Abstract
Evolutionary cytogenetic comparisons involved 5 species of birds (California condor, chicken, zebra finch, collared flycatcher and black stork) belonging to divergent taxonomic orders. Seventy-four clones from a condor BAC library containing 80 genes were mapped to condor chromosomes using FISH, and 15 clones containing 16 genes were mapped to the stork Z chromosome. Maps for chicken and finch were derived from genome sequence databases, and that for flycatcher from the published literature. Gene content and gene order were highly conserved when individual condor, chicken, and zebra finch autosomes were compared, confirming that these species largely retain karyotypes close to the ancestral condition for neognathous birds. However, several differences were noted: zebra finch chromosomes 1 and 1A are homologous to condor and chicken chromosomes 1, the CHUNK1 gene appears to have transposed on condor chromosome 1, condor chromosomes 4 and 9 and zebra finch chromosomes 4 and 4A are homologous to chicken chromosome arms 4q and 4p, and novel inversions on chromosomes 4, 12 and 13 were found. Condor and stork Z chromosome gene orders are collinear and differentiated by a series of inversions/transpositions when compared to chicken, zebra finch, or flycatcher; phylogenetic analyses suggest independent rearrangement along the chicken, finch, and flycatcher lineages.
Key Words: Avian molecular cytogenetics, California condor, Z chromosome in birds
Conventional cytogenetic analyses reveal that birds have relatively conserved karyotypes when compared with mammals. The vast majority of species contain diploid numbers from 2n = 66–86, with about 10 pairs of macrochromosomes and variously sized microchromosomes [Takagi and Sasaki, 1974; Griffin et al., 2007]. Although palaeognathous birds have homomorphic sex chromosomes [Shetty et al., 1999; Nishida-Umehara et al., 2007], those in neognathous species are morphologically distinct, with females ZW and males ZZ.
Molecular cytogenetic analyses, fueled largely by cross-species chromosome painting (Zoo-FISH), support these earlier observations by identifying whole chromosome conservation in a large number of comparisons. However, such analyses have also discovered a variety of rearrangements including inversions, fissions, and fusions among both closely and distantly related species. For example, the highly derived harpy eagle karyotype differs from that of chicken by at least 20 rearrangements [de Oliveira et al., 2005]. Changes involving each macrochromosome have occurred in one or more taxonomic orders [Nanda et al., 2006; Griffin et al., 2007]. The Z chromosome, although conserved in gene content among different species, has undergone a large number of intrachromosomal inversions and/or molecular transposition events [Backstrom et al., 2006; Itoh et al., 2006; Nanda et al., 2008]. The agriculturally important chicken has emerged as the model system for avian genomics. The chicken karyotype is conserved when compared with other birds, and its genome was the first among birds to be completely sequenced [Consortium, 2004; Masabanda et al., 2004].
This study extends the chromosome painting of Raudsepp et al. [2002] by using high-resolution bacterial artificial chromosome (BAC) hybridization to examine gene content and gene order of California condor (Gymnogyps californianus) chromosomes. A total of 74 BACs containing orthologues to 80 known chicken genes were localized using fluorescence in situ hybridization (FISH). In addition, 15 BACs were FISH mapped to the black stork (Ciconia nigra) Z chromosome. These 2 cytogenetic maps were compared to genome sequence based maps of chicken (Gallus gallus) and zebra finch (Taeniopygia guttata) and a linkage map of the collared flycatcher (Ficedula albicollis) Z chromosome, and results interpreted in a phylogenetic context.
The critically endangered California condor has become a symbol of success in conservation biology. Population numbers were drastically reduced by human impact; however, recent captive breeding has begun to restore them. The captive flock has produced severely deformed embryos caused by a long bone growth plate disorder called chondrodystrophy [Ralls et al., 2000]. In addition, various zoonotic diseases including West Nile virus have recently become threats for avian species in western North America [Reisen et al., 2009], although condors are vaccinated for this disease. Genomic resources such as those presented here could assist in the future management of California condors and other endangered species.
Materials and Methods
Preparation of BAC DNA
A BAC library of the California condor (CHORI-262) generated at the BACPAC Resources Center, Children's Hospital Oakland Research Institute (CHORI) (http://bacpac.chori.org/) was screened using over 380 overlapping oligonucleotide (overgo) probes corresponding to known vertebrate genes [Romanov et al., 2006, 2009]. A total of 88 positive condor clones were selected for FISH analyses. In addition, 6 clones from a chicken BAC library (CHORI-261) were also used for FISH (online supplementary table 1, www.karger.com/doi/10.1159/000272458).
