Abstract
We describe a novel method for producing homogeneous eukaryotic N-glycoproteins. The method involves the engineering and functional transfer of the C. jejuni glycosylation machinery in E. coli to express glycosylated proteins with the key GlcNAc-Asn linkage. The bacterial glycans were then trimmed and remodeled in vitro by enzymatic transglycosylation to fulfill a eukaryotic N-glycosylation. It provides a potentially general platform for producing eukaryotic N-glycoproteins.
N-linked glycosylation of proteins is one of the most prevalent posttranslational modifications in eukaryotes. The attached N-glycans are responsible for a number of important biological recognition processes such as intracellular sorting, cell adhesion, host-pathogen interaction, and immune response 1,2. Compelling evidence has shown that subtle changes in N-glycans confer significantly different functions to glycoproteins. However, a major challenge in defining the roles of glycans in biological processes comes from the remarkable structural micro-heterogeneity of natural and recombinant glycoproteins. The urgent need of homogeneous materials for functional studies and biomedical applications has stimulated a great interest in exploring new methods for making homogeneous glycoproteins. Major advances include: total or semi-chemical synthesis with native chemical ligation; site-directed chemo-selective glycosylation of proteins; engineering of glycosylation pathways in yeast Pichia pastoris; and convergent chemoenzymatic synthesis using enzymes for sugar chain elongation 3-5. While N-glycosylation was once considered to be restricted to eukaryotes, recent discovery of a protein N-glycosylation machinery in Campylobacter jejuni 6,7 and its successful functional transfer into Escherichia coli have raised an exciting opportunity to produce recombinant N-glycoproteins in bacteria 8,9. Nevertheless, the attached N-glycan, a unique heptasaccharide GalNAc-α1,4-GalNAc-α1,4-[Glc-β1,3-]GalNAc-α1,4-GalNAc-α1,4-GalNAc-α1,3-Bac-β1,N-Asn 7, is completely different from the eukaryotic N-glycans found in humans and other mammals. Moreover, the bacterial N-glycan is immunogenic, which limits its potential biomedical applications, and the sugar chain is linked to the asparagine (Asn) by an unusual deoxysugar, bacillosamine. We report here a new method for producing homogeneous glycoproteins carrying natural eukaryotic N-glycans, which combines bacterial glycoprotein expression with in vitro enzymatic glycosylation modification (Fig. 1). The method involves novel glycosylation pathway engineering in E. coli to introduce the key GlcNAc-Asn linkage found in eukaryotic N-glycoproteins, followed by glycan trimming and extension using the highly flexible in vitro chemoenzymatic approach that we have recently developed 10-15.
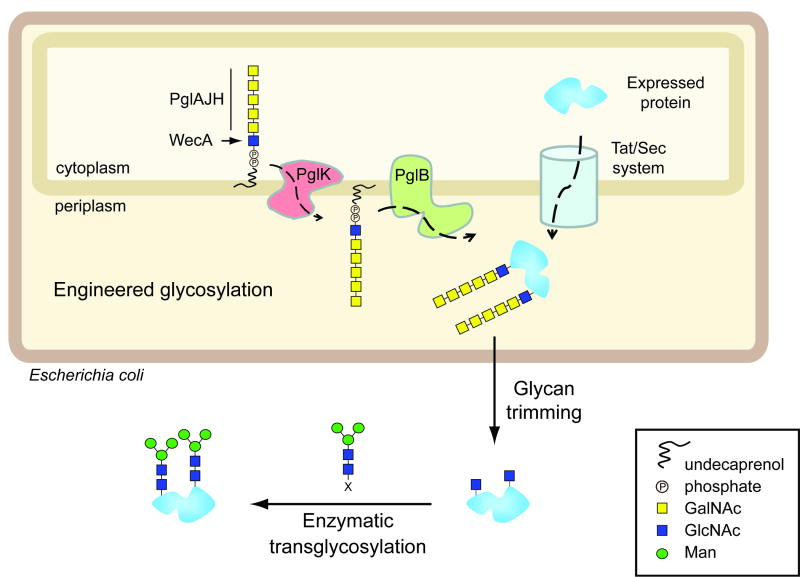
Figure 1.
