Abstract
Optical encoders are commonly used in macroscopic machines to make precise measurements of distance and velocity by translating motion into a periodic signal. Here we show how Förster resonance energy transfer can be used to implement this technique at the single-molecule scale. We incorporate a series of acceptor dye molecules into self-assembling DNA, and the periodic signal resulting from unhindered motion of a donor-labeled molecular motor provides nanometer-scale resolution in milliseconds.
In many macroscopic devices, such as desktop printers, disk drives, numerically-controlled machine tools, and astronomical telescopes, distance and velocity measurements are made using optical encoders. A typical encoder consists of a light source and sensor, the path between which is occluded by a patterned film having a series of transparent windows. As the source and sensor move with respect to the film, modulation of the sensor output by the windows produces a periodic signal. The distance traveled is then given by the product of the window spacing and the number of periods observed in the signal.
Förster resonance energy transfer1–3 (FRET) is a near-field electromagnetic interaction which, when monitored by photon counting, can be used to measure distances within and between single biomolecules. Two fluorescent dyes, one with relatively short-wavelength absorption and emission (the donor) and the other with longer-wavelength absorption and emission (the acceptor) are attached to the system of interest. A laser is tuned to excite only the donor, which will normally emit a fluorescence photon of its characteristic color within a few nanoseconds. If, however, an acceptor is close by, it can acquire the excitation energy via FRET, and the observed fluorescence will be at a longer wavelength. The probability of transfer (the “energy transfer efficiency”) is given approximately by
| (1) |
where r is the dye separation, and R0 (the Förster radius) is a characteristic distance at which E = 0.5. In typical single-molecule experiments, R0 ≈ 5 nm. An estimate of E, and hence r, can be obtained from the fraction of detected photons emitted by the acceptor. Owing to departures from Eq. 1 and inherently low signal-to-noise ratios, it is difficult to obtain absolute distances with much better than 10% precision using single-pair FRET.2,3
Molecular motors are essential components of the machinery of life, enabling processes including DNA replication, transcription and repair, protein synthesis, and muscle movement. Single-molecule measurements4–7 have yielded a wealth of otherwise inaccessible information about the mechanisms by which these biomolecules move and function. To achieve nanometer-precision tracking of individual motor proteins, available fluorescence techniques rely on long integration times, thousands of fluorophores, or