Abstract
Purpose
Detect tumor-related DNA using LigAmp in histologically clear margins and associate results with clinical outcome.
Experimental Design
Patients with head and neck cancer were registered for molecular analysis of surgical margins. Adequacy of resection was ensured using histologic margin analysis. Further margins were then harvested and DNA extracted. TP53 mutations in tumor were determined using Affymetrix p53 GeneChip. Margins were analyzed by Ligamp in comparison with standard curves for quantification of mutant DNA. Ligation employed 2 oligonucleotides to isolate DNA targeting the mutation. Ligated DNA was amplified using real-time PCR. The quantity of mutation in the margin was determined as percent of mutant species relative to plasmid (MRP) and relative to tumor (MRT). Cutpoints were identified and defined groups evaluated for local failure-free, cancer-specific, and overall survival. Study margins were examined for presence of tumor by light microscopy.
Results
Tissue from 95 patients with common mutations was analyzed. Fifteen experienced local recurrence. Cutpoints of 0.15% for MRP and 0.5% for MRT were chosen as most selective of recurrent cases. LigAmp had slightly better area under the ROC curve (p=0.09) than light microscopy correctly predicting 9 of 15 recurrent tumors. There were 6 false negative cases and 26 false positive results. No statistically significant distinctions were observed in cancer-specific or overall survival in this limited cohort.
Conclusions
Ligamp provides quantifiable, sensitive detection of mutant DNA in histologically normal margins. Detection of mutant species in margins may identify patients at risk of local recurrence.
INTRODUCTION
Despite significant improvement in loco-regional control by multimodal therapy, including surgery, radiotherapy and chemotherapy, only a modest change has been attained in the 5-year survival rates of head and neck squamous cell carcinoma (HNSCC) patients over recent decades. It is well established that during surgical treatment, clear margins represent one of the most important indicators of clinical outcome. However, even when surgical margins have been read as clear, the local recurrence rate is still 10–30% (1, 2). This observation suggests that occult tumor cells undetected by conventional histologic evaluation may be present surrounding resection margins, leading to treatment failure (3, 4). The detection of occult disease in margin tissue may be predictive of local recurrence and potentially serve as an indicator for consideration of further surgery and/or adjuvant chemo-radiotherapy (5).
Targets for molecular detection
Various targets have been proposed as detection markers of occult HNSCC (6, 7). Ideal candidates should have tumor specificity, high prevalence, and should be retained during tumor progression and metastasis. DNA molecular markers are robust due to the higher stability of DNA in tissue samples, rendering the possibility of using archived paraffin embedded materials. According to these criteria, TP53 mutations appear to be suitable markers for minimal residual disease detection in HNSCC (8). Numerous studies have attempted to assess the presence of cells with TP53 mutation by immunohistochemistry (IHC). However, IHC has substantial limitations as a surrogate for DNA sequence probing to detect TP53 mutations (9, 10). Not all mutations result in the production of a stable protein amenable to staining. Mis-sense mutations that introduce a “stop” sequence, for example, will result in no protein production. On the other hand, there are reasons other than gene mutation that may account for accumulation of p53 protein within a cell that is detected by IHC staining, such as binding to other cellular proteins (MDM2). Other problems include the use of different antibodies and unmasking techniques as well as an arbitrary threshold definition of “over-expression” in different reports.
Molecular detection methodology
Methods for detection of altered DNA include restriction fragment length polymorphism (RFLP) (11), allele-specific PCR (12), and oligonucleotide ligation (13, 14) Each of these assays has disadvantages, such as insufficient detection limits (0.1% - mutant/WT ratio of 1:1000) and labor-intensive procedures.
Our previous work has shown a sensitive and specific approach for molecular assessment of TP53 mutations in HNSCC margins using oligomer probe binding and radionucleotide detection (15). However, limitations of that method are the extensive time required and the need for radiolabelled probes. Recently a PCR-based method, called LigAmp (16) was described to assess single nucleotide genetic variants in the k-RAS gene and for HIV genotyping. This assay is able to detect 1 mutant species in the presence of 10,000 wild-type molecules (0.01%). LigAmp results are quantitative, allowing investigators to explore the threshold (percentage) of tumor species present associated with clinical outcome.
The aim of this study is to demonstrate the feasibility to detect and measure the presence of tumor-related DNA using LigAmp in histologically clear surgical resection margins and to compare results with clinical outcome, particularly local disease recurrence. The hypothesis is that submicroscopic levels of remaining malignant cells contribute to local treatment failure of HNSCC patients. Molecular assessment of risk of recurrence may help in the identification of patients who could benefit from additional surgery or chemo-radiotherapy, while avoiding inappropriate treatment of those subjects for which molecular assessment indicate the adequacy of surgical treatment.
MATERIALS AND METHODS
Study protocol
Between 1997 and 2002, 560 patients were accrued to a cooperative group study (ECOG 4393 and RTOG 9614). Eligible patients were diagnosed with HNSCC and planned for surgery with curative intent. Patients were identified at over 18 participating institutions and signed informed consent approved by the cooperative groups and each institutional investigational review board. Demographic patient information was submitted to the cooperative group data centers preoperatively. On the day of surgery, samples of tumor were harvested for study purposes. Tumor extirpation proceeded following standard surgical practice. Margin tissue was submitted to the institutional surgical pathology service for intra-operative frozen section analysis to confirm the adequacy of resection. Once adequate extirpation had been confirmed in this manner, further margin samples were taken from the presumed normal tissue at the perimeter of the surgical resection site for study purposes. All specimens were shipped on dry ice by overnight courier to the Johns Hopkins Head and Neck Cancer Research Laboratory.
Tumor and margin sample collection
Tumor and margin samples were rapidly frozen at −80°C. A series of 5-micron sections were cut from the primary tumor and margin specimens for hematoxylin-eosin staining to inspect for the presence of HNSCC or dysplasia. Additional 12-micron sections were cut and placed in 1% SDS and proteinase K at 48°C overnight followed by DNA extraction with phenol/chloroform and ethanol precipitation.
Tumor samples with at least 70% cancer cells were eligible for molecular studies. Tumors with low neoplastic cellularity (<70%) were microdissected to enrich the malignant cells. DNA was extracted from all margins samples and submitted for analysis. Margin tissue was processed in toto without microdissection. For margin processing, serial sectioning was performed, making thin section slides at the beginning, middle and end of the cut through the sample. These slides were stained with hematoxylin and eosin for light microscopic analysis by the study pathologist (WHW). Fifty sections of 12 microns thickness were harvested from each margin sample for DNA extraction.