Schematic representation of the protein expression and glycosylation engineering system. The protein is expressed in E. coli and secreted to the periplasm via the Sec or Tat pathway. A tailored lipid-linked oligosaccharide having a GlcNAc at the reducing end is built, re-oriented to the periplasmic side of the inner membrane by the flippase PglK, and attached to the Asn-side chain of the protein by the oligosaccharyltransferase PglB. The N-glycan is trimmed by α-N-acetyl-galactosaminidase to give a GlcNAc-tagged glycoprotein. Finally endoglycosidase-catalyzed transglycosylation using pre-assembled N-glycan species as substrates gives rise to a homogenous pool of glycoproteins.
First we sought to engineer the C. jejuni N-glycosylation pathway in E. coli to install the key GlcNAc-Asn linkage found in eukaryotic N-glycoproteins. In the C. jejuni glycosylation system, the glycan is first built sequentially by several glycosyltransferases (PglC,A,J,H,I) on a lipid anchor in the inner membrane, then flipped to the periplasmic side by the transporter PglK and finally transferred to an acceptor protein by the oligosaccharyltransferase PglB. This will yield a glycoprotein in which the glycan is attached to the Asn side chain through a rare deoxysugar, bacillosamine (Bac). To introduce a GlcNAc-Asn linkage to replace the bacterial Bac-Asn linkage, we altered the N-glycosylation pathway so as to produce a (GalNAc)5GlcNAc-PP-undecaprenyl. This was based on the finding that lipid-linked oligosaccharide (LLO) with a GlcNAc residue at the reducing end can serve as a substrate for PglK and PglB 16-18. We modified the pgl locus to delete the genes coding for the biosynthesis (pglDEF) and transfer (pglC) of bacillosamine, as well as the gene adding the glucose branch (pglI). The resulting glycosylation operon, named pgl2, was then transferred into E. coli. In the absence of PglC, the synthesis of the lipid-anchored glycan is expected to be initiated by the transfer of a GlcNAc 1-P to the lipid-P under the catalysis of WecA. The wecA gene is located in the wec cluster of the E. coli genome and links GlcNAc 1-P to undecaprenyl-P during the biosynthesis of both O-antigen lipopolysaccharide and enterobacterial common antigen (ECA) 19.
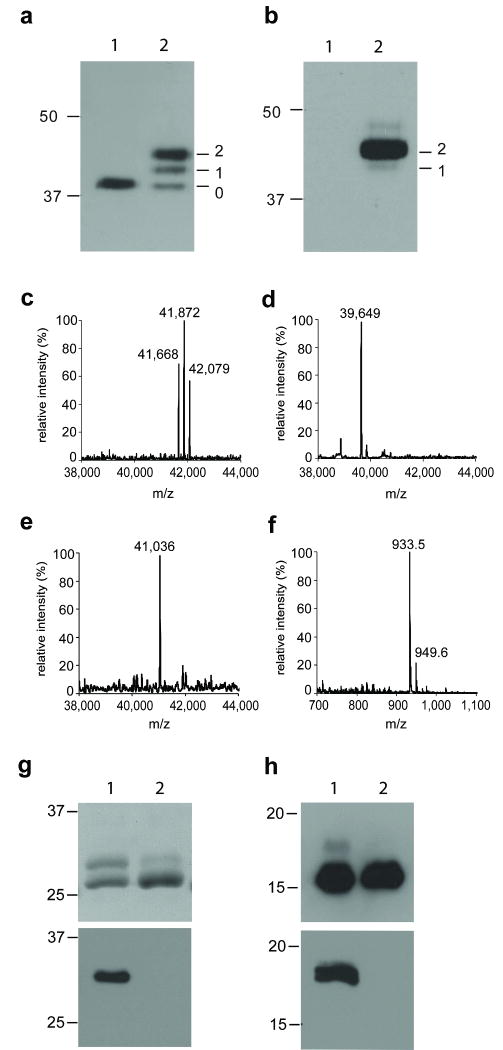
To examine the feasibility of the glycoengineered bacterial system to produce GlcNAc-Asn-linked glycoproteins, we expressed AcrA, a glycoprotein from C. jejuni, in E. coli SCM3 strain that carries the pgl2 cluster. AcrA contains two consensus sequences for glycosylation at positions N123 and N273 (Supplementary Fig. 1) 20. The expression of AcrA in E. coli SCM3 gave a major glycoprotein that was detected by anti-sera against the AcrA protein (Fig. 2a) and the C. jejuni N-glycan (Fig. 2b), respectively. We observed efficient secretion of AcrA to the periplasm in the SCM3 