large quantum dot labels.4 Magnetic and optical trapping assays5–7 require that significant tension be applied to the motor or its substrate. In a 1999 review,1 Weiss proposed using periodic arrays of FRET acceptors to monitor digestion of DNA by a single nuclease, and transcription by a tethered RNA polymerase. Prior to now, this suggestion had not been realized. In a related ensemble measurement, Yin et al.8 recently used sequential quenching to track the average motion of a population of synthetic molecular walkers as they passed three spectrally distinct DNA-bound fluorophores over a period of tens of minutes. Here we describe the synthesis and use of a “FRET encoder,” similar to the configuration envisioned by Weiss, that circumvents uncertainties in FRET distance determinations by producing a periodic signal, thereby enabling rapid and precise tracking of single molecular motors.
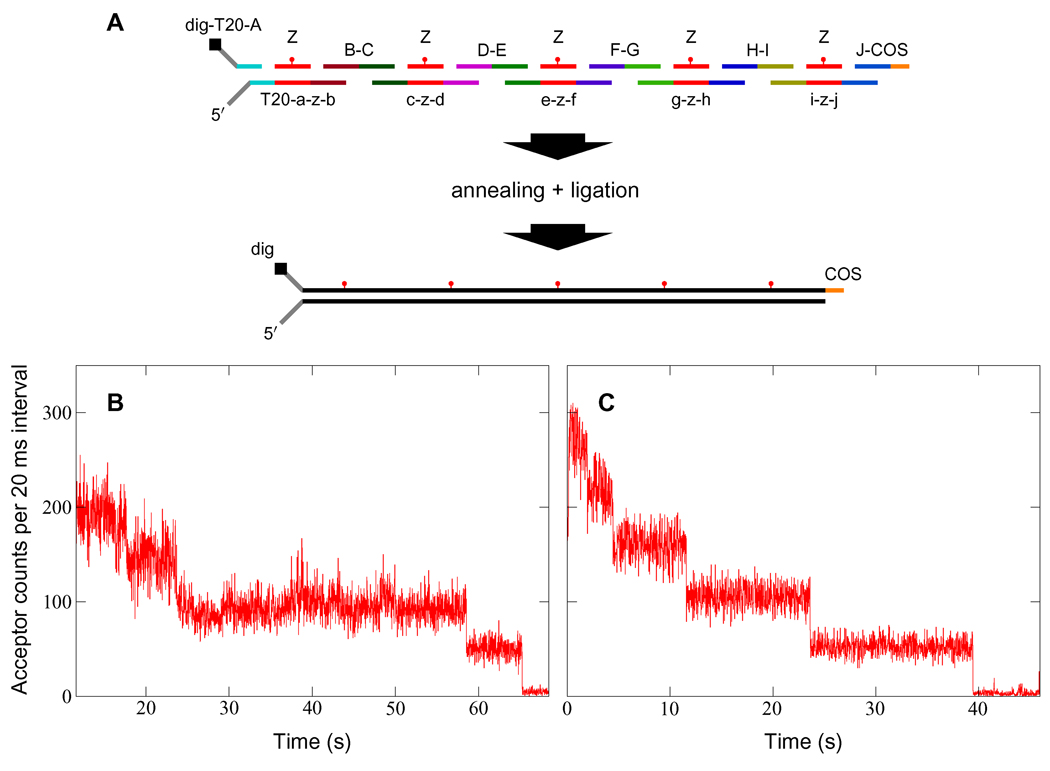
We synthesized FRET encoders from a set of phosphorylated single-stranded (ss) DNA oligonucleotides. These were designed to form, upon annealing and ligation, double-stranded (ds) DNA labeled with an acceptor fluorophore every 69 base pairs (bp) (see Fig. 1, Fig. 2 inset, and Supporting Table 1). Following synthesis and purification, encoders were ligated at one end to biotinylated λ DNA handles. The other end was terminated with two single-stranded poly(dT) tails, one of which was labeled with digoxigenin at the 3′ terminal end. Complete self-assembly was verified by continuously exciting sacrificial encoders with a 1 µW 633 nm laser, resulting in successive photobleaching of the dyes over a period of tens of seconds (Fig. 1). A discrete downward step in the fluorescence data indicates bleaching of a single dye, and we find that the number of photo-bleaching steps corresponds precisely to the number of dyes designed into the encoders.
Figure 1.
Synthesis and verification of FRET encoders. (A) DNA self-assembly scheme for 5-period encoders (see Supporting Table 1 for sequences). Each unique sequence is identified with a lower-case letter, and its complement with the corresponding upper-case letter. For example, oligonucleotide E-D (read from 5′ to 3′) pairs only with c-z-d and e-z-f, while e-z-f pairs only with G-F, Z, and E-D. Sequence Z is internally labeled with an acceptor dye. The free 5′ poly(dT) tail facilitates DnaB loading, while the digoxigenin label at the 3′ terminus allows the encoder to be immobilized by attachment to anti-digoxigenin on a fused-silica surface. The 5′ COS overhang is complementary to the 12 bp single-stranded cos site of the λ phage genome. (B and C) Sacrificial photobleaching of four- (B) and five-period (C) encoders by direct excitation at 633 nm verifies complete self-assembly. More than 80% of tested encoders produced the expected photobleaching pattern.
