, determined by silver staining, and in biotinylated proteins composition of the IP samples obtained from the DRM fractions of the two cell lines (Fig. 9, lanes 4 and 5). Thus, the lysosomal storage of SM in NPA cells influences the protein environment of membrane SM. As reported elsewhere, [3H]SM-N3 has the photoactivable group at the end of the Sph moiety; in this way, when the molecule is inserted into the membrane, it remains located at the center of membrane lipid core. Therefore, after illumination, glycosyl-phosphatidyl inositol-anchored proteins and transmembrane proteins can form complexes with photoactivated SM and also be biotinylated. Complexes formed with protein belonging to the membrane inner layer can also be formed, but these are distinguishable from the previous being not biotinylated.
To understand the topology of proteins involved in the SM-protein complexes, the IP from the low density fraction prepared from normal cells previously submitted to biotinylation was solubilized at 100°C with SDS. Under these conditions, the membrane is disrupted and only biotinylated proteins can be immunoprecipitated with streptavidin. In this way, we found that the SM-protein complexes were not immunoprecipitated and remained in the supernatant. Thus, the complexes involve proteins of the inner layer (Fig. 10B).
Altogether, our results suggest the following considerations. Precursors of SLs probed with a radionuclide and a photoactivable group enter in the SL metabolic pathways, allowing the biosynthesis of radioactive and photoactivable SLs that display a correct cell topology. In this work, using tritiated choline and Sph-N3 in normal and pathological cells, we were able to produce tritiated and photoactivable SM. The [3H]SM-N3 was highly enriched, as and together with the natural SM, into a low-density detergent-resistant membrane fraction often named DRM or “lipid rafts”. After illumination, SL-protein complexes are formed. The position of photoactivated group at the end of ceramide moiety of SM and the lipid-lipid interactions occurring in the membrane lipid core greatly favor the reactions within the hydrophobic chains. In confirmation of this, 99% of the total radioactivity associated with the cells was linked to lipids. The formation of SM-protein complexes requires proximity and direct interaction between SM and the protein. Many complexes were found in both normal and NPA fibroblasts. Nevertheless, the two patterns were different, indicating that the storage of SM due to the lack of acidic sphingomyelinase in NPA disease has repercussions on the membrane organization of both lipid and proteins. We did not find SM-protein complexes in DRM from NPA cells and, in DRM from normal cells, only a few were formed with acylated or very hydrophobic proteins of the inner layer (13). This suggests that SM in lipid rafts of NPA cells is largely or completely excluded from the lipid environment of proteins, thus the photoactivable probe is not enough closed to protein in order to yield a covalent linkage.
In conclusion, our data suggest that the DRM obtained from normal and NPA fibroblasts are not only different in lipid and protein composition, but also in their clustering within the plasma membrane, thus determining specific lipid-protein interactions. Future research will be aimed at characterizing proteins that interact with SM in normal fibroblasts and to understand if the lack of this interaction in NPA fibroblasts has any role in determining the pathology.
Supplementary Material
Footnotes
Abbreviations:
- DRM
- detergent-resistant membrane
- EMEM
- Minimal Essential Medium with Earle's salt
- GSL
- glycosphingolipid
- HD
- high density
- IP
- immunoprecipitate
- NPA
- Niemann-Pick type A
- PC
- phosphatidylcholine
- PVDF
- polyvinyldifluoride
- SL
- sphingolipid
- SM-N3
- photoactivable SM
- [3H]SM-N3
- radioactive and photoactivable SM
- Sph
- sphingosine
- Sph-N3
- photoactivable sphingosine
- Sph-NH2
- reduced photoactivable sphingosine
- THF
- tetrahydrofurane
This work was supported by Mizutani Foundation for Glycoscience Grant 070002 to A.P. and Associazione Italiana Ricerca sul Cancro (AIRC 2006-2008) and Cariplo Foundation Grants to S.S.
The online version of this article (available at http://www.jlr.org) contains supplementary data.