strain, when compared to other E. coli strains (Supplementary Fig. 2). Fully glycosylated AcrA was separated from the other minor glycoforms via hydroxylapatite chromatography. ESI-MS analysis of the doubly glycosylated AcrA revealed three species (41668, 41872, and 42079 Da, respectively), suggesting that it was a mixture of AcrA glycoforms containing the (GalNAc)5GlcNAc and (GalNAc)6GlcNAc N-glycans (Fig. 2c). These results imply that in the absence of PglI, the GalNAc transferase PglH has a more relaxed flexibility to extend the GalNAc sugar chain in this system, in contrast to the original C. jejuni expression that gives a single (GalNAc)2[Glc](GalNAc)3Bac glycoform 7,21. The successful expression of the glycosylated AcrA with a desired GlcNAc-Asn linkage also suggests that the engineered lipid-linked oligosaccharide (LLO) serves as an efficient substrate for PglK and PglB. As expected, deletion of wecA completely abolished the glycosylation of AcrA (Supplementary Fig. 3). Treatment of the glycosylated AcrA with the exo-α-N-acetylgalactosaminidase resulted in a stepwise removal of the GalNAc residues to give a pure AcrA glycoform that carries only two GlcNAc residues (ESI-MS: calculated, M = 39633 Da; found, M = 39640 Da) (Fig. 2d). Therefore, the expression of a heterologous glycoprotein in the engineered E. coli system, followed by purification and exo-α-N-acetylgalactosaminidase digestion, permits access to GlcNAc-tagged proteins, ready to serve as an acceptor substrate for further enzymatic sugar chain elongation.
Figure 2.
Production of homogenous N-glycoproteins. (a-b) Immunoblot analysis of the AcrA protein expressed in E. coli SCM3 carrying the pACYC184 (lanes 1) or the pACYC(pgl2) plasmid (lanes 2). Proteins are detected with anti-sera against AcrA (a) or against the glycan (b). Numbers on the left of the gel frame show the electrophoretic mobility of the molecular weight marker (kDa). The number of the N-glycans present on the different glycoforms of AcrA is indicated at the right of the gel frames. (c-f) MS analysis of AcrA glycoforms: (c) doubly glycosylated AcrA expressed in E. coli; (d) [GlcNAc]2-AcrA; (e) [Man3GlcNAc]2-AcrA; (f) N-glycan released from the product. (g-h) Expression of eukaryotic glycoproteins in E. coli: (g) Purified CH2-GI was incubated with (lane 2) or without (lane 1) exo-α-N-acetyl-galactosaminidase to remove terminal GalNAc residues of the glycan. The resulting protein was analyzed by immunoblot, using anti-His antibody (top) or anti-glycan serum (bottom); (h) Purified F8 was incubated with (lanes 2) or without (lanes 1) exo-α-N-acetyl-galactosaminidase. The resulting protein was analyzed by Coomassie-stained SDS-PAGE (top), and immunoblot using anti-glycan serum (bottom). Numbers on the left of the gel frame show the electrophoretic mobility of the molecular weight marker (kDa).