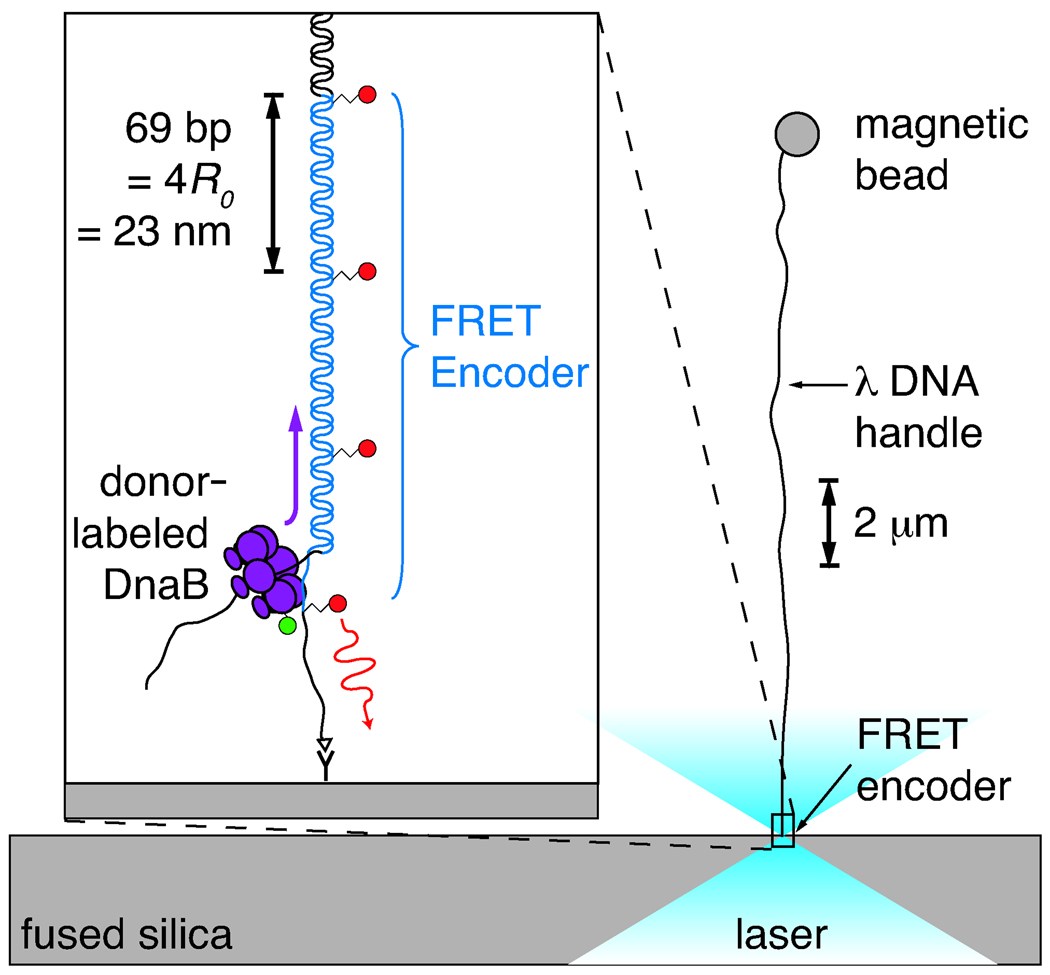
Figure 2.
Illustration of the experiment, approximately to scale. The FRET encoder is tethered via an anti-digoxigenin/digoxigenin linkage to a fused-silica cover slip. A biotinylated λ DNA handle, ligated to the untethered end of the encoder, is attached to a streptavidin-coated magnetic bead. A 0.5–3.0 pN vertical magnetic force is applied to the bead, pulling it away from the surface and aligning the encoder with the optical axis. Donor-labeled DnaB helicase diffuses into the focus and loads onto the free 5′ tail of the encoder. As the encoder is unwound, the moving laser-excited donor passes one acceptor after another, inducing long-wavelength fluorescence via FRET. The resulting periodic acceptor signal reports on the motion. A spacing of 4R0 between acceptor dyes was chosen so that the donor would be within 2R0 of only a single acceptor at any given time.
To test the FRET encoder, we chose DnaB, the primary replicative helicase from Escherichia coli.9–18 Single-molecule techniques have already revealed many properties of helicases, proteins that translocate along DNA, separating the double helix into its component strands. Stochastic behaviors that would be difficult to detect with ensemble methods, such as backtracking,19 strand switching20,21 and pausing,19,22 have all been observed. Direct application of force to DNA substrate molecules has been used to differentiate between possible unwinding mechanisms, and to examine the role played by ATP.20,22–24 In addition, single-molecule measurements of interactions between helicases and other replication proteins have demonstrated how coordination may arise.25,26 Myong et al.27 have used single-pair FRET to measure the hepatitis C virus NS3 helicase, proposing a “spring-loaded” unwinding mechanism. Ha and coworkers have also observed “repetitive shuttling” behavior for Escherichia coli Rep helicase along ssDNA,28 and strand switching of human BLM helicase.21
DnaB is an ATP-driven hexamer that encircles one strand of DNA and translocates in the 5′ to 3′ direction, displacing the non-encircled complementary strand.9 It is processive, completing several enzymatic cycles before dissociating from its substrate.18 While a number of studies have focused on the structure and function of DnaB9,13,14,16,17 and its interaction with other replisome components,10,12,15,26,29,30 there have been few direct measurements of the velocity and processivity (distance traveled prior to dissociation) of DnaB alone,12,18 more of which will improve our understanding of its unwinding mechanism and movement.