TP53 exon amplification
A second generation p53 GeneChip assay (Affymetrix, Santa Clara, CA) was used to screen TP53 mutations in exons 2–11 in Head and Neck primary tumors (17). The presence of mutation in primary tumor was confirmed by sequencing. For tumor and margins, each exon harboring a mutation was amplified in a single polymerase chain reaction (PCR). Reactions were carried out in a volume of 25 µl using 200 ng of template, 2mM of each primer (Invitrogen, CA), 5 units of Platinum Taq polymerase (Invitrogen, CA), 1.5 mM each dATP, dCTP, dGTP, dTTP, 16.6 mM ammonium sulfate; 67 mM Trizma; 6.7 mM magnesium chloride; 10 mM mercaptoethanol; 0.1% DMSO; PCR was performed using the following conditions: 95°C for 2 min, followed by 35 cycles at 95°C for 30 s, Melting Temperature specific for each exon for 1 minute, 72°C for 90 s, extension at 72°C for 2 min. After each gel run, PCR product was purified using a QIAquick Gel Extraction Kit (Qiagen), and the concentration was determined using a Nanodrop (ND-1000 Spectrophotometer, NanoDrop Technologies, Inc.).
TP53 mutant plasmids
A subset of common TP53 mutations that occurred in more than one tumor within the population were selected for Ligamp margin analysis. Plasmids containing the TP53 mutated sequences were manufactured to provide a source of positive control DNA for mutant species quantification. Site-direct mutagenesis was performed from Top Gene Technologies (Quebec, Canada) using pcDNA 3.1 p53 vector to obtain mutant TP53 plasmids for the following mutated codons: 157.1T, 179.1G, 192.1T, 193.2T, 193.2G, 196.1T. 205.2G, 206.2A, 213.1T, 238.2A, 272.1T, 272.1A, 273.2T, 278.1T, 280.2A, 282.1T, 298.1T, 342.1T. pCR259 plasmids (a gift from Dr. C. Ishioka) have been used as mutant plasmids for other TP53 mutations: 175.2A, 220.2G, 232.1T, 245.1A, 248.1T, 248.2A, 273.1T, 273.2A (Fig. 1 Supplementary). Each plasmid was amplified for the exon containing the mutation (primer sequences are available on request). Concentration was determined using a Nanodrop (ND-1000 Spectrophotometer, NanoDrop Technologies, Inc.). 1010 copies of PCR product were serially diluted to generate standard curves. Tumor, normal and margin samples have been analyzed in comparison with the standard curve in order to get an accurate quantification of mutant DNA.
TP53 LigAmp
After DNA amplification, dilutions of each PCR product were made to give 1010 amplicon copies/5 µl calculated from the size of the amplicon for each exon and assuming 1 bp of ddDNA = 660pg/pmol and 1 pmol = 6.02 × 1011 copies. According to this calculation the LigAmp assay was performed in two steps (Fig 2 Supplementary). First, the Ligation Step was conducted using two different oligonucleotides (upstream and downstream) designed to specifically match the mutation (Ligation primer sequences are available on request). DNA was incubated in a final volume of 25 µl with 1 pmol of mutant oligonucleotide, 0.5 pmol of downstream oligonucleotide, 4 U of Pfu DNA ligase in 1X Pfu ligase buffer (Stratagene). Ligation conditions were the following: 95°C for 3 min, 90 cycles at 95°C for 30 sec. and 64°C for 4 min. In the presence of mutation the two oligonucleotides are ligated to one other. Each contains a region specific to target the mutation and a tail (M13F and M13R) that permit amplification of the ligated product in a subsequent Q-PCR (Quantitative PCR) reaction. In order to increase the specificity of the assay, a mismatch has been inserted 2 base pairs upstream from the mutant base. For all TP53 mutant upstream oligonucleotides, we tested all possible mismatches and then we chose the one with best performance (r2 values ranging from 0.95 to 0.98). The upstream oligonucleotide also contains a region of unique foreign DNA (16rSDNA) that serves as the binding region for a probe in the Q-PCR reaction. The second step, the Q-PCR amplification, was performed using the 7500 Real-Time PCR System (Applied Biosystem). The ligated DNA was amplified using 5 pmol of upstream (5’-ACTGTAAAACGACGGCCAGTG-3 ’) and downstream M13 primers (5 ’-TGCAGGAAACAGCTATGACCA-3 ’), and 2.5 pmol of a probe (16rSDNA FAM-5’-CGTATTACCGCGGCTGCTGGCAC-3’-TAMRA), 12.5 µl of Taqman Universal PCR Master Mix (Applied Biosystems) in a final volume of 25 µl. The M13 primers hybridize to the M13 tails and the probe hybridizes to the 16rSDNA sequence. This step is independent of the specific mutation targeted in the ligation step.
The binding of the probe requires ligation of the two oligonucleotides and subsequent polymerization of the complementary strand of DNA in the Q-PCR step. The Q-PCR probe binds to the bottom strand of the amplified DNA. Extension from the M13 forward primer allows the probe to be cleaved and the released fluorophore to be detected by 7500 SDS software. If no ligation occurs in the first step because of mispairing, there is no template for amplification in the Q-PCR step. The quantity of mutations in margins has been expressed as two ratios: the percent of mutant species in margins relative to plasmid (MRP, absolute quantity) and the percent of mutant species relative to tumor (MRT, relative quantity).
Statistical methods
Local failure events (local tumor recurrence) were documented during follow-up at treating institutions by physical examination and/or radiographic imaging confirmed with biopsy when possible. Patients who died of other causes or who are still alive were censored as of the date last known alive. Cause-specific survival was estimated by considering death from disease to be an event and censoring patients who died of other or unknown causes and those still alive at the date of last reported follow-up . Overall survival was counted from the date of surgery to the date of death or last date known alive. Mutations were classified as disruptive if they were nonconservative mutations located inside the key DNA-binding domain (L2–L3 region), or if they were stop codons in any region (17).
Descriptive statistics were used to characterize patients and clinical features. Fisher’s exact test (18) and the Wilcoxon rank sum test (19) were used to compare characteristics between patients with and without positive margins and with and without local recurrence. Recursive partitioning was used to explore possible cutpoints (20). ROC curves were used to compare the relative utility of microscopy and molecular margin status. The logrank test was used to estimate one-year local failure-free survival, cause-specific survival, and overall survival rates (21). Gray’s method was also used to examine competing causes of failure, considering local failure, regional/distant failure, and death without recurrence as the types of failure (22).
