Abstract
Optical biosensors for short peptide motifs, an important class of biomarkers, have been developed based on “affinity clamps”, a new class of recombinant affinity reagents. Affinity clamps are engineered by linking a peptide-binding domain and an antibody mimic domain based on the fibronectin type III scaffold, followed by optimizing the interface between the two. This two-domain architecture allows for the design of allosteric coupling of peptide binding to fluorescence energy transfer between two fluorescent proteins attached to the affinity clamp. Coupled with high affinity and specificity of the underlying affinity clamps and rationally designed mutants with different sensitivity, peptide concentrations in crude cell lysate were determined with a low nanomolar detection limit and over three orders of magnitude. Because diverse affinity clamps can be engineered, our strategy provides a general platform to generate a repertoire of genetically encoded, label-free sensors for peptide motifs.
Biosensors exploiting molecular recognition by proteins and nucleic acids represent an important strategy for specific and facile detection of a target molecule in a complex solution, with many applications in chemistry, biology, medicine and biotechnology. A biosensor requires a recognition module, a signal output module and coupling between the two. Most protein-based biosensors designed to date utilize natural protein as the recognition module. The limited repertoire of natural binding proteins is an impediment to the generation of biosensors for a broader range of target molecules. Hellinga et al. designed new ligand binding functions into a protein involved in sugar sensing, demonstrating the possibility of significantly expanding the variety of biosensors (1). However, their system based on bacterial sugar-binding proteins appears to be limited to recognizing small molecules. Bacterial periplasmic peptide-binding proteins serve as sensors for peptide-based signaling (2). They may be a promising platform for developing peptide sensors, but to date no such sensors have been reported (3, 4).
Recent advances have enabled one to produce recombinant antibody fragments such as the Fab and single-chain Fv for diverse types of antigens (5). Similarly, binding proteins based on non-antibody scaffolds show considerable promise as alternatives to antibodies (6). Although these engineered binding proteins can serve as recognition modules, there is no general strategy for converting them into sensors. The antibody and other commonly used scaffolds are highly rigid, and consequently they do not undergo large conformational changes upon target binding. These characteristics make them ill suited as platforms for sensor design through conformational coupling and consequently necessitates laborious development (7, 8). Antibody-based sandwich ELISA (enzyme-linked immunosorbent assay) is widely used for detecting and quantifying a target molecule, but it requires a pair of antibodies that can simultaneously bind to a given target, a requirement that is difficult to meet particularly for a small target (9).
Short peptide motifs containing a chemical signature (e.g. the free C-terminus and phosphorylation) are important biomarkers involved in signal transduction and protein assemblies (10, 11). Such motifs are often recognized by the so-called modular interaction domains, and strategies have been developed to convert modular interaction domains into biosensors by coupling binding and folding (12, 13). While these strategies can be generalized, the repertoire of natural peptide-binding domains is limited, and they usually have low levels of affinity and specificity, limiting the utility of biosensors constructed in this manner. Furthermore, these design strategies require a sensor to be marginally stable so that its folding can be triggered by ligand binding, which reduces its sensitivity and in vivo stability. Also, while one could envision adopting existing sensor technologies such as nucleotide aptamers, molecular beacons and bacterial sensor proteins (1, 14, 15), it is not clear whether one can engineer high levels of binding specificity for peptide motifs into these systems. Together there are urgent needs for a technology to generate highly specific and sensitive sensors for peptide motifs.
Here we reported an integrated approach to generate high-performance affinity reagents for short peptide motifs and then convert them into label-free optical biosensors. Our strategy is based on a new type of binding proteins termed “affinity clamps” that have high affinity and high specificity to a short peptide motif (16). We found that target binding induces a large conformational change of an affinity clamp, which was exploited to construct genetically encoded, ratiometric sensors.