Next, we examined the transglycosylation of the GlcNAc-containing AcrA by the endo-β-N-acetylglucosaminidase from Arthrobacter protophormiae (Endo-A), using Man3GlcNAc oxazoline as the model donor substrate 10. We have previously reported that Endo-A is able to perform glycosylation remodeling of ribonuclease B and human IgG1-Fc using N-glycan oxazolines as the donor substrates 11,13, but it was not clear whether this approach could be generally expanded to other proteins having more than one glycosylation site. We found that Endo-A could simultaneously transfer two synthetic N-glycans to form the doubly glycosylated AcrA (Fig. 1). When an excess Man3GlcNAc-oxazoline was used, the transglycosylation went to completion, giving the homogeneous AcrA glycoform carrying two N-glycans. The obtained glycoprotein was subjected to MS analyses. Deconvolution of the ESI-MS spectrum of the doubly glycosylated product gave a single species with a mass of 41036 Da, which is consistent with the AcrA glycoprotein carrying two Man3GlcNAc2 glycans (calculated, M = 41011 Da) (Fig. 2e). For further confirmation, the N-glycans in the product were released by PNGase F treatment. MALDI-TOF MS analysis indicated that the released N-glycans were the pentasaccharide Man3GlcNAc2 [calculated for Man3GlcNAc2, M =910.30; found (m/z), 933.5 (M+Na) and 949.6 (M+K)] (Fig. 2f). These results suggest that the Endo-A could successfully transglycosylate the GlcNAc-containing AcrA to form the doubly glycosylated AcrA carrying the eukaryotic core N-glycans. Similarly, an AcrA variant (AcrAN273Q), which contains a single glycosylation site at N123, was also expressed in the engineered E. coli SCM3 system and the glycan was successfully remodeled to the core N-pentasaccharide Man3GlcNAc2 (Supplementary Fig. 4a). The identity of the initial glycoprotein (GalNAc)5,6GlcNAc-AcrAN273Q, the trimmed intermediate GlcNAc-AcrAN273Q, and the final remodeled product Man3GlcNAc2-AcrAN273Q was confirmed by ESI-MS analysis (Supplementary Fig. 4b-e). The purity of various glycoforms of AcrA and AcrAN273Q, including those before and after glycan remodeling was confirmed by the SDS-PAGE analysis (Supplementary Fig. 5).
The usefulness of the new method for incorporating natural mammalian N-glycans at a specific site of the E. coli expressed protein was exemplified by the synthesis of the AcrAN273Q glycoforms carrying a natural high-mannose type glycan (Man9GlcNAc2) and a bi-antennary complex type N-glycan (Supplementary Fig. 6a). This was made possible by the novel glycosynthases that can use activated sugar oxazolines for transglycosylation, but lack product hydrolysis activity 14,15. For example, The EndoA-N171A mutant can efficiently transfer high-mannose type N-glycan oxazolines without product hydrolysis 15, while the EndoM-N175A mutant can transfer both high-mannose and complex type N-glycan oxazolines 14. Thus, transglycosylation of GlcNAc-AcrAN273Q with the Man9GlcNAc-oxazoline under the catalysis of glycosynthase EndoA-N171A gave the AcrAN273Q glycoform carrying a natural high-mannose type N-glycan (Supplementary Fig. 6b). Similarly, the enzymatic reaction between GlcNAc-AcrAN273Q and the sugar oxazoline corresponding to the bi-antennary complex type N-glycan (CT-oxazoline) under the catalysis of EndoM-N175A resulted in the formation of the complex type AcrAN273Q glycoform (Supplementary Fig. 6c). In comparison, the reaction catalyzed by EndoM-N175A was found to be relatively slow, partly because of the relatively low specific activity of the Endo-M mutant. These results attest to the potential of the combined method to produce glycoproteins carrying natural, full-size eukaryotic N-glycans.
With the successful introduction of natural eukaryotic N-glycans into the bacterial protein AcrA, we asked whether the method could be extended to production of eukaryotic N-glycoproteins. Previous studies have demonstrated that the C. jejuni oligosaccharyltransferase PglB requires an extended consensus sequon D/E-Z-N-X-S/T, where X and Z can be any amino acid but proline, for efficient N-glycosylation 20. We have previously shown that PglB could glycosylate, in vitro, the eukaryotic protein ribonuclease A, when the original glycosylation sequon is extended to the bacterial type, and cholera toxin, when the glycosylation sequon is introduced at a relatively flexible region 9,20. Here we selected two eukaryotic proteins, the CH2 domain of human IgG-Fc and the single chain antibody F8, to test the in vivo glycosylation in the glycoengineered