RESULTS
Demographic, clinical and histopathologic features of the population
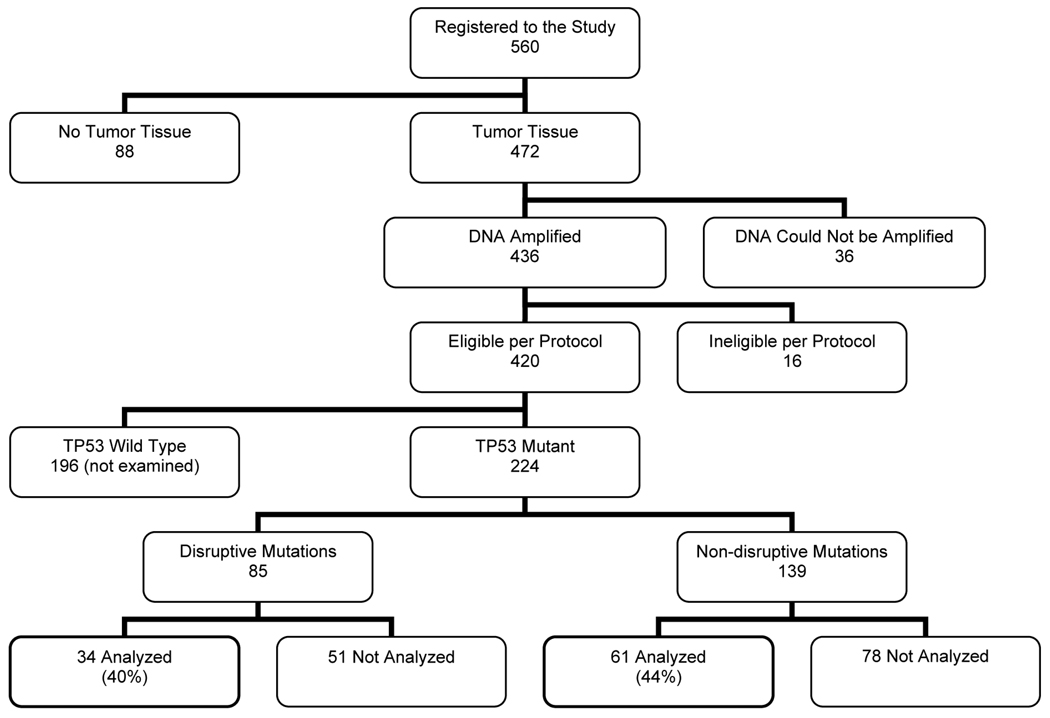
A cohort of 560 cases was enrolled in over 18 member institutions of ECOG and RTOG. As shown in Figure 1, 140 cases were eliminated for a variety of reasons (no tumor tissue available (n=88), DNA could not be amplified (n=36), ineligible per protocol (n=16)). TP53 mutation status was determined in the remaining 420 cases. 224 (53.3%) had mutation of TP53 and thus were potentially available for molecular margin analysis. We selected cases for development of the LigAmp approach that had a specific missense point mutation also found in at least one other case so as to maximize the utility of each plasmid control that was manufactured (Fig. 1 Supplementary). A total of 26 different TP53 mutations shared in at least two of 95 subjects have been analyzed and form the study cohort. 34 of these cases have a disruptive mutation resulting in amino acid substitution predicted to disrupt DNA binding (17). The remaining 61 subjects were found to have a non-disruptive (conservative) TP53 genetic alteration. Characteristics of the selected patients are shown in Table 1. The histologic status of submitted study margin tissue (harvested after clinical margins were judged to be free of tumor) was evaluated centrally (WHW) by light microscopy. Among 95 patients, there were 10 with tumor (severe dysplasia or invasive carcinoma) visible in at least one margin sample and 85 patients for whom all margin specimens were negative. The distribution of the number of slides/samples evaluated did not differ between these two groups (p=0.76) indicating that there was no improvement in detection by supplemental light microscopy with more margin samples available.
Figure 1.
Disposition of Cases.
Table 1.
Demographic and Clinical Characteristics
| Sex | Male | 65 | 68.4% |
| Female | 30 | 31.6% | |
| Race | White | 81 | 85.3% |
| Hispanic | 6 | 6.3% | |
| African-American | 8 | 8.4% | |
| Age | <55 | 29 | 30.5% |
| 55–64 | 27 | 28.4% | |
| >64 | 39 | 41.1% | |
| Cell Differentiation | Well differentiated | 20 | 22.2% |
| Moderately differentiated | 54 | 60.0% | |
| Poorly differentiated | 16 | 17.8% | |
| Undifferentiated | - | - | |
| Unknown | 5 | ||
| Primary Tumor Site | Oral Cavity | 56 | 58.9% |
| Oropharynx | 12 | 12.6% | |
| Hypopharynx | 8 | 8.4% | |
| Larynx | 13 | 13.7% | |
| Other | 5 | 5.3% | |
| >1 Primary | 1 | 1.1% | |
| Pathologic T Stage | pT1 | 24 | 25.5% |
| pT2 | 32 | 34.0% | |
| pT3 | 15 | 16.0% | |
| pT4 | 22 | 23.4% | |
| pTx/is | 1 | 1.1% | |
| missing | 1 | 1.1% | |
| Pathologic N Stage | pNo/Nx | 46 | 48.9% |
| pN+ | 48 | 51.1% | |
| missing | 1 | ||
| Status of Primary Tumor |
Eradicated but Local Recurrence |
9 | 9.7% |
| Residual Disease After Primary Surgery |
14 | 15.1% | |
| Untreated | 70 | 75.3% | |
| Unknown | 2 | ||
| Clinical TNM Stage | I | 8 | 9.9% |
| II | 15 | 18.5% | |
| III | 23 | 28.4% | |
| IV | 35 | 43.2% | |
| Unknown | 14 | ||
| Treatment | Surgery Alone | 33 | 34.7% |
| Surgery & Post-Operative Treatment |
45 | 47.4% | |
| Salvage Treatment | 17 | 17.9% | |
| Smoking History | Never Smoked | 16 | 17.4% |
| Pipe/Cigar Only | 4 | 4.3% | |
| <20 Pack Years | 9 | 9.8% | |
| 20–40 Pack Years | 26 | 28.3% | |
| ≥40 Pack Years | 37 | 40.2% | |
| Unknown | 3 | ||
| Average Alcohol Consumption |
<10 oz/wk | 52 | 59.8% |
| 10–20 oz/wk | 16 | 18.4% | |
| >20 oz/wk | 19 | 21.8% | |
| Unknown | 8 | ||
| Weight Loss in Previous 6 months |
<5% | 61 | 72.6% |
| 5–9.9% | 12 | 14.3% | |
| 10–19.9% | 8 | 9.5% | |
| ≥20% | 3 | 3.6% | |
| Unknown | 11 | ||
| Performance Status | 0 | 68 | 73.9% |
| 1 | 20 | 21.7% | |
| 2 | 4 | 4.3% | |
| Unknown | 3 | ||
| Did Primary Extend to Adjacent Structures? |
No | 53 | 56.4% |
| Yes | 41 | 43.6% | |
| Unknown | 1 | ||
| Clinical T Stage | T1 | 17 | 18.1% |
| T2 | 27 | 28.7% | |
| T3 | 27 | 28.7% | |
| T4 | 23 | 24.5% | |
| Unknown | 1 | ||
| Clinical N Stage | N0 | 56 | 60.2% |
| N1 | 17 | 18.3% | |
| N2 | 17 | 18.3% | |
| N3 | 3 | 3.2% | |
| Unknown | 2 | ||
| Mutation Status | Disruptive | 34 | 36% |
| Non-Disruptive | 61 | 64% | |
Table 2 shows mean proportions of TP53 mutant species in the margins, analyzed both as absolute levels (MRP) and relative to tumor concentration (MRT), among patients with and without local recurrence.