RESULTS AND DISCUSSION
Ligand-induced conformational change of affinity clamps
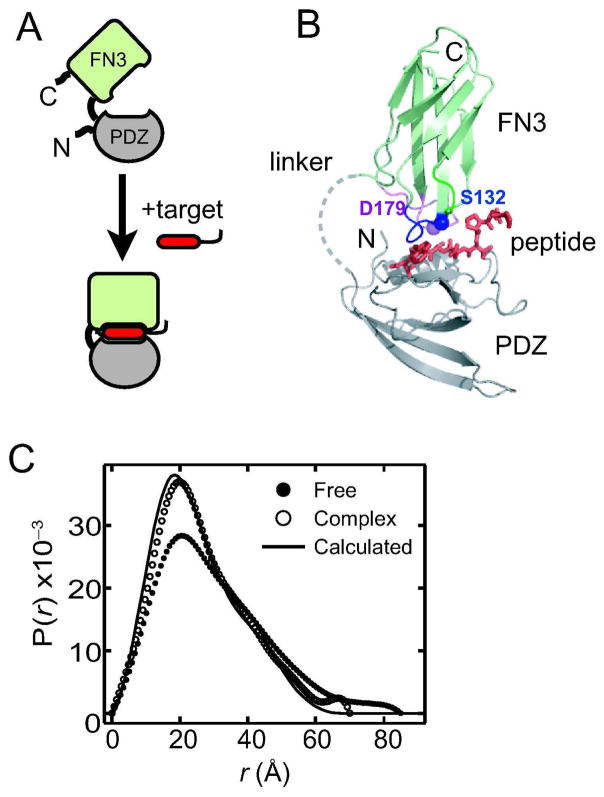
We have recently established a new protein-engineering strategy termed “directed domain-interface evolution” to generate affinity reagents to short peptide motifs with high affinity and exquisite specificity (16, 17). This strategy first genetically attaches an inert protein scaffold (“enhancer” domain) to a natural peptide-binding domain (“capture” domain). Subsequently, a combinatorial phage-display library is constructed in which portions of the enhancer domain are diversified. High-affinity variants with an optimized enhancer domain are then selected. Using the fibronectin type III (FN3) domain as the enhancer, we have dramatically enhanced the binding affinity and specificity of the erbin PDZ domain (18), an interaction domain with weak affinity to the C-terminal extreme of target proteins (Figure 1A and B). We have termed these two-domain proteins affinity clamps. The architecture of affinity clamps is distinctly different from those of common affinity reagents (e.g. antibodies) in that the target recognition site is constructed at the interface between two domains that are loosely connected with a short linker. Consequently, target binding may induce a large change in the relative orientation of the two domains (Figure 1A), which could be allosterically coupled to a read-out mechanism for a biosensor.
Figure 1.
The architecture and ligand-induced conformational change of an affinity clamp. (A) Schematic drawing of an affinity clamp. PDZ corresponds to the capture domain and FN3 to the enhancer domain. The ePDZ affinity clamps were developed by optimizing the binding interface of the FN3 domain through phage-display library selection. Ligand-induced conformational change of the affinity clamp is also indicated. (B) The crystal structure of the ePDZ-a affinity clamp in complex with the ARVCF peptide (16). The PDZ domain, FN3 domain and the peptide are shown in gray, pale green and red, respectively. The linker connecting the PDZ and FN3 domains was disordered. The mutation sites are also indicated. (C) The P(r) curves derived of free ePDZ-a and of the ePDZ-a/peptide complex. A P(r) curve is the Fourier transformation of the SAXS data and indicates the distribution of mass-weighted atom-atom distances. The line shows a prediction from the crystal structure of the ePDZ-a/peptide complex.
Small-angle X-ray scattering (SAXS) analysis of an affinity clamp, termed ePDZ-a, revealed large conformational change induced by ligand binding. This affinity clamp binds to the C-terminal peptide motif, PQPVDSWV-COOH, derived from a human protein, ARVCF (Armadillo repeat gene deleted in Velo-cardio-facial syndrome) (16, 18). Upon an addition of a ten-residue peptide that contains the binding motif, GGPQPVDSWV, we observed a significant change in the SAXS profile (Figure 1C), suggesting a large change in inter-domain orientation of the affinity clamp.