Bacterial stab cultures were streaked on LB agar plates containing 40 μg/ml chloramphenicol. Two colonies were picked for each clone and grown separately in overnight cultures following standard procedures. The Plasmid Midi kit (Qiagen Inc.) was used following standard alkaline lysis.
Cell Cultures
Fibroblast cultures were established from tracheae and cornea biopsies from one male and 2 female California condors and one male black stork. Biopsies were cleaned, minced, and grown as standard cell cultures. Metaphase chromosomes were harvested from early passages following standard colcemid treatment, exposure to hypotonic solution and fixation with methanol:acetic acid. Slides were aged at room temperature at least 14 days before hybridization. One female tracheae cell clone developed a spontaneously transformed phenotype [Romanov et al., 2009]. Characteristics included rounded cellular morphology, rapid growth rate, and karyotype heteroploidy with many cells appearing tetraploid.
Fluorescent in Situ Hybridization
The FISH protocol followed established methods [Pinkel et al., 1986; Lawrence et al., 1988]. Probes were labeled with either biotin-14-dATP or digoxigenin-11-dUTP and detected with fluoroescein avidin DCS or Cy3-conjugated anti-digoxigenin, respectively. All images were captured with a cooled CCD camera mounted on an Olympus microscope equipped with Genus System 3.93 software from Applied Imaging Systems (San Jose, Calif., USA).
In addition, conventional Giemsa-stained and C-banded chromosomes from several males and females of both species were photographed to assess overall chromosome quality, to confirm diploid numbers, and to assist in chromosome identification.
Chromosome Identification
Chromosome identification for both species was based upon size and morphology as opposed to banding pattern. A comprehensive cytogenetic analysis of the California condor has been published [Raudsepp et al., 2002]. This species has 2n = 80 with 10 pairs of macrochromosomes, a graded series of acrocentric microchromosomes, and a medium-sized submetacentric Z chromosome. Chromosomes 1–3 and 9 present unique size and morphology. The Z is discernable from 4 and 5 based upon larger relative size of the short arm. Pair 4 is typically larger than pair 5. Acrocentric pairs 6, 7, 8 and 10 are often distinct. The remaining autosomes are a graded series of acrocentrics that are not possible to differentiate and were identified based upon gene assignment. Condor microchromosomes were given numeric designations corresponding to those in the chicken following localization of homologous genes in both species.
The black stork has 2n = 52 with 10 pairs of biarmed macrochromosomes, including the sex chromosomes [Belterman and de Boer, 1990]. The Z is the largest of 5 medium-sized pairs of submetacentrics; although in this study, it was not unambiguously identified in all metaphase preparations.
Results and Discussion
A total of 88 positive clones were selected after screening the California condor BAC library. In addition, 6 clones from a chicken BAC library were also used for FISH (online supplementary table 1). When hybridized to metaphase chromosomes of the condor, 74 of 88 (84%) condor clones produced an interpretable hybridization signal, while 0 of 6 chicken clones yielded a positive result. Further, 15 of 35 (43%) condor clones were localized to the stork Z chromosome. The decreased hybridization efficiency of the heterologous clones can be explained by the sequence divergence among non-coding regions. Representative hybridization images are shown in figures 1 and 2.
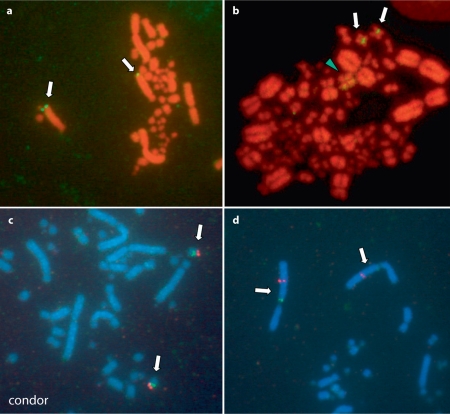
Fig. 1.
Gene localization on California condor metaphase chromosomes following FISH using BAC clones labeled with biotin and counterstained with propidium iodide (a, b), or 2-color FISH using biotin- and digoxigenin-labeled clones withHoechst 33258 counterstaining (c, d). a SUPT3H and RUNX2 (clone 13G5) on chromosome 3q, b BTK (1N8) near the centromere of chromosome 9 with hybridization of an uncharacterized repeat to the W chromosome (green arrowhead), c OVM (5J24, biotin) and NR3C1 (29N19, digoxigenin) on chromosome 14, d USP5 (1D23, biotin) and OCA2 (5D3, digoxigenin) on chromosome 1. In figures 1 and 2, biotin-FITC signals appear yellow or green, while digoxigenin-CY3 probes are red.