Table 2.
Margin Status by Local Recurrence – All Patients
| No Local Recurrence | Recurrence | ||||
|---|---|---|---|---|---|
| Number of Patients | 80 | 15 | |||
| Plasmid %< 0.15 | 49 | 61.3% | 5 | 33.3% | |
| ≥ 0.15 | 31 | 38.8% | 10 | 66.7% | |
| Tumor % < 0.5 | 54 | 67.5% | 6 | 40.0% | |
| ≥ 0.5 | 26 | 32.5% | 9 | 60.0% | |
| Positive Margins by Light Microscopy |
|||||
| No | 72 | 90.0% | 13 | 86.7% | |
| Yes | 8 | 10.0% | 2 | 13.3% | |
| Plasmid % | |||||
| mean | 5.77 | 1.28 | |||
| sd | 20.11 | 2.54 | |||
| median | 0.12 | 0.22 | |||
| range | 0–100 | 0.01–9.55 | |||
| IQR | 0.06–0.395 | 0.03–1.58 | |||
| Tumor % | |||||
| mean | 19.26 | 10.01 | |||
| sd | 95.48 | 24.15 | |||
| median | 0.26 | 0.75 | |||
| range | 0.01–793.53 | 0.03–79.00 | |||
| IQR | 0.12–1.05 | 0.15–2.28 | |||
| Disruptive Mutations | |||||
| No | 52 | 65.0% | 9 | 60.0% | |
| Yes | 28 | 35.0% | 6 | 40.0% | |
| Adjuvant Therapy | |||||
| No | 43 | 53.8% | 7 | 46.7% | |
| Yes | 37 | 46.3% | 8 | 53.3% | |
Evaluation of an optimal threshold for margin analysis
We used recursive partitioning to explore possible cutpoints for distinguishing patients with and without local recurrence, using both ratios described above. A survival model was used for the partitioning, with local failure as the endpoint of interest. Figure 2 shows the main partitions for the two ratios. Since the MRT was the model with the best discriminant power at the first split, this was selected for further exploration. A mutation threshold of 0.5% characterized this split. A mutation threshold of 0.15% was the optimal cutpoint for MRP.
Figure 2.
Recursive partitioning trees to determine optimal cutpoint for percentage of margin cells relative to tumor (left) and relative to plasmid (right). The height of the vertical lines is proportional to the error in the fit.
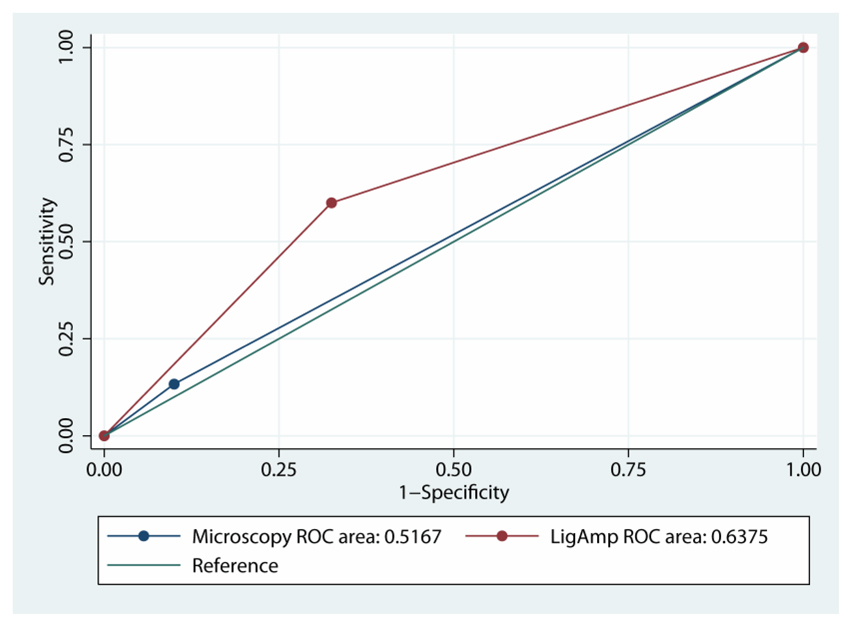
Figure 3 shows the receiver operator characteristic (ROC) curves for light microscopy of study margins and the LigAmp metric assessed at the 0.5% MRT cutpoint as predictors of local recurrence status. There is some evidence in this limited sample for the LigAmp evaluation to have greater area under the ROC curve than supplemental light microscopy (p=0.09).
Figure 3.
ROC (Receiver Operator Characteristic) curves for light microscopy and LigAmp analysis. The LigAmp method had a somewhat higher area under the ROC curve than light microscopy (p=0.09).
Relationships among histopathological margin status, molecular assessment and clinical outcome
Among the 95 patients analyzed by LigAmp, 15 experienced local recurrence. Table 2 shows the distribution of the metrics, the proportion of patients above and below each LigAmp cutpoint, the proportion with and without positive margins, and the proportion with and without disruptive mutations among patients who did and did not experience local recurrence. There was no association between margin status or mutation category and local recurrence.