Sensor design
To couple the observed conformational change with optical read out, the affinity clamp was genetically fused to a pair of FRET-optimized fluorescent proteins (FPs), YPet and CyPet (19), with YPet at the N-terminus of ePDZ-a and CyPet at the C-terminus (Figure 2A). This FP pair exhibits a large FRET change upon dissociation, because surface mutations in them increase their tendency to heterodimerize when they are linked within a single polypeptide and achieve a high FRET efficiency (20–22). A similar set of “sticky” FPs has been used to develop a zinc sensor, “eCALWY-1”, in which zinc binding to a zinc-recognition module disrupts heterodimerization of the FP pair (23). We envisioned that the conformational change of the affinity clamp could be similarly exploited to develop FRET sensors.
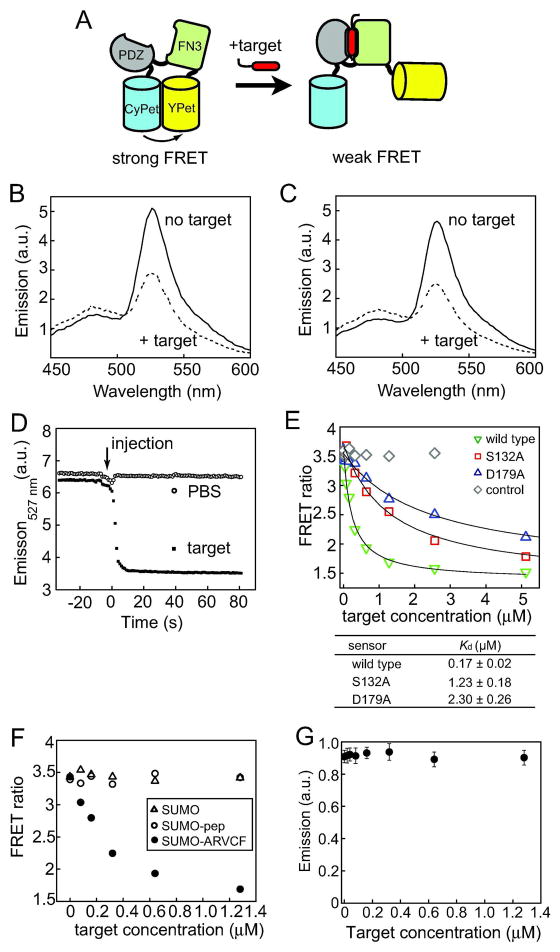
Figure 2.
Clamp sensor design and characterization. (A) Schematic drawing of the design principle. The closure of the affinity clamp alters interactions between the two fluorescent proteins, CyPet and YPet, affecting the FRET efficiency. (B) Fluorescence emission spectra of the clamp sensor in PBS in the presence and absence of the target peptide (1.2 μM) with excitation at 415 nm. (C) Fluorescence emission spectra of the clamp sensor in the presence and absence of the SUMO-ARVCF fusion protein (1.2 μM). (D) Response time course of the clamp sensor. Fluorescence emission intensity at 527 nm with excitation at 415 nm is plotted as a function of time, as a saturating concentration of peptide was injected. The intensity was monitored every second. (E) FRET ratios (emission at 527 nm over emission at 475 nm) of clamp sensors in PBS plotted as a function of SUMO-ARVCF concentration. Responses of three different sensors are shown. “Control” is a nonfunctional sensor containing an FN3 domain that does not bind to the target. The data points at 10 and 20 μM are omitted for clarity. The curves show the best fit of a 1:1 binding model. The Kd values are also shown. (F) Responses of the wild-type clamp sensor to SUMO-ARVCF, SUMO with no attached peptide and a SUMO fusion with an unrelated peptide (“SUMO-pep”; GGDVHVNATYVNVKSVA). The FRET ratios are plotted as a function of ligand concentration. (G) Fluorescence emission intensity at 527 nm of YPet with excitation at 514 nm plotted as a function of SUMO-ARVCF concentration, showing that the quantum yield of YPet is not affected by peptide binding.