DnaB monomer, expressed and purified as described by Yuzhakov et al.,31 was sparsely and nonspecifically labeled with an amine-reactive form of the FRET donor dye Alexa Fluor 488 (Invitrogen), and dialyzed exhaustively. For this first experiment, nonspecific labeling provided several advantages: it was not necessary to mutate the protein, there was no possibility that an unfortunate choice of label site would prevent us from collecting data, and we were able to observe a variety of signal forms. It should be noted that a change in label position parallel to the helicase axis would be expected to cause only an overall shift in the phase of the FRET encoder signal. In order to decrease the likelihood that a single hexamer would carry more than one donor fluorophore, at most 5% of monomers were labeled.
Using the FRET encoder, we measured donor-labeled DnaB as it unwound DNA (Fig. 2). By means of the 3′ digoxigenin labels, FRET encoders were tethered to a fused-silica coverglass, which formed the bottom wall of a flow cell. A magnetic bead was attached to the distal end of each encoder via the biotinylated λ DNA handle. The flow cell was mounted on a combined confocal detection system and magnetic tweezer (Supporting Fig. 2). A constant force32 between 0.5 and 3.0 pN was applied to the magnetic beads, pulling them away from the surface and aligning the encoders parallel to the optical axis.
A buffered solution containing between 32 and 200 nM DnaB hexamer and 5 mM ATP was added to the flow cell, after which a pre-selected encoder was automatically centered using the detectors (see supporting information). A 488 nm laser at between 11 and 15 µW was then focused on the sample in order to excite the FRET donor attached to the helicase.
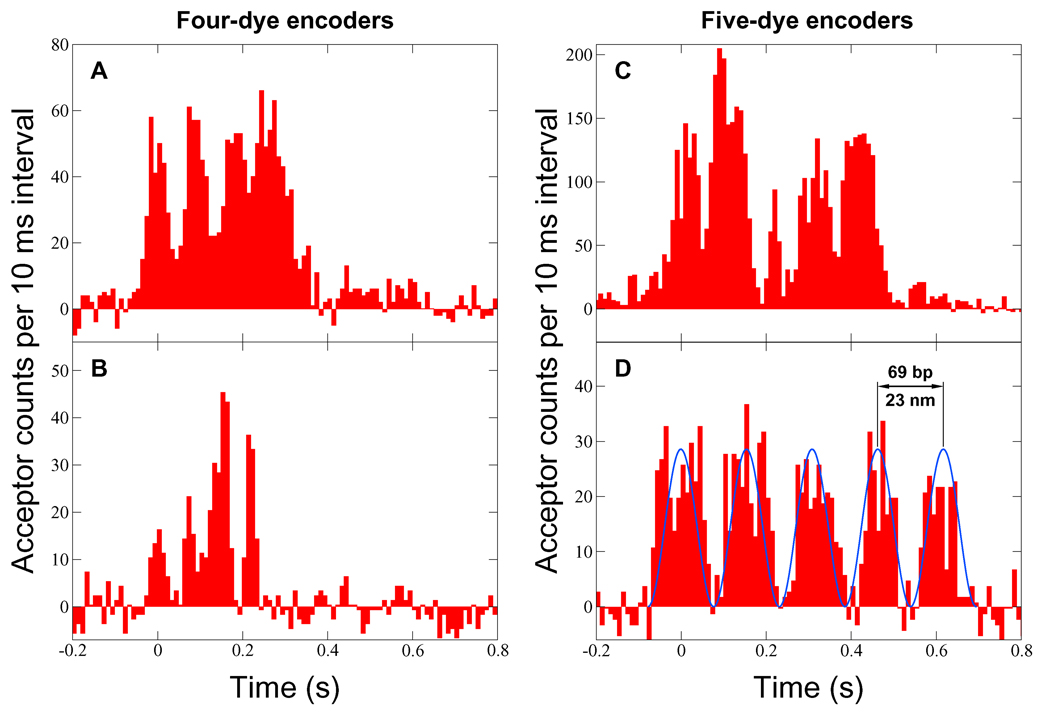
The resulting acceptor fluorescence data (Fig. 3) consist of a series of peaks, with the number of peaks in each complete event correlating precisely with the number of dyes in the corresponding encoder. No events were detected in control experiments without DnaB, nor with unlabeled DnaB. Measurement of the donor signal was precluded by relatively high concentrations of labeled DnaB, required to promote hexamer stability, achieve unwinding activity,11 and produce a sufficient number of visible events. At the lowest concentration used in these experiments, there were on average approximately three labeled hexamers per confocal detection volume.