Nine of 15 (60%) cases with documented local recurrence had MRT above the 0.5% threshold. These are cases in which molecular margin analysis was “true positive” indicating likely disease recurrence despite clear margins as judged by standard histopathologic evaluation at the treating institution. In two of these cases, tumor was also detected by light microscopic evaluation of the study (supplemental) margins submitted for molecular evaluation. Using the same threshold criteria, 26 of 80 cases that have not demonstrated disease recurrence were found to contain tumor cells by molecular analysis (32.5% false positive rate), and six cases have recurred in which LigAmp did not demonstrate molecular evidence of tumor cells (false negatives). Light microscopy evaluation of supplemental margins also found no evidence of tumor in any of these 6 cases.
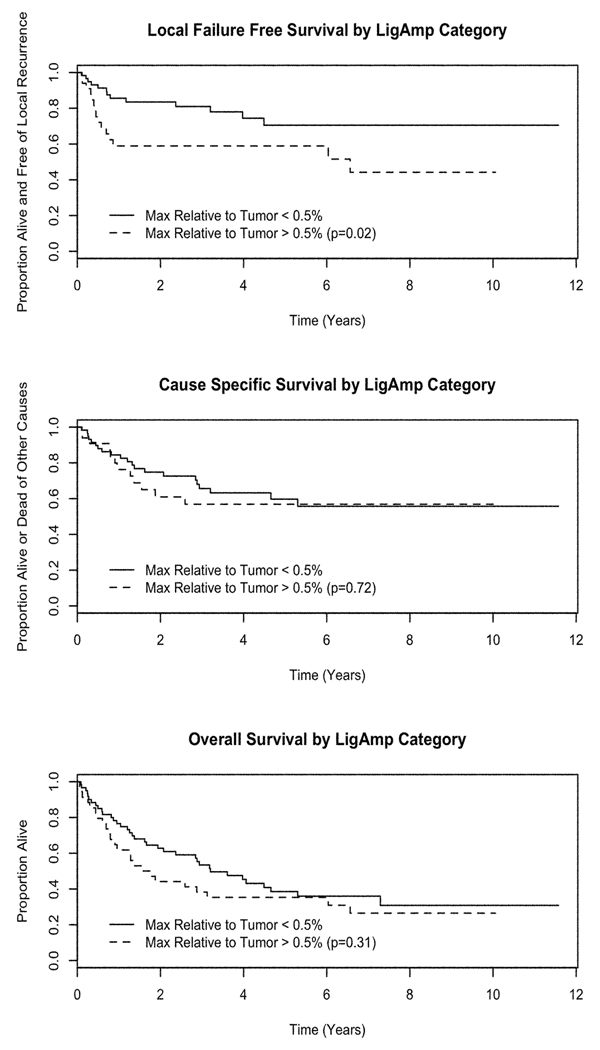
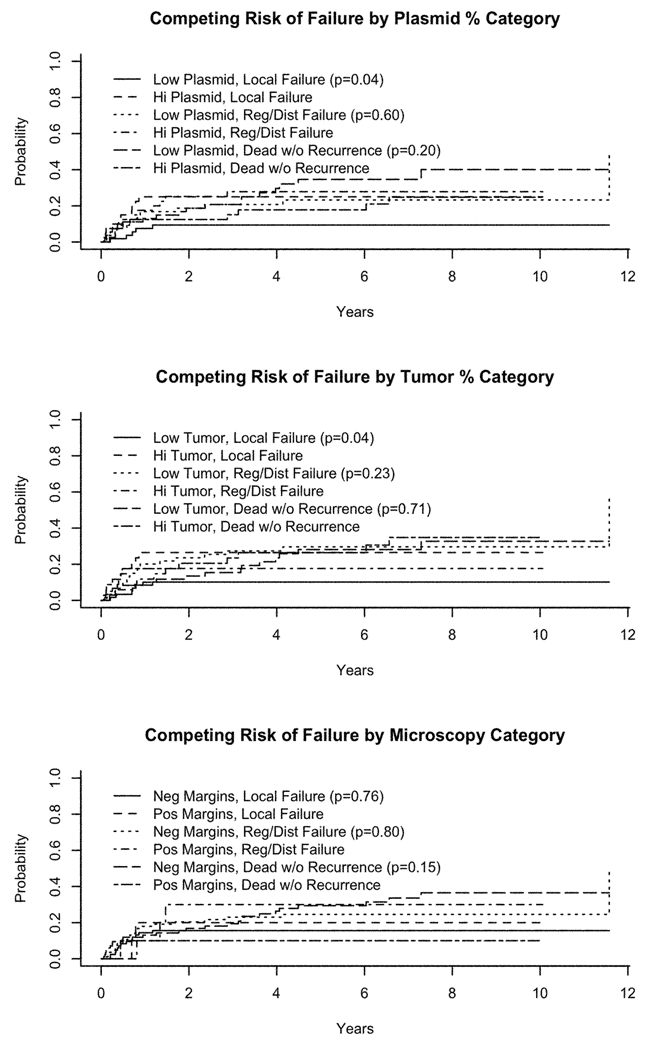
Median follow-up among patients still alive is 6.7 years. Table 3 provides estimates of two-year and five-year local failure-free survival (FFS), cause-specific survival (CSS), and overall survival (OS) by each of the predictive metrics. There was a statistically significant prolongation in local FFS for patients below the MRT cutoff compared to those above the cutoff (p=0.02). Since the cutpoints were optimized based on the presence of local recurrence, the local FFS metrics would be expected to perform well. While the p-value provides an estimate of the relative value of each metric, it does not represent the true Type I error for the test and should be interpreted with caution. No other statistically significant associations were identified. The analysis of competing causes of death and failure yielded no statistically significant differences by either light microscopy or LigAmp metrics (data not shown). It should be noted that these are exploratory analyses and statistical power is extremely limited.
Table 3.