The resulting fusion protein YPet–ePDZ-a–CyPet was produced in E. coli. In the absence of an added peptide, the fluorescence emission spectrum of the fusion protein with excitation of CyPet at 415 nm exhibited an intense peak at 527 nm (Figure 2B), similar to the spectrum of CyPet and YPet linked with a short peptide (19). As the target peptide was added, the spectrum of the fusion protein showed large changes. The overall decrease in the intensity of the YPet emission peak at 527 nm was ~50% corresponding to a FRET ratio change of ~250% (Figure 2B), a magnitude similar to those observed for other FP-based biosensors (23–25). Similar results were observed when the target peptide was attached to a small protein, yeast SUMO via a short linker (referred to as SUMO-ARVCF hereafter; Figure 2C), indicating that the sensor can recognize the ARVCF peptide motif attached to a protein equally well. Because of the ease of production and quantification of SUMO-ARVCF fusion protein, we used SUMO-ARVCF as a target for subsequent experiments. The sensor showed rapid response to a change in peptide concentration with an apparent relaxation time of a few seconds (Figure 2D). The titration data (Figure 2E) gave a Kd value of 170 ± 20 nM, ~3 fold larger than the Kd of isolated ePDZ-a (56 ±5 nM). As expected, a control construct, with an FN3 domain with no appreciable binding to the target peptide, did not change the emission upon target addition (Figure 2E). Similarly, SUMO without an attached peptide or SUMO fused with an unrelated peptide had no significant effect on the FRET ratio of the wild-type sensor (Figure 2F). These results validate our strategy in converting an affinity reagent into a biosensor. We name this class of biosensors “clamp sensors”.
Several lines of evidence support a model for the mechanism of the clamp sensor (Figure 2A) in which conformational changes of the affinity clamp disrupt optimal dimerization of CyPet and YPet necessary for efficient FRET. Our sensor exhibited a decrease, rather than increase, in YPet emission upon peptide binding. Peptide addition did not affect the YPet emission when YPet was directly excited (Figure 2G), indicating that the peptide binding to the affinity clamp did not affect the quantum yield of YPet. The Kd of the clamp sensor was increased relative to that of the underlying affinity clamp, similar to a ten-fold Kd increase observed for the eCALWY-1 zinc sensor designed using the same principle (23). Together, these results strongly support that the observed changes in YPet emission is due to changes in FRET efficiency between CyPet and YPet. The relatively small changes in the CyPet emission at 475 nm appear to be characteristic of sensors constructed with the CyPet-YPet pair, as observed previously (19). It is notable that a large level of emission change was achieved without optimizing the linkers between the affinity clamp and the fluorescent proteins, consistent with a view that disruption of a sticky FP pair may be an efficient strategy for developing FRET sensors (22, 23).
Rational modulation of clamp sensor sensitivity
A series of sensors with different sensitivity would expand the detection range and thus can increase their utility. Based on the crystal structure of ePDZ-a in complex with the ARVCF peptide (16), we identified S132 and D179 located in the loops of FN3 that form the peptide-binding interface as potential sites for mutation (Figure 1B). S132 forms a hydrogen bond with the target peptide. D179 forms multiple hydrogen bonds with other FN3 residues, which appeared to stabilize the loop conformation for binding. The S132A and D179A mutants of the clamp sensor respectively showed 7.2 and 13.5-fold reduced affinity relative to the wild-type sensor (Figure 2E). Importantly, they maintained the sensor characteristics in terms of the FRET ratios in the absence of the target and the total change upon saturation estimated from curve fitting (Figure 2E). Thus, we have successfully altered the sensitivity of the clamp sensor without diminishing its signal transduction property.