REFERENCES
- 1.Prinetti A., Chigorno V., Prioni S., Loberto N., Marano N., Tettamanti G., Sonnino S. 2001. Changes in the lipid turnover, composition, and organization, as sphingolipid-enriched membrane domains, in rat cerebellar granule cells developing in vitro. J. Biol. Chem. 276: 21136–21145 [DOI] [PubMed] [Google Scholar]
- 2.Prinetti A., Chigorno V., Tettamanti G., Sonnino S. 2000. Sphingolipid-enriched membrane domains from rat cerebellar granule cells differentiated in culture. A compositional study. J. Biol. Chem. 275: 11658–11665 [DOI] [PubMed] [Google Scholar]
- 3.Valsecchi M., Mauri L., Casellato R., Prioni S., Loberto N., Prinetti A., Chigorno V., Sonnino S. 2007. Ceramide and sphingomyelin species of fibroblasts and neurons in culture. J. Lipid Res. 48: 417–424 [DOI] [PubMed] [Google Scholar]
- 4.Scandroglio F., Venkata J. K., Loberto N., Prioni S., Schuchman E. H., Chigorno V., Prinetti A., Sonnino S. 2008. Lipid content of brain, brain membrane lipid domains, and neurons from acid sphingomyelinase deficient mice. J. Neurochem. 107: 329–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prinetti A., Loberto N., Chigorno V., Sonnino S. 2009. Glycosphingolipid behaviour in complex membranes. Biochim. Biophys. Acta. 1788: 184–193 [DOI] [PubMed] [Google Scholar]
- 6.Bremer E. G., Schlessinger J., Hakomori S. 1986. Ganglioside-mediated modulation of cell growth. Specific effects of GM3 on tyrosine phosphorylation of the epidermal growth factor receptor. J. Biol. Chem. 261: 2434–2440 [PubMed] [Google Scholar]
- 7.Song W. X., Vacca M. F., Welti R., Rintoul D. A. 1991. Effects of gangliosides GM3 and De-N-acetyl GM3 on epidermal growth factor receptor kinase activity and cell growth. J. Biol. Chem. 266: 10174–10181 [PubMed] [Google Scholar]
- 8.Kabayama K., Sato T., Saito K., Loberto N., Prinetti A., Sonnino S., Kinjo M., Igarashi Y., Inokuchi J. 2007. Dissociation of the insulin receptor and caveolin-1 complex by ganglioside GM3 in the state of insulin resistance. Proc. Natl. Acad. Sci. USA. 104: 13678–13683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levade T., Jaffrezou J. P. 1999. Signalling sphingomyelinases: which, where, how and why? Biochim. Biophys. Acta. 1438: 1–17 [DOI] [PubMed] [Google Scholar]
- 10.Goni F. M., Alonso A. 2002. Sphingomyelinases: enzymology and membrane activity. FEBS Lett. 531: 38–46 [DOI] [PubMed] [Google Scholar]
- 11.Huitema K., van den Dikkenberg J., Brouwers J. F., Holthuis J. C. 2004. Identification of a family of animal sphingomyelin synthases. EMBO J. 23: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prioni S., Mauri L., Loberto N., Casellato R., Chigorno V., Karagogeos D., Prinetti A., Sonnino S. 2004. Interactions between gangliosides and proteins in the exoplasmic leaflet of neuronal plasma membranes: a study performed with a tritium-labeled GM1 derivative containing a photoactivable group linked to the oligosaccharide chain. Glycoconj. J. 21: 461–470 [DOI] [PubMed] [Google Scholar]
- 13.Mauri L., Prioni S., Loberto N., Chigorno V., Prinetti A., Sonnino S. 2004. Synthesis of radioactive and photoactivable ganglioside derivatives for the study of ganglioside-protein interactions. Glycoconj. J. 20: 11–23 [DOI] [PubMed] [Google Scholar]
- 14.Thiele C., Hannah M. J., Fahrenholz F., Huttner W. B. 2000. Cholesterol binds to synaptophysin and is required for biogenesis of synaptic vesicles. Nat. Cell Biol. 2: 42–49 [DOI] [PubMed] [Google Scholar]
- 15.Schwarzmann G., Hoffmann-Bleihauer P., Schubert J., Sandhoff K., Marsh D. 1983. Incorporation of ganglioside analogues into fibroblast cell membranes. A spin-label study. Biochemistry. 22: 5041–5048 [DOI] [PubMed] [Google Scholar]
- 16.Wiegandt H., Baschang G. 1965. Die Gewinnung des Zuckeranteiles der Glykosphingolipide durch Ozonolyse und Fragmentierung. Z. Naturforsch. [B]. 20: 164–166 [PubMed] [Google Scholar]
- 17.Brady R. O., Kanfer J. N., Mock M. B., Fredrickson D. S. 1966. The metabolism of sphingomyelin. II. Evidence of an enzymatic deficiency in Niemann-Pick diseae. Proc. Natl. Acad. Sci. USA. 55: 366–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leroy J. G., Ho M. W., MacBrinn M. C., Zielke K., Jacob J., O'Brien J. S. 1972. I-cell disease: biochemical studies. Pediatr. Res. 6: 752–757 [DOI] [PubMed] [Google Scholar]
- 19.Kozikowski A. P., Qingjie D., Spiegel S. 1996. Synthesis of erythro-[omega]-aminosphingosine and preparation of an affinity column for sphingosine kinase purification. Tetrahedron Lett. 37: 3279–3282 [Google Scholar]
- 20.Lu X., Cseh S., Byun H-S., Tigyi G., Bittman R. 2003. Total synthesis of two photoactivatable analogues of the growth-factor-like mediator sphingosine 1-phosphate: differential interaction with protein targets. J. Org. Chem. 68: 7046–7050 [DOI] [PubMed] [Google Scholar]