Status Two and Five Years Post-Surgery
| Plasmid % | Tumor % | Margins by Light Microscopy | Type of Mutation | |||||
|---|---|---|---|---|---|---|---|---|
| All Negative | Any Positive | Conservative | Disruptive | |||||
| <0.15 | ≥0.15 | <0.5 | ≥0.5 | |||||
| Patients | 54 | 41 | 60 | 35 | 85 | 10 | 61 | 34 |
| Local Failure-free Survival Events | 13 | 15 | 13 | 15 | 25 | 3 | 15 | 13 |
| 2-Yr Local Failure-free Survival | 81.7% | 65.3% | 83.5% | 58.9% | 75.4% | 70.0% | 78.0% | 68.7% |
| 95% CI | 71.5 – 93.3% | 51.7 – 82.6% | 74.2 – 94.0% | 44.0 – 79.0% | 66.3 – 85.7% | 46.7 – 100.0% | 67.7 – 89.9% | 54.3 – 86.9% |
| 5-Yr Local Failure-free Survival | 68.4% | 65.3% | 70.5% | 58.9% | 66.3% | 70.0% | 68.2% | 64.1% |
| 95% CI | 54.8 – 85.4% | 51.7 – 82.6% | 57.4 – 86.7% | 44.9 – 79.0% | 55.3 – 79.4% | 46.7 – 100.0% | 55.3 – 84.1% | 48.9 – 84.1% |
| Logrank p | 0.13 | 0.02 | 0.80 | 0.18 | ||||
| Deaths from Head & Neck Cancer | 17 | 16 | 21 | 12 | 29 | 4 | 17 | 16 |
| 2-Yr Cancer-specific Survival | 75.7% | 62.3% | 74.8% | 60.9% | 69.4% | 77.8% | 77.1% | 58.6% |
| 95% CI | 64.5 – 88.7% | 47.8 – 81.0% | 64.2 – 87.2% | 45.1 – 82.3% | 59.6 – 80.7% | 54.9 – 100.0% | 66.4 – 89.5% | 43.6 – 78.7% |
| 5-Yr Cancer-specific Survival | 63.8% | 51.7% | 59.7% | 56.9% | 60.0% | 51.9% | 66.1% | 48.3% |
| 95% CI | 50.5 – 80.7% | 36.8 – 72.4% | 46.9 – 76.0% | 40.9 – 79.1% | 49.2 – 73.3% | 26.7 – 100.0% | 53.3 – 81.9% | 33.4 – 69.7% |
| Logrank p | 0.28 | 0.72 | 0.88 | 0.69 | ||||
| Deaths from Any Cause | 32 | 28 | 36 | 24 | 54 | 6 | 36 | 24 |
| 2-Yr Overall Survival | 62.6% | 47.2% | 62.8% | 44.2% | 55.5% | 60.0% | 59.4% | 50.0% |
| 95% CI | 50.8 – 77.0% | 33.9 – 65.6% | 51.6 – 76.4% | 30.3 – 64.4% | 45.7 – 67.3% | 36.2 – 99.5% | 48.1 – 73.4% | 35.7 – 70.0% |
| 5-Yr Overall Survival | 40.1% | 34.1% | 38.6% | 35.3% | 37.6% | 40.0% | 39.9% | 34.6% |
| 95% CI | 28.2 – 56.9% | 22.0 – 52.7% | 27.3 – 54.4% | 22.4 – 55.7% | 28.2 – 50.2% | 18.7 – 85.5% | 28.8 – 55.3% | 21.6 – 55.3% |
| Logrank p | 0.25 | 0.31 | 0.69 | 0.36 | ||||
DISCUSSION
Histopathological evaluation of surgical margins is the standard approach to assess the adequacy of surgical treatment. In light of a significant recurrence rate despite clear margins (1), several groups have investigated alternative approaches to detect occult tumor cells in surgical margins of HNSCC (14, 15).
Typically, a limited number of samples are submitted during surgery in order to evaluate resection margin status, both for immediate (frozen section) evaluation and for permanent (fixed tissue) histologic evaluation. The surgeon and pathologist select how many margins will be analyzed and from what locations based on the physical features of the tumor and resection site. In most HNSCC cases, 0–10 frozen section specimens are submitted, and 6–10 fixed tissue slides evaluated. In this study, all cases had been determined to have clear margins using these standard clinical procedures before additional margins were taken for molecular analysis. The process of margin evaluation by surgical pathology is limited both by the economy of sampling (number of slides evaluated), the skill and experience of the pathologist, and the sensitivity of light microscopy.
No method to detect TP53 alterations has proven to be amenable to wide-spread clinical use. Markers of high quality for molecular detection must be specific to fully transformed cancer cells and not merely indicative of a field effect of precancerous potential. As we have already suggested, immunohistochemical analysis of p53 expression in surgical margins (6) is limited in its ability to detect the presence of minimal residual cancer cells. Furthermore, IHC, like standard histopathologic evaluation of thin sections taken from margin samples analyzes only a small portion of the total perimeter around a tumor. So laboratory assays have been developed to explore other methods to detect the presence of tumor-specific molecular alterations (13, 14, 15, 23) indicative of occult cancer cells in surgical margins of HNSCC. Submission of the entire margin sample for DNA extraction allows for a rigorous evaluation of the entire margin volume.
The TP53 plaque assay used in our earliest study is a laborious method requiring 3 days and using radio-labelled oligonucleotides. Alternative PCR-based approaches have been described, with a similar detection limit of around 0.1% of mutant/wild-type species. However, in some of these studies, quantification of mutant cells has been performed using tumor DNA to generate the standard curve, leading to an inaccurate assessment of the mutant fraction present in the margin (tumor samples are not comprised of 100% cancer cells). In this study, we sought to overcome the limitations and disadvantages of these assays. The LigAmp assay (16) has been shown to be a reliable method for detecting single nucleotide variants in cancer. By using TP53 mutant plasmids to generate the standard curve we were able to derive an absolute quantification of mutant DNA in tumor and margins. Accurate quantification of mutation in primary tumors allows exclusion of cases with subclinical passenger mutations.
The aim of this study was to demonstrate the usefulness of LigAmp for the evaluation of TP53 mutation in surgical margins. To that end, we selected a subset of 95 of the 224 cases with mutation in order to maximize utility of effort to create plasmids for quantification control and optimize conditions for assaying each mutation. This subset was reflective of the entire cohort and the entire mutation group in all demographic and tumor-related parameters (supplemental table 2). Cases with TP53 mutation are more common in tumors of the oral cavity and less common in the oropharynx where HPV is an etiologic factor. Furthermore, this cohort allowed for the exploration of threshold cutpoints of clinically relevant mutant species detection setting the stage for a full scale evaluation of the entire cohort.
We evaluated all study margins by light microscopy to explore the relative sensitivity of molecular analysis to supplemental focused histologic evaluation. Light microscopy also served as a means of internal verification of molecular findings. Of note, the light microscopy was performed by an expert pathologist who was cognizant of the goal of identifying minute clusters of suspicious cells in these samples taken after the local institutional pathologic diagnosis had already deemed margins to be clear. The molecular approach performed better than supplemental histopatological evaluation in identifying cases with subclinical positive margins. LigAmp thresholds correctly identified 60% of all cases that went on to recur locally (9 of 15), while light microscopy confirmed the presence of visible tumor cells in only two of these cases. Both methods failed to predict recurrence in the other six local failure cases.