Figure 3.
Acceptor fluorescence signal as a function of time (background subtracted). A signal peak is produced each time the donor-labeled helicase passes an acceptor dye on its encoder. For each event, the unwinding speed was determined by fitting a function of the form A cos2 [πf(t − t0)] to the data using least squares. One such fit is illustrated in (D). The speeds were (A) 759 ± 13 bp/s, (B) 950 ± 24 bp/s, (C) 665 ± 12 bp/s, and (D) 447 ± 6 bp/s. A complete event indicates that a single labeled helicase traveled at least 276 bp (A and B) or 345 bp (C and D). This implies processivity much greater than has previously been measured for DnaB alone.
The variation in overall signal level among the Fig. 3 eventsmost likely results from suboptimal sample centration and/or focal drift (the risk of encoder photobleaching makes repeated adjustment impractical). Control experiments using a donor-acceptor pair on DNA (data not shown) indicate that with optimal alignment, the maximum count rate is approximately 20 ms−1. Thus a single donor label could account for the signal shown in Fig. 3C. Multiple donors on the helicase would be expected to reduce signal contrast and produce additional peaks unless the attachment sites happened to be in close proximity. The gradually increasing background in Fig. 3A may have been caused by a growing cluster of acceptor dyes on single-stranded DNA behind the helicase. Because of the time required, it is not possible to calibrate the tension applied to each encoder.
Since donor labeling was not site-specific, and the radius of the DnaB hexamer is approximately equal to R0,13 the position of the donor (or possibly multiple donors) on the protein could significantly affect the form of the observed signal. In particular, if the label is on the outer surface, a 180° rotation of the protein about its central axis could change the donor-acceptor distance by as much as 2R0. Figures 3B and 3C (see also Supporting Fig. 3D) exhibit modulation of peak heights that could be caused by rotation of the protein with respect to the acceptor-labeled strand, or possibly by multiple donors. A low-frequency envelope may indicate gradual rotation of the helicase, or might instead result from a nonlinear combination, similar to aliased sampling or inter-frequency beating, of the oscillations associated with translation and rapid rotation. Future measurements employing longer encoders and site-specific protein labeling will better indicate the causes of the observed modulation.
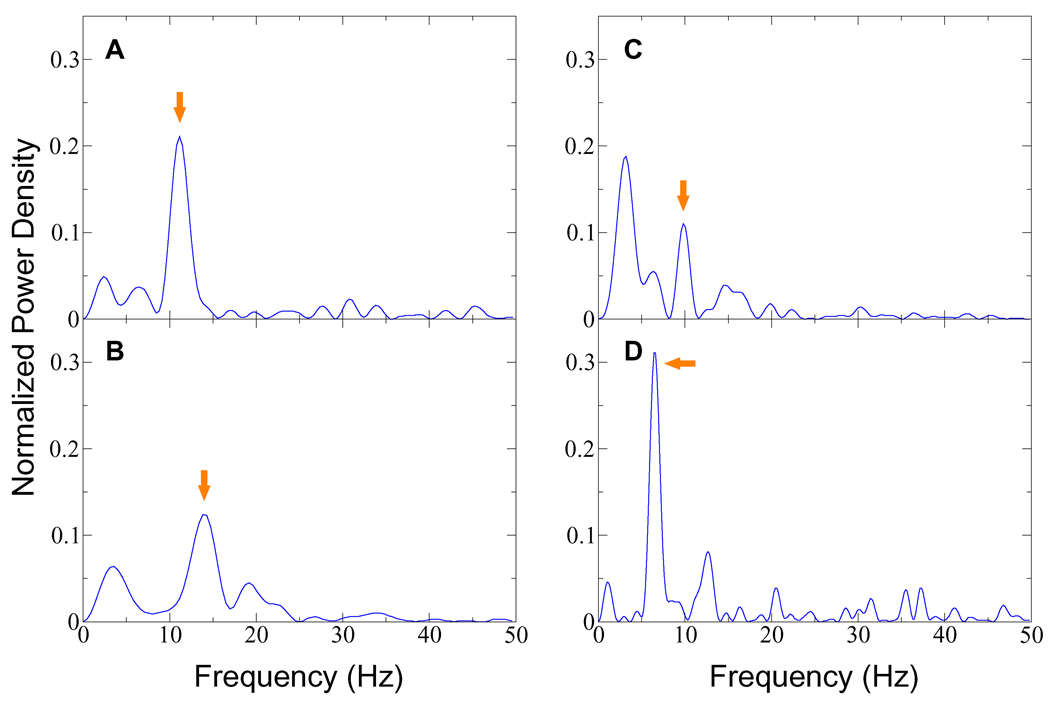
Consistent correspondence between the maximum number of contiguous periodic peaks and the number of encoder dyes strongly suggests that the observed signals result from translation of DnaB. In order to separate the primary translational frequency from modulation, noise, and background drift, we computed the power spectra of the four events shown in Fig. 3. For each event, the mean count rate was subtracted from all bins to remove the zero-frequency component. The signal was then padded with zeros to eight times its original length, and a discrete Fourier transform was calculated with no windowing. Fig. 4 shows the squared moduli of the positive-frequency portions of the four transforms.