E. coli system (SCM3). Glycosylation of the CH2 domain of human IgG-Fc at the conserved N297 glycosylation site is critical for antibody's effector functions and the anti-inflammatory activity of IgG-Fc 22,23. We cloned the CH2 ORF downstream to a pelB signal sequence that targets the protein to the periplasm. Then, we altered the original glycosylation region QYNST (aa 295-299) into a potential bacterial glycosylation site DFNST (Supplementary Fig. 7). Expression of the engineered CH2 domain (CH2-GI) in E. coli SCM3 carrying the pgl2 machinery resulted in production of glycosylated and non-glycosylated CH2 domain (Fig. 2g). The glycosylated CH2 domain was about 1 kDa larger in size than the non-glycosylated CH2 and was recognized by the anti-glycan serum. Treatment of the glycoprotein with exo-α-N-acetyl-galactosaminidase abolished recognition by the anti-glycan serum, releasing the GlcNAc-tagged CH2 domain. However, it was found that only less than 5% of the CH2 domain was glycosylated under these conditions. We also tested the glycosylation of another eukaryotic protein, a single chain antibody F8, in the E. coli system. F8 is a clinical-stage human monoclonal antibody specific to the alternatively spliced extra-domain A (EDA) of fibronectin, a marker of tumor angiogenesis 24. Glycosylation may provide an efficient means to fine-tune protein's properties such as its half-life in vivo 25. Thus, we engineered a portable glycosylation sequence at the C-terminus of the F8 single-chain antibody (Supplementary Fig. 8). Expression of the F8 protein in the glycoengineered E. coli cells resulted in glycosylated F8 (Fig. 2h). MS analysis of purified glycosylated F8 revealed the presence of two major peaks corresponding to the glycoforms HexNAc6 and HexNAc7 (Supplementary Fig. 9), as in the case of AcrA. Exo-α-N-acetyl-galactosaminidase removed the GalNAc residues from the N-glycan and abolished reactivity to the anti-glycan serum. In contrast to the low glycosylation of CH2 domain, the glycosylation of the F8 protein in the engineered system was much more efficient, reaching approx. 40%. Several factors, such as the local conformations of the glycosylation site, may affect the glycosylation efficiency of a given eukaryotic protein.
In summary, we described a potentially general platform for producing homogeneous glycoproteins carrying eukaryotic N-glycans. This method combines the convenience of protein expression in E. coli with the remarkable flexibility of in vitro chemoenzymatic glycan remodeling. We provided proof of concept data showing that this method can be extended to production of eukaryotic N-glycoproteins. Future studies will be directed to optimizing the engineered E. coli expression system to enhance the efficiency of glycosylation of biomedically important eukaryotic proteins. In addition, mutagenesis and directed evolution of PglB, as well as mining the diversity of bacterial oligosaccharyltransferases, might provide variants of PglB that are able to glycosylate proteins at the short consensus sequence N-X-S/T found in natural human glycoproteins.
Supplementary Material
Acknowledgments
We thank members of the Aebi and Wang labs for fruitful discussions. This work was supported in part by the Swiss National Science Foundation (grant 3100AQ-105541 to M.A.), the ETH Zurich, and the US National Institutes of Health (NIH grant GM080374 to L.X.W.). We thank Prof. Kaoru Takegawa for providing the pGEX-2T/Endo-A plasmid, and Prof. Kenji Yamamoto for providing the pET23b-Endo-M plasmid that were used to express the endo-enzymes. F.S. and Ch. L. are members of the Zurich Ph.D. Program in Molecular Life Sciences.
Footnotes
Author Contributions: F.S., engineering glycosylation pathway, protein characterization, and writing the manuscript; W.H., glycan trimming, glycosylation remodeling, and product characterization; C.L. glycosylation remodeling and product characterization; B.L.S., preliminary MS analyses of glycosylated AcrA; Ch.L., engineering the CH2 domain; A. P., characterization of the F8 protein; S.N., conceiving the original idea and performing preliminary experiments; D.N., supervising the research and development of F8; M.A., supervising the research and writing the manuscript; L.X.W., conceiving the original idea, supervising the research, and writing the manuscript. All authors contributed to editing the manuscript.
Competing Financial Interests Statement: Dario Neri is a co-founder and shareholder of Philogen, the company which owns the F8 antibody.