The rate of false positive and false negative molecular analyses was somewhat disappointing - particularly the latter, for which increased concern for recurrence and potential need for supplemental therapy would not have been predicted. There are several possible reasons that the LigAmp assay may have underestimated recurrence risk. The sample size is modest (n=95), yet three times larger than in our pilot study. The number of events (local recurrences) is low (n=15), perhaps due to improvement in loco-regional disease control with modern adjuvant therapy or rigorous surgical management in this prospective trial focused on adequacy of tumor extirpation. It is also possible that the sampling of margins failed to include critical deep tissue which, if positive, would have predicted some of the local recurrences. Finally, the threshold of 0.5% may be too high to include some true positive cases with tumor cells in margins.
A recent report from analysis of TP53 mutations in this cohort revealed a decrement in overall survival among patients whose tumor contained TP53 alterations predicted to disrupt DNA binding. Analysis of clinical endpoints in this subset of 95 patients failed to demonstrate any association between clinical outcome and type of p53 mutation. Use of post-operative adjuvant radiation also did not affect the rate of survival. Among those patients who did not have tumor recurrence, 46% had received postoperative radiation compared with 53% of those whose tumor recurred (Table 2).
Given the limited number of recurrence events and the availability of a TP53 mutation amenable to Ligamp in less than half of all cases, alternate markers must be utilized for the expanded evaluation of this cohort. Furthermore, LigAmp still requires several hours of laboratory work to evaluate a margin sample even when the tumor has been analyzed for mutation previously and a pertinent plasmid is on hand. Optimally, molecular evaluation of margins should be completed during the time frame of surgical resection and reconstruction in order to permit further resection of identified positive margins. Tumor-specific promoter hypermethylation has shown promise as a marker of rare tumor cells (24) and results applying this approach to molecular detection in our margin cohort are forthcoming. Data from the current study provides the first accurate quantification of tumor cells in recurrent cases, and this cohort of tumors and margins for which TP53 results are known will permit comparison of results using novel markers with that of TP53 analysis.
We conclude that TP53 LigAmp analysis represents a rapid, quantitative and reproducible method to investigate the clinical significance of detecting occult tumor cells in margins of HNSCC. Accurate prediction of recurrent cases may lead to an improvement in early detection of residual cancer and overall management of HNSCC patients.
STATEMENT OF TRANSLATIONAL RELEVANCE
Clear surgical resection margins represent one of the most important prognostic indicators for patients with squamous cell carcinoma of the head and neck (HNSCC). However, the local recurrence rate is still 10% to 30% among patients with microscopically clear margins, suggesting that occult tumor cells may be present surrounding resection boundaries. Better detection and subsequent removal or other treatment of occult disease in adjacent tissue may result in improved outcomes.
Supplementary Material
Supplementary Figure 1. pcDNA 3.1 Mutant TP53 plasmids were manufactured from Top Gene Technologies (Quebec Canada). pCR259 Mutant TP53 plasmids (blue codons) have been kindly donated from Dr. C. Ishioka. All plasmids have been used to generate standard curve in LigAmp Assay.
Supplementary Figure 2. LigAmp assay. In the Ligation Step two different oligonucleotides specifically designed for each single mutant are ligated to one other in presence of mutation. In the Q-PCR Step, amplification occurs only if ligated product has been generated in the first step.
Figure 4.
Kaplan-Meier plots of local failure-free survival, cancer specific survival, and overall survival by optimal cutpoint of LigAmp percentage of mutant species relative to tumor. Logrank p-values for differences in local failure-free, cancer specific and overall survival by cutpoint category are 0.02, 0.72, and 0.31, respectively. Because the cutpoint was selected on the basis of local failure, the p-value for local failure-free survival does not represent the probability of observing the results by chance alone and thus should be interpreted with caution.
Acknowledgments
Supported by NIH/NIDCR R01 DE013152
This study was coordinated by the Eastern Cooperative Oncology Group (Robert L. Comis, M.D., Chair) and supported in part by Public Health Service Grants CA23318, CA66636, CA21115, CA16116, CA27525 and from the National Cancer Institute, National Institutes of Health and the Department of Health and Human Services. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Cancer Institute.
REFERENCES
- 1.van Houten VM, Leemans CR, Kummer JA, Dijkstra J, Kuik DJ, van den Brekel MW, Snow GB, Brakenhoff RH. Molecular diagnosis of surgical margins and local recurrence in head and neck cancer patients: a prospective study. Clin Cancer Res. 2004 Jun 1;10(11):3614–3620. doi: 10.1158/1078-0432.CCR-03-0631. [DOI] [PubMed] [Google Scholar]
- 2.Leemans CR, Tiwari R, Nauta JJ, van der Waal I, Snow GB. Recurrence at the primary site in head and neck cancer and the significance of neck lymph node metastases as a prognostic factor. Cancer (Phila) 1994;73:187–190. doi: 10.1002/1097-0142(19940101)73:1<187::aid-cncr2820730132>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 3.Partridge M, Li SR, Pateromichelakis S, Francis R, Phillips E, Huang XH, Tesfa-Selase F, Langdon JD. Detection of minimal residual cancer to investigate why oral tumors recur despite seemingly adequate treatment. Clin Cancer Res. 2000 Jul;6(7):2718–2725. [PubMed] [Google Scholar]