Applications of the clamp sensors
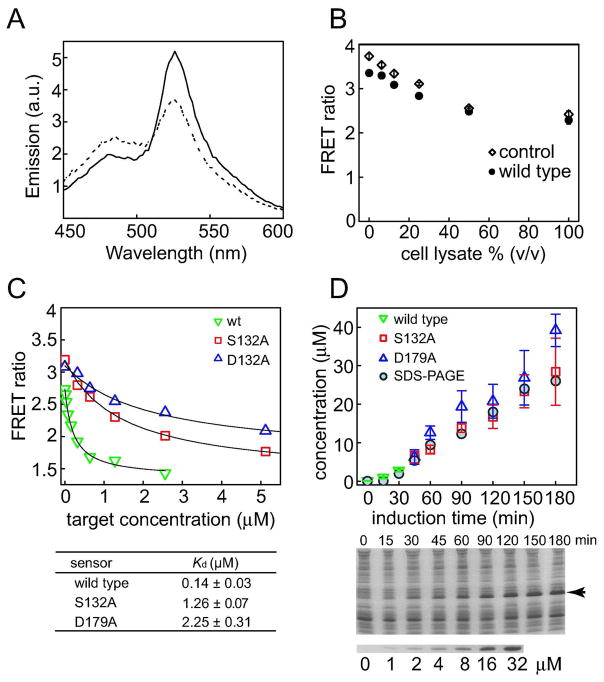
The utility of these clamp sensors was further illustrated by determining the concentration of the target peptide motif in crude cell lysate. The responses of the clamp sensor in cell lysate were similar to those observed in PBS, indicating that the clamp sensor was not significantly influenced by other molecules in cell lysate, as expected from exquisite specificity of the affinity clamp used (Figure 3A and C). These results further support high specificity of the affinity clamp used. We noted that the absolute values for the FRET ratio were different in cell lysate from those in PBS (Figure 2E). This difference was caused by the cell lysate altering the FRET profile between YPet and CyPet, probably by affecting their association (Figure 3B). However, this effect can easily be accounted for by establishing a calibration curve in the presence of cell lysate. Then, we used the clamp sensors to determine the concentration of SUMO-ARVCF expressed in E. coli at different time points after induction of protein expression. The protein concentrations determined with the clamp sensors agreed well with those estimated from SDS-PAGE (Figure 3C). These results demonstrate that the clamp sensor provides a robust and highly sensitive method to measure the absolute protein concentrations in a complex mixture.
Figure 3.
Peptide quantification in cell lysate using clamp sensors. (A) Fluorescence emission spectra of the clamp sensor in crude cell lysate in the presence and absence of SUMO-ARVCF (2 μM). (B) Effects of cell lysate on the FRET ratio of the clamp sensor. The FRET ratios of the wild-type sensor and a negative control sensor (as described in Figure 2E) are plotted versus the concentration of cell lysate. Measurements were taken in the absence of a target peptide. (C) FRET signals of clamp sensors in cell lysate plotted as a function of SUMO-ARVCF concentrations. (D) Measurements of SUMO-ARVCF expression in E. coli cell lysates with the clamp sensors. The concentrations of SUMO-ARVCF at different time points after the initiation of expression were determined. The protein levels were also estimated from SDS-PAGE (middle panel; the arrow indicates the band for SUMO-ARVCF) of cell lysates using SUMO-ARVCF of known concentrations as standards (lower panel).
In conclusion, we have described a rational strategy to convert affinity clamps into label-free, ratiometric fluorescence sensors for peptide biomarkers. Affinity clamps with different binding properties can be generated by replacing and/or mutating the capture domain and the overall architecture of affinity clamps is conserved regardless of their target. Therefore, we speculate that we can establish an integrated pipeline to simultaneously generate high-quality affinity reagents and label-free biosensors for diverse peptide motifs, which would make large impacts on cell biology, biotechnology and medicine. Furthermore, because clamp sensors can be genetically encoded and expressed in the functional form in cells, we speculate that they could be used to monitor real-time changes in biomarker concentration and localization in living cells.
METHODS
Protein Expression and Purification
The ePDZ-a and SUMO-ARVCF proteins were prepared as described previously (16). The amino acid sequence for SUMO-ARVCF is: mkhhhhhhssdykddddkgenlyfqg-SDSEVNQEAKPEVKPEVKPETHINLKVSDGSSEIFFKIKKTTPLRRLMEAFAKRQG KEMDSLRFLYDGIRIQADQTPEDLDMEDNDIIEAHREQIGG-GGPQPVDSWV, where the segment in lower case encodes His6 and FLAG tags, and the ARVCF peptide is underlined. Its molecular weight is 15.3 kDa. BL21(DE3) cell lysates were prepared by treating cell suspension with 0.5mg/ml lysozyme containing 1 mM PMSF in 50 mM Tris-HCl buffer (pH 8) followed by sonication. Lysates were cleared with centrifugation and protein concentration was estimated with SDS-PAGE using PhastSystem (Amersham) and CBB staining. The lysates were diluted 10–15x in PBS prior to fluorescence measurement.