References
- 1.Dwek RA. Glycobiology: Toward understanding the function of sugars. Chem Rev. 1996;96:683–720. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 2.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–9. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 3.Gamblin DP, Scanlan EM, Davis BG. Glycoprotein synthesis: an update. Chem Rev. 2009;109:131–63. doi: 10.1021/cr078291i. [DOI] [PubMed] [Google Scholar]
- 4.Rich JR, Withers SG. Emerging methods for the production of homogeneous human glycoproteins. Nat Chem Biol. 2009;5:206–15. doi: 10.1038/nchembio.148. [DOI] [PubMed] [Google Scholar]
- 5.Bennett CS, Wong CH. Chemoenzymatic approaches to glycoprotein synthesis. Chem Soc Rev. 2007;36:1227–38. doi: 10.1039/b617709c. [DOI] [PubMed] [Google Scholar]
- 6.Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol Microbiol. 1999;32:1022–30. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 7.Young NM, et al. Structure of the N-linked glycan present on multiple glycoproteins in the Gram-negative bacterium, Campylobacter jejuni. J Biol Chem. 2002;277:42530–9. doi: 10.1074/jbc.M206114200. [DOI] [PubMed] [Google Scholar]
- 8.Wacker M, et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–3. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- 9.Kowarik M, et al. N-linked glycosylation of folded proteins by the bacterial oligosaccharyltransferase. Science. 2006;314:1148–50. doi: 10.1126/science.1134351. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Zeng Y, Hauser S, Song H, Wang LX. Highly efficient endoglycosidase-catalyzed synthesis of glycopeptides using oligosaccharide oxazolines as donor substrates. J Am Chem Soc. 2005;127:9692–3. doi: 10.1021/ja051715a. [DOI] [PubMed] [Google Scholar]
- 11.Li B, Song H, Hauser S, Wang LX. A highly efficient chemoenzymatic approach toward glycoprotein synthesis. Org Lett. 2006;8:3081–4. doi: 10.1021/ol061056m. [DOI] [PubMed] [Google Scholar]
- 12.Ochiai H, Huang W, Wang LX. Expeditious chemoenzymatic synthesis of homogeneous N-glycoproteins carrying defined oligosaccharide ligands. J Am Chem Soc. 2008;130:13790–803. doi: 10.1021/ja805044x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei Y, et al. Glycoengineering of human IgG1-Fc through combined yeast expression and in vitro chemoenzymatic glycosylation. Biochemistry. 2008;47:10294–304. doi: 10.1021/bi800874y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umekawa M, et al. Mutants of Mucor hiemalis endo-beta-N-acetylglucosaminidase show enhanced transglycosylation and glycosynthase-like activities. J Biol Chem. 2008;283:4469–79. doi: 10.1074/jbc.M707137200. [DOI] [PubMed] [Google Scholar]
- 15.Huang W, et al. Glycosynthases enable a highly efficient chemoenzymatic synthesis of N-glycoproteins carrying intact natural N-glycans. J Am Chem Soc. 2009;131:2214–23. doi: 10.1021/ja8074677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman MF, et al. Engineering N-linked protein glycosylation with diverse O antigen lipopolysaccharide structures in Escherichia coli. Proc Natl Acad Sci USA. 2005;102:3016–21. doi: 10.1073/pnas.0500044102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wacker M, et al. Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems. Proc Natl Acad Sci USA. 2006;103:7088–93. doi: 10.1073/pnas.0509207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alaimo C, et al. Two distinct but interchangeable mechanisms for flipping of lipid-linked oligosaccharides. EMBO J. 2006;25:967–76. doi: 10.1038/sj.emboj.7601024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J Bacteriol. 2007;189:2618–28. doi: 10.1128/JB.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowarik M, et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–66. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troutman JM, Imperiali B. Campylobacter jejuni PglH is a single active site processive polymerase that utilizes product inhibition to limit sequential glycosyl transfer reactions. Biochemistry. 2009;48:2807–16. doi: 10.1021/bi802284d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jefferis R. Glycosylation of recombinant antibody therapeutics. Biotechnol Prog. 2005;21:11–6. doi: 10.1021/bp040016j. [DOI] [PubMed] [Google Scholar]
- 23.Anthony RM, et al. Recapitulation of IVIG anti-inflammatory activity with a recombinant IgG Fc. Science. 2008;320:373–6. doi: 10.1126/science.1154315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villa A, et al. A high-affinity human monoclonal antibody specific to the alternatively spliced EDA domain of fibronectin efficiently targets tumor neo-vasculature in vivo. Int J Cancer. 2008;122:2405–13. doi: 10.1002/ijc.23408. [DOI] [PubMed] [Google Scholar]
- 25.Holliger P, Hudson PJ. Engineered antibody fragments and the rise of single domains. Nat Biotechnol. 2005;23:1126–36. doi: 10.1038/nbt1142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.