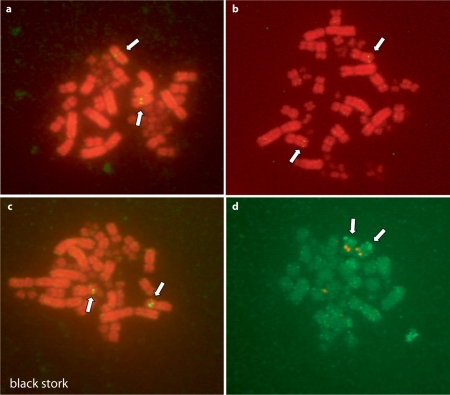
Fig. 2.
Gene localization on black stork Z chromosomes following one-color (biotin) (a–c), or 2-color (d) FISH. a TAL2 (3O23) on Zq near centromere, b UHRF2 (6C12) on Zq, c PALM2 (42P15) on Zp, d UBE2R2 (37I2, biotin) on Zq near telomere and SNCAIP (184F6, digoxigenin) on Zp near centromere.
Ancestral Gene Content and Gene Order of Avian Autosomes
The localization of 80 condor genes contained within 74 clones is shown by their placement on ideograms and comparisons with chicken and zebra finch. Fifty clones map to autosomal positions and 24 to the Z chromosome (fig. 3, 4). Zebra finch, chicken, and condor reveal extensive conservation in gene content and gene order for all autosomes. Among the macrochromosomes (pairs 1–9), zebra finch, chicken, and condor chromosomes 2, 3, 5, 6, 7, 8 are highly conserved with no differences detected. Also, genes that map to microchromosomes in zebra finch and chicken map to microchromosomes in the condor, and 2-color hybridization revealed conserved order in several cases such as centromere-DBC-FBXW2 on condor chromosome 18 (= finch and chicken chromosomes 17) (fig. 3). Another pair of genes, PAFAH1B2 and TAGLN, was previously identified as closely linked on condor BAC clone 43E17 (online supplementary table 1) [Romanov et al., 2006], and are only 9.5 kb apart in the latest chicken assembly, and 13.6 kb apart in the latest zebra finch assembly. This clone was localized to a condor microchromosome, and was previously assigned to microchromosome 24 in both finch and chicken.
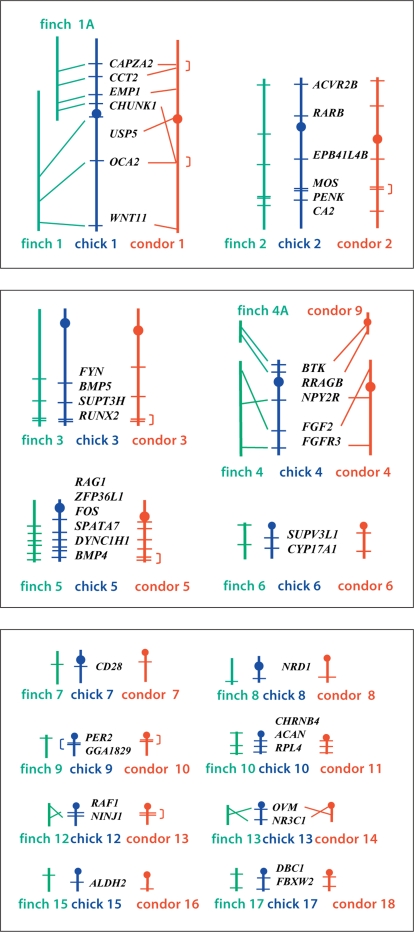
Fig. 3.
Ideogrammatic comparisons between autosomes of zebra finch, chicken and California condor. Maps for finch and chicken are from the UCSC (finch July 2008; chicken May 2006) and NCBI (finch build 1.1; chicken build 2.1) genome browsers, those for condor are present FISH results. Unresolved gene orders are indicated by brackets (]).
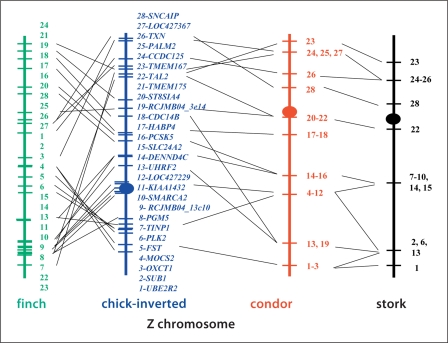
Fig. 4.