- 21.Nieuwenhuizen W. F., v. Leeuwen S., Götz F., Egmond M. R. 2002. Synthesis of a novel fluorescent ceramide analogue and its use in the characterization of recombinant ceramidase from Pseudomonas aeruginosa PA01. Chem. Phys. Lipids. 114: 181–191 [DOI] [PubMed] [Google Scholar]
- 22.Staros J. V., Bayley H., Standring D. N., Knowles J. R. 1978. Reduction of aryl azides by thiols: implications for the use of photoaffinity reagents. Biochem. Biophys. Res. Commun. 80: 568–572 [DOI] [PubMed] [Google Scholar]
- 23.Sonnino S., Chigorno V., Tettamanti G. 2000. Preparation of radioactive gangliosides, 3H or 14C isotopically labeled at oligosaccharide or ceramide moieties. Methods Enzymol. 311: 639–656 [DOI] [PubMed] [Google Scholar]
- 24.Chung T., Huang J. S., Mukherjee J. J., Crilly K. S., Kiss Z. 2000. Expression of human choline kinase in NIH 3T3 fibroblasts increases the mitogenic potential of insulin and insulin-like growth factor I. Cell. Signal. 12: 279–288 [DOI] [PubMed] [Google Scholar]
- 25.Altin J. G., Pagler E. B. 1995. A one-step procedure for bio tinylation and chemical cross-linking of lymphocyte surface and intracellular membrane-associated molecules. Anal. Biochem. 224: 382–389 [DOI] [PubMed] [Google Scholar]
- 26.Cole S. R., Ashman L. K., Ey P. L. 1987. Biotinylation: an alternative to radioiodination for the identification of cell surface antigens in immunoprecipitates. Mol. Immunol. 24: 699–705 [DOI] [PubMed] [Google Scholar]
- 27.Sonnino S., Chigorno V., Acquotti D., Pitto M., Kirschner G., Tettamanti G. 1989. A photoreactive derivative of radiolabeled GM1 ganglioside: preparation and use to establish the involvement of specific proteins in GM1 uptake by human fibroblasts in culture. Biochemistry. 28: 77–84 [DOI] [PubMed] [Google Scholar]
- 28.Sonnino S., Chigorno V., Valsecchi M., Pitto M., Tettamanti G. 1992. Specific ganglioside-cell protein interactions: a study perfor med with GM1 ganglioside derivative containing photoactivable azide and rat cerebellar granule cells in culture. Neurochem. Int. 20: 315–321 [DOI] [PubMed] [Google Scholar]
- 29.Chigorno V., Valsecchi M., Acquotti D., Sonnino S., Tettamanti G. 1990. Formation of a cytosolic ganglioside-protein complex following administration of photoreactive ganglioside GM1 to human fibroblasts in culture. FEBS Lett. 263: 329–331 [DOI] [PubMed] [Google Scholar]
- 30.Prinetti A., Marano N., Prioni S., Chigorno V., Mauri L., Casellato R., Tettamanti G., Sonnino S. 2000. Association of Src-family protein tyrosine kinases with sphingolipids in rat cerebellar granule cells differentiated in culture. Glycoconj. J. 17: 223–232 [DOI] [PubMed] [Google Scholar]
- 31.Phillips H. J.1973. Tissue culture methods and applications. Kruse P.F., Patterson M.K., Jr. Academic Press, New York [Google Scholar]
- 32.Prinetti A., Prioni S., Chigorno V., Karagogeos D., Tettamanti G., Sonnino S. 2001. Immunoseparation of sphingolipid-enriched membrane domains enriched in Src family protein tyrosine kinases and in the neuronal adhesion molecule TAG-1 by anti-GD3 ganglioside monoclonal antibody. J. Neurochem. 78: 1162–1167 [DOI] [PubMed] [Google Scholar]
- 33.Brown D. A., Rose J. K. 1992. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 68: 533–544 [DOI] [PubMed] [Google Scholar]
- 34.Kasahara K., Watanabe Y., Yamamoto T., Sanai Y. 1997. Association of Src family tyrosine kinase Lyn with ganglioside GD3 in rat brain. Possible regulation of Lyn by glycosphingolipid in caveolae-like domains. J. Biol. Chem. 272: 29947–29953 [DOI] [PubMed] [Google Scholar]
- 35.Scandroglio F., Loberto N., Valsecchi M., Chigorno V., Prinetti A., Sonnino S. 2009. Thin layer chromatography of gangliosides. Glycoconj. J. 26: 961–973 [DOI] [PubMed] [Google Scholar]
- 36.Vaskovsky V. E., Kostetsky E. Y. 1968. Modified spray for the detection of phospholipids on thin-layer chromatograms. J. Lipid Res. 9: 396. [PubMed] [Google Scholar]
- 37.Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193: 265–275 [PubMed] [Google Scholar]
- 38.Loberto N., Prioni S., Prinetti A., Ottico E., Chigorno V., Karagogeos D., Sonnino S. 2003. The adhesion protein TAG-1 has a ganglioside environment in the sphingolipid-enriched membrane domains of neuronal cells in culture. J. Neurochem. 85: 224–233 [DOI] [PubMed] [Google Scholar]
- 39.Chigorno V., Palestini P., Sciannamblo M., Dolo V., Pavan A., Tettamanti G., Sonnino S. 2000. Evidence that ganglioside enriched domains are distinct from caveolae in MDCK II and human fibroblast cells in culture. Eur. J. Biochem. 267: 4187–4197 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.