Figure 4.
Power spectra for the four events shown in Fig. 3. The frequency generated by translation of the helicase can be estimated for a complete event by dividing the number of encoder dyes by the event duration. Peaks corresponding to the estimated frequencies are indicated above by arrows. Significant low-frequency components seen in (B) and (C) may be caused by rotational modulation (see text). Side lobes appearing in these power spectra do not exceed one twentieth the height of the associated peaks.
Since acceptor intensity is determined by Eq. 1, the leading and trailing edges of the measured peaks are steep, and the observed signals will not be sinusoidal, even in the absence of modulation. Some power will consequently appear in harmonics above the translational frequency. The finite duration of an event sets a lower bound on its power spectrum peak widths. Widths in excess of this fundamental limit can be caused by nonuniformity of helicase motion (for example, a slight slowing can be seen in the third and fourth peaks of Fig. 3D), and by the small amount of noise that happens to be in a frequency band near the peak.
Precisely estimating the frequency of a noisy, limited-duration signal is a complicated problem, particularly if the underlying functional form is not known.33,34 In addition to locating peaks in the power spectra, we computed least-squares fits to the acceptor fluorescence data using functions of the form A cos2 [πf(t − t0)]. One such fit, corresponding to the translational frequency component in the power spectrum, is illustrated in Fig. 3D. Frequencies from the fits were multiplied by the 69 bp encoder period to obtain helicase speeds (see Fig. 3 caption). Single standard deviation uncertainties were determined from the fit parameters.35 For the event shown in Fig. 3C, the translational frequency at the power spectrum peak differed by 1.2 standard deviations from that found by fitting. For events A, B, and D, the differences were less than one standard deviation. The largest fractional uncertainty was only 2.5%, indicating that the two frequency estimates are consistent, and that these events exhibit nearly constant-speed unwinding.
For a sample consisting of the events shown in Fig. 3 and eight additional events shown in Supporting Fig. 3, helicase speeds were determined to be between 230 and 1060 bp/s, with a mean of 660 bp/s and a standard deviation of 250 bp/s. This large variation is consistent with other single-molecule helicase measurements.19 The highest unwinding velocities we observe exceed those previously reported in room-temperature ensemble studies of DnaB alone (our measurements were made at 22 ± 1 °C), but are comparable to the average speed of a complete E. coli replication fork in vivo.12,18 The speed of DnaB is known to vary over a wide range,36 being strongly dependent on interactions with other proteins involved in DNA replication, and on temperature.18
A complete five-period event with constant phase advance indicates that a single labeled helicase has unwound a length of DNA equal to or greater than 345 base pairs (an event begins and ends with the donor approximately one half period from the closest encoder dye). The probability is low that more than one labeled helicase is involved in the production of a single periodic event. The peak intervals in Fig. 3 are nearly uniform, exhibiting no pauses for dissociation of one helicase and replacement by another. For more than one labeled helicase to line up and produce a signal with the observed contrast would require that they maintain a fixed spacing close to a multiple of the encoder dye interval (69 bp), with the speeds of the trailing helicases equal to that of the lead, or some unlikely combination of fixed spacing and synchronized, phase-staggered rotational motion. Furthermore, there is no reason to expect a trailing helicase on the separated unlabeled strand to remain in contact with the encoder strand. Multiple labeled helicases would also produce more peaks per event than the number of encoder dyes.
These results contrast significantly with a prior ensemble study reporting a processivity of approximately 10 bp for DnaB.18 Tanner et al.26 recently found that interactions between DnaB and other replisome components increase its processivity to 10.5 ± 0.9 kilobases, but did not detect unwinding activity of DnaB alone.