Ideogrammatic comparisons of the Z chromosomes of zebra finch, chicken, California condor and black stork. Maps for zebra finch and chicken were based upon genome sequences, those for condor and stork are current FISH results.
On the other hand, differences between species were noted on condor chromosomes 1, 4, 9, 13 and 14 (fig. 3). Zebra finch chromosomes 1 and 1A are homologous to chicken and condor chromosomes 1, while finch chromosomes 4 and 4A are homologous to condor chromosomes 4 and 9 and chicken chromosome 4. The CHUNK1 and OCA2 genes are on finch chromosomes 1A and 1, respectively, located over 70 Mb apart on chicken chromosome 1, but contained within 150 kb on condor BAC clone 5D3 on chromosome 1 (online supplementary table 1) [Romanov et al., 2006]. The order of these 2 and 5 other genes is collinear when chicken chromosome 1 and zebra finch chromosomes 1 and 1A are compared (fig. 3). These results argue that the gene arrangement seen in chicken is ancestral, that CHUNK1 has transposed in the condor, and that finch chromosomes 1 and 1A are derived fission products.
Two genes on condor chromosome 9 (BTK, RRAGB) map to chicken chromosome arm 4p and finch chromosome 4A. Three additional genes on condor chromosome 4 map to chicken 4q and finch 4. The order of these genes in chicken (NPY2R-FGF2-FGFR3) differs from that in finch and condor (FGF2-NPY2R-FGFR3) (fig. 3). The existence of chromosomes 4 and 9 in condor and 4 and 4A in finch have been proposed to be ancestral, while chicken 4 is thought to be a derived fusion product. If this is correct, then the gene order appearing in zebra finch and condor may be ancestral, while that of chicken is the result of a derived paracentric inversion. Our data confirm earlier Zoo-FISH findings of homology between chicken chromosome 4 with condor chromosomes 4 and 9 [Raudsepp et al., 2002]. This relationship has been observed in other birds [Shetty et al., 1999; Schmid et al., 2000; Guttenbach et al., 2003], and phylogenetic analyses suggest that fission/fusion events involving these 2 chromosomes may be occurring independently in different avian lineages [Griffin et al., 2007].
When zebra finch and chicken chromosomes 12 are compared, the order of RAF1 and NINJ1 is reversed, while the order of these genes on condor chromosome 13 was not resolved. Similarly, the gene order of OVM and NR3C1 on chicken 13 was reversed relative to that of finch 13 and condor 14. These 2 differences could have originated equally parsimoniously on either the chicken lineage or in the common ancestor of Passeriformes and Cathartidae.
The ancestral avian karyotype probably contained a diploid number of 2n = 80, with 10 pairs of macrochromosomes (including the sex chromosomes) and variously sized microchromosomes. Pairs 1 and 2 were biarmed, while all others were acrocentric. This ancestral karyotype has been retained by a variety of avian taxa [Takagi and Sasaki, 1974; Belterman and de Boer, 1990; Shetty et al., 1999; Fillon et al., 2007; Nishida et al., 2008]. The California condor possesses the ancestral diploid number 2n = 80 with 10 pairs of macrochromosomes; however, chromosomes 3, 4, 5, 9 and the Z are biarmed as a result of pericentric inversions and/or centromeric repositioning.
Ancestral Gene Order of the Avian Z Chromosome
Thirty-five BAC clones containing genes located on the chicken Z chromosome were hybridized to both California condor and black stork chromosomes. Twenty-four clones produced a positive signal in the condor, while only 15 were successfully mapped in the stork. All clones mapped to the Z in both species with no evidence of hybridization to the condor W (the stork W was not examined). The order between the 28 genes mapped in the condor and the 16 in the stork appears collinear, and differs from that of chicken by several inversions/transpositions and even more so from zebra finch (fig. 4).