Small Angle X-Ray Scattering
Data were collected at the BioCAT beamline at the Advanced Photon Source, Argonne National Laboratory. Samples were prepared at a final protein concentration of 1 mg/ml in TBS pH 7.5. For complex formation, the peptide was added at 1.2:1 ratio to the protein. Buffer blanks and samples were flowed through a 1.5 mm capillary at a rate of 2 μL/s and 15 exposures of ~ 1 s duration were collected. The exposures were averaged, buffer blanks subtracted, and Guinier analysis performed using IGOR Pro (Wave Metrics). I(Q) data were converted to P(r) data using GNOM (26). P(r) data were normalized to an area proportional to (molecular weight)2.
Clamp Sensor Construction and Purification
The vectors for CyPet and YPet were made available by Dr. P. Daugherty through Addgene (19). The CyPet gene was cloned between the NheI and BamHI sites of pET24a (Novagen) and the Ypet gene was cloned between the EcoRI and XhoI sites of the resulting plasmid to yield a plasmid, pCypetYpet. The ePDZ-a gene was then inserted between the BamHI and XhoI sites of pCypetYpet. There was no linker between CyPet and ePDZ-a, and a four-residue linker (GGLE) was used between ePDZ-a and YPet. The CyPet-ePDZ-a-YPet fusion gene was expressed in BL21(DE3) cells and purified using Ni affinity chromatography. Mutant sensors were constructed using Kunkel mutagenesis (27).
Fluorescence Measurements
The clamp sensor was used at 140 nM unless otherwise noted. For titration, samples were placed in a black flat-bottom 96-well plate (Corning COSTAR), degassed and incubated at 25°C for 30 minutes in the dark prior to measurement in using a Tecan Safire2 plate reader. An excitation wavelength of 415 nm was used unless otherwise stated. The Kd was determined by fitting a 1:1 binding model to the emission ratio (emission at 527 nm/emission at 475 nm) as a function of peptide concentration:
where F is the observed fluorescence intensity, F0 is the fluorescence intensity in the absence of a target peptide, ΔF is the change in fluorescence upon sensor saturation, S0 is the total sensor concentration, and L0 is the total target concentration.
To determine the SUMO-ARVCF concentration in crude lysates, a sensor calibration curve was made using cell lysates containing known concentrations of SUMO-ARVCF. The intensities of protein bands in SDS-PAGE were analyzed using ImageJ (http://rsbweb.nih.gov/ij/).
The relaxation time of the clamp sensor was measured using a Aminco-Bowman AB-2 fluorometer with excitation at 415 nm and emission at 527 nm with one second interval. To a cuvette containing 0.23 μM sensor in PBS, SUMO-ARVCF was manually injected at a final concentration of 2 μM) while the solution was vigorously mixed with a magnetic stirrer.
Acknowledgments
We thank T. Sosnick and D. Strickland for SAXS analysis and P. Daugherty for the CyPet and YPet genes. This work was supported by the National Institutes of Health grants R21-CA132700, R21-DA025725 and R01-GM090324 to SK.