Owing to the form of Eq. 1, the position resolution of the FRET encoder varies over the course of each cycle, peaking at r ≈ R0. Using the observed signal and background levels, it is possible to estimate the signal-to-noise ratio SNRr=R0 (see supporting information), which will depend on the laser excitation power, the integration time, and the change in dye separation (the step size). For the 4.5 bp (1.5 nm) translation in a 10 ms bin of Fig. 3D, we expect SNRr=R0 ≈ 2.6. This is consistent with the abrupt non-random transitions seen on the leading and trailing edges of the peaks, and compares well with other single-molecule techniques using similar integration times.4–7
Under the conditions used to collect the data in Fig. 3D, SNRr=R0 ≈ 1800 for a single base pair (0.34 nm) step, where P is the laser power in watts and τ is the measurement time in seconds. Presently, P = 11 µW and τ = 10 ms, giving SNRr=R0 ≈ 0.6. While this is clearly not adequate to resolve stepping motion, one can envision an experiment on a somewhat slower molecular motor, for example a DNA polymerase,37 which allows translocation to be triggered. This would enable the use of much higher excitation power without fear of bleaching the encoders. With P = 200 µW, we would expect SNRr=R0 ≈ 5 for a 1 bp step in 40 ms. Thus the prospects appear good for high-resolution FRET encoder measurements of full-speed processive molecular motion.
The technique we have demonstrated could be extended in several directions. We have recently synthesized longer FRET encoders using polymerization and rolling-circle DNA amplification38 (to be published elsewhere). These will enable increased precision through signal averaging over many periods. A single-molecule quadrature encoder, which would report both speed and direction by producing two signals of the same frequency with a ±π/2 phase difference, could be synthesized by replacing the acceptor-labeled oligonucleotide with one containing two labels of different colors. Absolute molecular location could be determined using a bicolor FRET encoder with two different acceptor periods, and rotary FRET encoders could be used for measuring flagellar motors and other rotating biomolecular assemblies.
Encoders with vastly improved photostability could be constructed using quantum dots,39,40 and rigid encoders fabricated on inorganic substrates could be used to monitor translation, rotation, and flexure of future engineered nanostructures.
Because FRET encoders have high resolution, are independent of force, require no calibration beyond a priori knowledge of acceptor dye spacing, and are potentially applicable to a wide range of molecular motors and other nanoscale objects, we expect this technique to find use in many new measurements.
Supplementary Material
Acknowledgement
Omar Saleh and Noah Ribeck provided us with essential advice and software for using magnetic tweezers, many useful suggestions regarding DnaB helicase, and comments on the manuscript. We thank Megan Valentine and Keir Neuman for comments on the manuscript, Deborah Fygenson for discussions of FRET encoder synthesis, and Benjamin Schuler for advice regarding protein labeling. This work was supported by the Hellman Family Foundation, the Human Frontier Science Program, and the California Nanosystems Institute. K.J.C. and K.W.P. were supported by National Institutes of Health grant GM062958-01. E.A.L. is an Alfred P. Sloan research fellow.