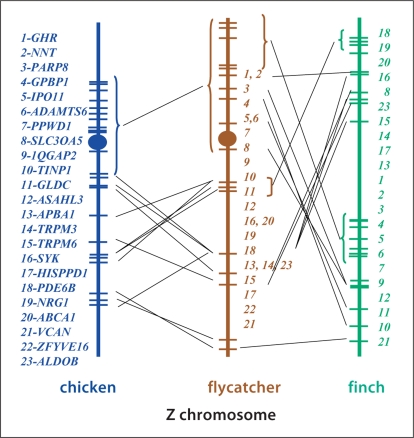
In addition, comparisons involving a different set of genes were made between chicken, collared flycatcher and zebra finch (fig. 5). The chicken and flycatcher contain 12 genes (GHR-NNT-PARP8-GPBP1-IPO11-ADAMTS6-PPWD1-SLC30A5-IQGAP2-TINP1- GLDC-ASAH3L)whose order is conserved between both species; however the remainder of the chromosome shows a number of rearrangements between the 2 species. The order of 7 (GHR-NNT-PARP8-GPBP1-IPO11-ADAMTS6-PPWD1) of these 12 genes are also conserved between flycatcher and finch, although these genes have been translocated to the middle of the finch Z chromosome. Another group of 4 genes (PDE6B-NRG1-ABCA1-SYK) also show a conserved order between flycatcher and finch, although the genes are located at different positions on the Z chromosome in the 2 species. The remaining genes show significant differences in gene order when flycatcher and finch are compared. This confirms earlier studies that showed the rearranged nature of the Z chromosome in finch [Itoh et al., 2006] and flycatcher [Backstrom et al., 2006] when compared with chicken; however, zebra finch and flycatcher Z chromosomes have apparently not previously been compared to each other.
Fig. 5.
Comparisons of Z chromosomes from chicken, zebra finch, and collared flycatcher using different genes than those presented in figure 4. Data for chicken and finch are based upon genome sequences, those for flycatcher are from the linkage map [Backstrom et al., 2007].
The present information is valuable since condor (Falconiformes) and stork (Ciconiiformes) belong to different taxonomic orders than chicken (Galliformes) or finch and flycatcher (Passeriformes). Therefore, it is useful to evaluate autosomal and Z chromosome inversions for these 5 species in a phylogenetic context. Condor and stork Z chromosomes appear collinear, suggesting they retain the ancestral gene order and that inversions have occurred independently along the finch, flycatcher, and chicken lineages. This indicates reorganization of the avian Z both between and within taxonomic orders. This is significant since zebra finch and flycatcher are both passerines and share a common ancestor more recently with each other than with the remaining taxa in the study. Additional Z chromosome rearrangements have been documented in other taxonomic orders of birds [Fillon et al., 2007; Nanda et al., 2008], and more extensive mapping data involving larger numbers of genes in avian and outgroup reptilian taxa are needed to more accurately establish ancestral and derived conditions.
A recent study of chromosome breakpoints in mammals found that they are enriched for genes with adaptive functions, segmental duplications, copy number variants, indels, retrotransposed genes, and single nucleotide polymorphisms [Larkin et al., 2009]. Given the interspecific variation in gene order on the avian Z, this chromosome is an excellent candidate for future high-throughput sequencing from different species in order to better understand the molecular architecture of vertebrate genomes and to identify sequences promoting chromosomal rearrangements.
Acknowledgements
S. Charter, J. Fronczek, D. Gallagher, and M. Houck offered technical assistance. This work was supported in part by the Intramural Research Program of the National Human Genome Research Institute and the Seaver Institute.