- 4.Haque R, Contreras R, McNicoll MP, Eckberg EC, Petitti DB. Surgical margins and survival after head and neck cancer surgery. BMC Ear Nose Throat Disord. 2006 Feb 17;6:2 doi: 10.1186/1472-6815-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradley PJ, MacLennan K, Brakenhoff RH, Leemans CR. Status of primary tumour surgical margins in squamous head and neck cancer: prognostic implications. Curr Opin Otolaryngol Head Neck Surg. 2007 Apr;15(2):74–81. doi: 10.1097/MOO.0b013e328058670f. [DOI] [PubMed] [Google Scholar]
- 6.van Houten VM, Tabor MP, van den Brekel MW, Kummer JA, Denkers F, Dijkstra J, Leemans R, van der Waal I, Snow GB, Brakenhoff RH. Mutated p53 as a molecular marker for the diagnosis of head and neck cancer. J Pathol. 2002 Dec;198(4):476–486. doi: 10.1002/path.1242. [DOI] [PubMed] [Google Scholar]
- 7.Tunca B, Erisen L, Coskun H, Cecener G, Ozuysal S, Egeli U. P53 gene mutations in surgical margins and primary tumor tissues of patients with squamous cell carcinoma of the head and neck. Tumori. 2007 Mar–Apr;93(2):182–188. doi: 10.1177/030089160709300212. [DOI] [PubMed] [Google Scholar]
- 8.van Houten VM, Tabor MP, van den Brekel MW, Denkers F, Wishaupt RG, Kummer JA, Snow GB, Brakenhoff RH. Molecular assays for the diagnosis of minimal residual head-and-neck cancer: methods, reliability, pitfalls, and solutions. Clin Cancer Res. 2000 Oct;6(10):3803–3816. Review. [PubMed] [Google Scholar]
- 9.Taylor D, Koch WM, Zahurak M, Shah K, Sidransky D, Westra WH. Immunohistochemical detection of p53 protein accumulation in head and neck cancer: correlation with p53 gene alterations. Hum Pathol. 1999 Oct;30(10):1221–1225. doi: 10.1016/s0046-8177(99)90041-2. [DOI] [PubMed] [Google Scholar]
- 10.Sjögren S, Inganäs M, Norberg T, Lindgren A, Nordgren H, Holmberg L, Bergh J. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. J Natl Cancer Inst. 1996 Feb 21;88(3–4):173–182. doi: 10.1093/jnci/88.3-4.173. [DOI] [PubMed] [Google Scholar]
- 11.Whetsell LH, Ringer DP, Schaefer FV. Molecular approach to rapid assessment of p53 tumor suppressor mutations in esophageal tumors from stained histological slides. Diagn Mol Pathol. 1994 Jun;3(2):132–141. doi: 10.1097/00019606-199406000-00010. [DOI] [PubMed] [Google Scholar]
- 12.Vary CP, Carmody M, LeBlanc R, Hayes T, Rundell C, Keilson L. Allele-specific hybridization of lipoprotein lipase and factor-V Leiden missense mutations with direct label alkaline phosphatase-conjugated oligonucleotide probes. Genet Anal. 1996 Sep;13(3):59–65. doi: 10.1016/1050-3862(95)00149-2. [DOI] [PubMed] [Google Scholar]
- 13.Harden SV, Thomas DC, Benoit N, Minhas K, Westra WH, Califano JA, Koch W, Sidransky D. Real-time gap ligase chain reaction: a rapid semiquantitative assay for detecting p53 mutation at low levels in surgical margins and lymph nodes from resected lung and head and neck tumors. Clin Cancer Res. 2004 Apr 1;10(7):2379–2385. doi: 10.1158/1078-0432.ccr-03-0405. [DOI] [PubMed] [Google Scholar]
- 14.Huang X, Pateromichelakis S, Hills A, Sherriff M, Lyons A, Langdon J, Odell E, Morgan P, Harrison J, Partridge M. p53 mutations in deep tissues are more strongly associated with recurrence than mutation-positive mucosal margins. Clin Cancer Res. 2007 Oct 15;13(20):6099–6106. doi: 10.1158/1078-0432.CCR-07-1369. [DOI] [PubMed] [Google Scholar]
- 15.Brennan JA, Mao L, Hruban RH, Boyle JO, Eby YJ, Koch WM, Goodman SN, Sidransky D. Molecular assessment of histopathological staging in squamous-cell carcinoma of the head and neck. N Engl J Med. 1995 Feb 16;332(7):429–435. doi: 10.1056/NEJM199502163320704. [DOI] [PubMed] [Google Scholar]
- 16.Shi C, Eshleman SH, Jones D, Fukushima N, Hua L, Parker AR, Yeo CJ, Hruban RH, Goggins MG, Eshleman JR. LigAmp for sensitive detection of single-nucleotide differences. at Methods. 2004 Nov;1(2):141–147. doi: 10.1038/nmeth713. Epub 2004 Oct 21. [DOI] [PubMed] [Google Scholar]
- 17.Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, Ridge JA, Goodwin J, Kenady D, Saunders J, Westra W, Sidransky D, Koch WM. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007 Dec 20;357(25):2552–2561. doi: 10.1056/NEJMoa073770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox DR. Analysis of Binary Data. London, England: Methuen & Co, Ltd; 1970. [Google Scholar]
- 19.Wilcoxon F. Individual comparisons by ranking methods. Biometrics. 1945;1:80–83. [Google Scholar]
- Breiman L, Friedman J, Stone CJ, Olshen RA. Classification and Regression Trees. Pacific Grove, California: Wadsworth, Inc.; 1984. [Google Scholar]
- 21.Peto R, Peto J. Asymptotically efficient rank invariant test procedures. J R Stat Soc Ser A. 1972;135(2):185–206. [Google Scholar]
- 22.Gray RJ. A class of k-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16(3):1141–1154. [Google Scholar]
- 23.Zhou S, Kachhap S, Sun W, Wu G, Chuang A, Poeta L, Grumbine L, Mithani SK, Chatterjee A, Koch W, Westra WH, Maitra A, Glazer C, Carducci M, Sidransky D, McFate T, Verma A, Califano JA. Frequency and phenotypic implications of mitochondrial DNA mutations in human squamous cell cancers of the head and neck. Proc Natl Acad Sci U S A. 2007 May 1;104(18):7540–7545. doi: 10.1073/pnas.0610818104. Epub 2007 Apr 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carvalho AL, Jeronimo C, Kim MM, Henrique R, Zhang Z, Hoque MO, Chang S, Brait M, Nayak CS, Jiang WW, Claybourne Q, Tokumaru Y, Lee J, Goldenberg D, Garrett-Mayer E, Goodman S, Moon CS, Koch W, Westra WH, Sidransky D, Califano JA. Evaluation of promoter hypermethylation detection in body fluids as a screening/diagnosis tool for head and neck squamous cell carcinoma. Clin Cancer Res. 2008 January;14(1):97–107. doi: 10.1158/1078-0432.CCR-07-0722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. pcDNA 3.1 Mutant TP53 plasmids were manufactured from Top Gene Technologies (Quebec Canada). pCR259 Mutant TP53 plasmids (blue codons) have been kindly donated from Dr. C. Ishioka. All plasmids have been used to generate standard curve in LigAmp Assay.
Supplementary Figure 2. LigAmp assay. In the Ligation Step two different oligonucleotides specifically designed for each single mutant are ligated to one other in presence of mutation. In the Q-PCR Step, amplification occurs only if ligated product has been generated in the first step.