References
- 1.Looger LL, Dwyer MA, Smith JJ, Hellinga HW. Computational design of receptor and sensor proteins with novel functions. Nature. 2003;423:185–190. doi: 10.1038/nature01556. [DOI] [PubMed] [Google Scholar]
- 2.Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci U S A. 1997;94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tame JR, Murshudov GN, Dodson EJ, Neil TK, Dodson GG, Higgins CF, Wilkinson AJ. The structural basis of sequence-independent peptide binding by OppA protein. Science. 1994;264:1578–1581. doi: 10.1126/science.8202710. [DOI] [PubMed] [Google Scholar]
- 4.Levdikov VM, Blagova EV, Brannigan JA, Wright L, Vagin AA, Wilkinson AJ. The structure of the oligopeptide-binding protein, AppA, from Bacillus subtilis in complex with a nonapeptide. J Mol Biol. 2005;345:879–892. doi: 10.1016/j.jmb.2004.10.089. [DOI] [PubMed] [Google Scholar]
- 5.Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J Immunol Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Binz HK, Amstutz P, Pluckthun A. Engineering novel binding proteins from nonimmunoglobulin domains. Nat Biotechnol. 2005;23:1257–1268. doi: 10.1038/nbt1127. [DOI] [PubMed] [Google Scholar]
- 7.Hudson PJ, Souriau C. Engineered antibodies. Nat Med. 2003;9:129–134. doi: 10.1038/nm0103-129. [DOI] [PubMed] [Google Scholar]
- 8.Renard M, Belkadi L, Hugo N, England P, Altschuh D, Bedouelle H. Knowledge-based design of reagentless fluorescent biosensors from recombinant antibodies. J Mol Biol. 2002;318:429–442. doi: 10.1016/S0022-2836(02)00023-2. [DOI] [PubMed] [Google Scholar]
- 9.Hamaguchi Y, Kato K, Fukui H, Shirakawa I, Okawa S. Enzyme-linked sandwich immunoassay of macromolecular antigens using the rabbit antibody-coupled glass rod as a solid phase. Eur J Biochem. 1976;71:459–467. doi: 10.1111/j.1432-1033.1976.tb11134.x. [DOI] [PubMed] [Google Scholar]
- 10.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. doi: 10.1146/annurev.biochem.75.103004.142710. [DOI] [PubMed] [Google Scholar]
- 12.Kohn JE, Plaxco KW. Engineering a signal transduction mechanism for protein-based biosensors. Proc Natl Acad Sci U S A. 2005;102:10841–10845. doi: 10.1073/pnas.0503055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stratton MM, Mitrea DM, Loh SN. A Ca2+-sensing molecular switch based on alternate frame protein folding. ACS Chem Biol. 2008;3:723–732. doi: 10.1021/cb800177f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 15.Nutiu R, Li Y. Aptamers with fluorescence-signaling properties. Methods. 2005;37:16–25. doi: 10.1016/j.ymeth.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Huang J, Koide A, Makabe K, Koide S. Design of protein function leaps by directed domain interface evolution. Proc Natl Acad Sci U S A. 2008;105:6578–6583. doi: 10.1073/pnas.0801097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang J, Makabe K, Biancalana M, Koide A, Koide S. Structural basis for exquisite specificity of affinity clamps, synthetic binding proteins generated through directed domain-interface evolution. J Mol Biol. 2009;392:1221–1231. doi: 10.1016/j.jmb.2009.07.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laura RP, Witt AS, Held HA, Gerstner R, Deshayes K, Koehler MF, Kosik KS, Sidhu SS, Lasky LA. The Erbin PDZ domain binds with high affinity and specificity to the carboxyl termini of delta-catenin and ARVCF. J Biol Chem. 2002;277:12906–12914. doi: 10.1074/jbc.M200818200. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 20.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinkenborg JL, Evers TH, Reulen SW, Meijer EW, Merkx M. Enhanced sensitivity of FRET-based protease sensors by redesign of the GFP dimerization interface. Chembiochem. 2007;8:1119–1121. doi: 10.1002/cbic.200700109. [DOI] [PubMed] [Google Scholar]
- 22.Campbell RE. Fluorescent-Protein-Based Biosensors: Modulation of Energy Transfer as a Design Principle. Anal Chem. 2009 doi: 10.1021/ac802613w. [DOI] [PubMed] [Google Scholar]
- 23.Vinkenborg JL, Nicolson TJ, Bellomo EA, Koay MS, Rutter GA, Merkx M. Genetically encoded FRET sensors to monitor intracellular Zn2+ homeostasis. Nat Methods. 2009;6:737–740. doi: 10.1038/nmeth.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 25.Awais M, Sato M, Sasaki K, Umezawa Y. A genetically encoded fluorescent indicator capable of discriminating estrogen agonists from antagonists in living cells. Anal Chem. 2004;76:2181–2186. doi: 10.1021/ac030410g. [DOI] [PubMed] [Google Scholar]
- 26.Semenyuk AV, Svergun DI. GNOM - a program package for small-angle scattering data processing. J Appl Cryst. 1991;24:537–540. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-directed mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]