References
- Backstrom N, Brandstrom M, Gustafsson L, Qvarnstrom A, Cheng H, Ellegren H. Genetic mapping in a natural population of collared flycatchers (Ficedula albicollis): conserved synteny but gene order rearrangements on the avian Z chromosome. Genetics. 2006;174:377–386. doi: 10.1534/genetics.106.058917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belterman RHR, de Boer LEM. A miscellaneous collection of bird karyotypes. Genetica. 1990;83:17–29. [Google Scholar]
- Consortium ICGS. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- de Oliveira EH, Habermann FA, Lacerda O, Sbalqueiro IJ, Wienberg J, Muller S. Chromosome reshuffling in birds of prey: the karyotype of the world's largest eagle (Harpy eagle, Harpia harpyja) compared to that of the chicken (Gallus gallus) Chromosoma. 2005;114:338–343. doi: 10.1007/s00412-005-0009-5. [DOI] [PubMed] [Google Scholar]
- Fillon V, Vignoles M, Crooijmans RP, Groenen MA, Zoorob R, Vignal A. FISH mapping of 57 BAC clones reveals strong conservation of synteny between Galliformes and Anseriformes. Anim Genet. 2007;38:303–307. doi: 10.1111/j.1365-2052.2007.01578.x. [DOI] [PubMed] [Google Scholar]
- Griffin DK, Robertson LB, Tempest HG, Skinner BM. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 2007;117:64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- Guttenbach M, Nanda I, Feichtinger W, Masabanda JS, Griffin DK, Schmid M. Comparative chromosome painting of chicken autosomal paints 1–9 in nine different bird species. Cytogenet Genome Res. 2003;103:173–184. doi: 10.1159/000076309. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Kampf K, Arnold AP. Comparison of the chicken and zebra finch Z chromosomes shows evolutionary rearrangements. Chromosome Res. 2006;14:805–815. doi: 10.1007/s10577-006-1082-1. [DOI] [PubMed] [Google Scholar]
- Larkin DM, Pape G, Donthu R, Auvil L, Welge M, Lewin HA. Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res. 2009;19:770–777. doi: 10.1101/gr.086546.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JB, Villnave CA, Singer RH. Sensitive, high-resolution chromatin and chromosome mapping in situ: presence and orientation of two closely integrated copies of EBV in a lymphoma line. Cell. 1988;52:51–61. doi: 10.1016/0092-8674(88)90530-2. [DOI] [PubMed] [Google Scholar]
- Masabanda JS, Burt DW, O'Brien PC, Vignal A, Fillon V, et al. Molecular cytogenetic definition of the chicken genome: the first complete avian karyotype. Genetics. 2004;166:1367–1373. doi: 10.1534/genetics.166.3.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I, Karl E, Volobouev V, Griffin DK, Schartl M, Schmid M. Extensive gross genomic rearrangements between chicken and Old World vultures (Falconiformes: Accipitridae) Cytogenet Genome Res. 2006;112:286–295. doi: 10.1159/000089883. [DOI] [PubMed] [Google Scholar]
- Nanda I, Schlegelmilch K, Haaf T, Schartl M, Schmid M. Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet Genome Res. 2008;122:150–156. doi: 10.1159/000163092. [DOI] [PubMed] [Google Scholar]
- Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, et al. The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res. 2007;15:721–734. doi: 10.1007/s10577-007-1157-7. [DOI] [PubMed] [Google Scholar]
- Nishida C, Ishijima J, Kosaka A, Tanabe H, Habermann FA, et al. Characterization of chromosome structures of Falconinae (Falconidae, Falconiformes, Aves) by chromosome painting and delineation of chromosome rearrangements during their differentiation. Chromosome Res. 2008;16:171–181. doi: 10.1007/s10577-007-1210-6. [DOI] [PubMed] [Google Scholar]
- Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralls K, Ballou JD, Rideout BA, Frankham R. Genetic management of chondrodystrophy in California condors. Anim Conserv. 2000;3:145–153. [Google Scholar]
- Raudsepp T, Houck ML, O'Brien PC, Ferguson-Smith MA, Ryder OA, Chowdhary BP. Cytogenetic analysis of California condor (Gymnogyps californianus) chromosomes: comparison with chicken (Gallus gallus) macrochromosomes. Cytogenet Genome Res. 2002;98:54–60. doi: 10.1159/000068532. [DOI] [PubMed] [Google Scholar]
- Reisen WK, Carroll BD, Takahashi R, Fang Y, Garcia S, et al. Repeated West Nile virus epidemic transmission in Kern County, California, 2004–2007. J Med Entomol. 2009;46:139–157. doi: 10.1603/033.046.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov MN, Koriabine M, Nefedov M, de Jong PJ, Ryder OA. Construction of a California condor BAC library and first-generation chicken-condor comparative physical map as an endangered species conservation genomics resource. Genomics. 2006;88:711–718. doi: 10.1016/j.ygeno.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Romanov MN, Tuttle EM, Houck ML, Modi WS, Chemnick LG, et al. The value of avian genomics to the conservation of wildlife. BMC Genomics. 2009;10(Suppl 2):S10. doi: 10.1186/1471-2164-10-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Nanda I, Guttenbach M, Steinlein C, Hoehn M, et al. First report on chicken genes and chromosomes 2000. Cytogenet Cell Genet. 2000;90:169–218. doi: 10.1159/000056772. [DOI] [PubMed] [Google Scholar]
- Shetty S, Griffin DK, Graves JA. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 1999;7:289–295. doi: 10.1023/a:1009278914829. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. A phylogenetic study of bird karyotypes. Chromosoma. 1974;46:91–120. doi: 10.1007/BF00332341. [DOI] [PubMed] [Google Scholar]