Footnotes
Supporting Information Available
Detailed methods, DNA sequences, supporting figures and data, data analysis, and signal-to-noise calculation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 2.Dolghih E, Ortiz W, Kim S, Krueger BP, Krause JL, Roitberg AE. J. Phys. Chem. A. 2009;113:4639–4646. doi: 10.1021/jp811395r. [DOI] [PubMed] [Google Scholar]
- 3.Camley BA, Brown FLH, Lipman EA. J. Chem. Phys. 2009;131:104509. [Google Scholar]
- 4.Park H, Toprak E, Selvin PR. Q. Rev. Biophys. 2007;40:87–111. doi: 10.1017/S0033583507004611. [DOI] [PubMed] [Google Scholar]
- 5.Greenleaf WJ, Woodside MT, Block SM. Annu. Rev. Biophys. Biomolec. Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moffitt JR, Chemla YR, Smith SB, Bustamante C. Annu. Rev. Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 7.Neuman KC, Nagy A. Nat. Methods. 2008;5:491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 9.LeBowitz JH, McMacken R. J. Biol. Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 10.Mok M, Marians KJ. J. Biol. Chem. 1987;262:16644–16654. [PubMed] [Google Scholar]
- 11.Bujalowski W, Klonowska MM, Jezewska MJ. J. Biol. Chem. 1994;269:31350–31358. [PubMed] [Google Scholar]
- 12.Kim SS, Dallmann HG, McHenry CS, Marians KJ. Cell. 1996;84:643–650. doi: 10.1016/s0092-8674(00)81039-9. [DOI] [PubMed] [Google Scholar]
- 13.San Martin C, Radermacher M, Wolpensinger B, Engel A, Miles CS, Dixon NE, Carazo JM. Structure. 1998;6:501–509. doi: 10.1016/s0969-2126(98)00051-3. [DOI] [PubMed] [Google Scholar]
- 14.Jezewska MJ, Rajendran S, Bujalowska D, Bujalowski W. J. Biol. Chem. 1998;273:10515–10529. doi: 10.1074/jbc.273.17.10515. [DOI] [PubMed] [Google Scholar]
- 15.Barcena M, Ruiz T, Donate LE, Brown SE, Dixon NE, Radermacher W, Carazo JM. Embo J. 2001;20:1462–1468. doi: 10.1093/emboj/20.6.1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan DL, O’Donnell M. Mol. Cell. 2002;10:647–657. doi: 10.1016/s1097-2765(02)00642-1. [DOI] [PubMed] [Google Scholar]
- 17.Yang SX, Yu XO, VanLoock MS, Jezewska MJ, Bujalowski W, Egelman EH. J. Mol. Biol. 2002;321:839–849. doi: 10.1016/s0022-2836(02)00711-8. [DOI] [PubMed] [Google Scholar]
- 18.Galletto R, Jezewskax MJ, Bujalowski W. J. Mol. Biol. 2004;343:83–99. doi: 10.1016/j.jmb.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 19.Perkins TT, Li HW, Dalal RV, Gelles J, Block SM. Biophys. J. 2004;86:1640–1648. doi: 10.1016/S0006-3495(04)74232-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Proc. Natl. Acad. Sci. U. S. A. 2004;101:6439–6444. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yodh JG, Stevens BC, Kanagaraj R, Janscak P, Ha T. EMBO J. 2009;28:405–416. doi: 10.1038/emboj.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Pyle AM, Bustamante C. Nature. 2006;439:105–108. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lionnet T, Spiering MM, Benkovic SJ, Bensimon D, Croquette V. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. Nature. 2006;439:621–624. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 26.Tanner NA, Hamdan SM, Jergic S, Schaeffer PM, Dixon NE, van Oijen AM. Nature Struct. Biol. 2008;15:170–176. doi: 10.1038/nsmb.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myong S, Bruno MM, Pyle AM, Ha T. Science. 2007;317:513–516. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Nature. 2005;437:1321–1325. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 29.Fang LH, Davey MJ, O’Donnell M. Mol. Cell. 1999;4:541–553. doi: 10.1016/s1097-2765(00)80205-1. [DOI] [PubMed] [Google Scholar]
- 30.Schaeffer PM, Headlam MJ, Dixon NE. IUBMB Life. 2005;57:5–12. doi: 10.1080/15216540500058956. [DOI] [PubMed] [Google Scholar]
- 31.Yuzhakov A, Turner J, O’Donnell M. Cell. 1998;86:877–886. doi: 10.1016/s0092-8674(00)80163-4. [DOI] [PubMed] [Google Scholar]
- 32.Gosse C, Croquette V. Biophys. J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kay SM. Modern Spectral Estimation: Theory and Application. Prentice Hall; 1988. [Google Scholar]
- 34.Quinn BG, Hannan EJ. The Estimation and Tracking of Frequency. Cambridge University Press; 2001. [Google Scholar]
- 35.Wolberg J. Data Analysis Using the Method of Least Squares. Springer Verlag; 2006. [Google Scholar]
- 36.Indiani C, Langston LD, Yurieva O, Goodman MF, O’Donnell M. Proc. Natl. Acad. Sci. U. S. A. 2009;106:6031–6038. doi: 10.1073/pnas.0901403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Christian TD, Romano LJ, Rueda D. Proc. Natl. Acad. Sci. U. S. A. 2009;106:21109–21114. doi: 10.1073/pnas.0908640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beyer S, Nickels P, Simmel FC. Nano Lett. 2005;5:719–722. doi: 10.1021/nl050155a. [DOI] [PubMed] [Google Scholar]
- 39.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin Y, Alivisatos AP. Nature. 2005;437:664–670. doi: 10.1038/nature04165. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.